Abstract
The most prominent mechanism of molecular evolution is believed to have been duplication and divergence of genes. Proteins that belong to sequence-related groups in any one organism are candidates to have emerged from such a process and to share a common ancestor. Groups of proteins in Escherichia coli having sequence similarity are mostly composed of proteins with closely related function, but some groups comprise proteins with unrelated functions. In order to understand how function can change while sequences remain similar, we have examined some of these groups in detail. The enzymes analyzed in this work include representatives of amidotransferases, phosphotransferases, decarboxylases, and others. Most sequence-related groups contain enzymes that are in the same classes of Enzyme Commission (EC) numbers. We have concentrated on groups that are heterogeneous in that respect, and also on groups containing more than one enzyme of any pathway. We find that although the EC number may differ, the reaction chemistry of these sequence-related proteins is the same or very similar. Some of these families illustrate how diversification has taken place in evolution, using common features of either reaction chemistry or ligand specificity, or both, to create catalysts for different kinds of biochemical reactions. This information has relevance to the area of functional genomics in which the activities of gene products of unknown reading frames are attributed by analogy to the functions of sequence-related proteins of known function.
Groups of sequence-related proteins of Escherichia coli have been assembled that seem likely to have arisen by duplication and divergence of genes in the ancestral genomes, some arising recently, some in early evolutionary times (Labedan and Riley 1995, 1999; Riley and Labedan 1997). Most of the groups are composed of proteins that all have the same reaction chemistry but differ by substrate specificity. Examples are groups of similar-sequence kinase enzymes that act on different substrates, groups of sequence-related acyltransferases that act on different substrates, sets of transcriptional regulators with similar reaction chemistry, and sets of transport proteins that use the same type of mechanism. These and other similar examples are likely to be instances of duplication in which the progeny proteins maintain the reaction chemistry performed, but change the identity of the specific substrate or ligand.
However, there are a few examples of sets of sequence-related enzymes within E. coli that one would not a priori expect to be related: those that seem to catalyze different reactions and those that occur within the same pathway. We collected such examples from among all sequence-related groups or paralogs (for definition, see Fitch 1970) in the E. coli genome (P. Liang, B. Labedan, and M. Riley, in prep.) to find out the biochemical basis of the observed sequence similarity. We found that in the pairs of proteins selected there are examples of (1) similar reaction chemistry but different substrate/ligand specificity, (2) similar substrate/ligand specificity but different reaction chemistry, and (3) both together. These are examples of how divergence by recruitment occurs in molecular terms.
RESULTS
All sequence-related groups of E. coli metabolic enzymes were examined for any examples of proteins having Enzyme Commission (EC) designations (Webb 1992) that differed in the first or second place. These could be examples of divergence of function from common ancestors. Enzymes performing different steps in the same pathways were also collected. Pairwise alignments were checked to locate the regions of sequence similarity within the proteins, confirming that the respective enzyme reactivities were present in the homologous regions of sequence similarity. Nine sets of enzymes and their genes in E. coli met these criteria and are listed in Table 1.
Table 1.
Groups of Sequence-Related Enzymes
| Gene | b number | Accession number | Enzyme | Length (aa) |
|---|---|---|---|---|
| entE | b0594 | P10378 | subunit of 2,3-dihydroxybenzoate-AMP ligase | 536 |
| entF | b0586 | P11454 | apo-serine activating enzyme | 1293 |
| purK | b0522 | P09029 | CO2 subunit of phosphoribosylaminoimidazole carboxylase | 355 |
| purT | b1849 | P33221 | GAR transformylase 2 | 391 |
| guaB | b2508 | P06981 | subunit of IMP dehydrogenase | 488 |
| guaC | b0104 | P15344 | subunit of GMP reductase | 346 |
| murC | b0091 | P17952 | UDP-N-acetylmuramate-alanine ligase | 491 |
| murD | b0088 | P14900 | UDP-N-acetylmuramoylalanine-D-glutamate ligase | 437 |
| murE | b0085 | P22188 | UDP-N-acetylmuramoylalanyl-D-glutamate 2,6-diaminopimelate ligase | 494 |
| murF | b0086 | P11880 | D-alanyl-D-alanine-adding enzyme | 452 |
| ansB | b0674 | P22106 | asparagine-synthase-(glutamine-hydrolysing) | 553 |
| glmS | b3729 | P17169 | L-glutamine:D-fructose-6-phosphate aminotransferase | 608 |
| purF | b2312 | P00496 | amidophosphoribosyl transferase | 504 |
| menF | b2265 | P38051 | isochorismate synthase, menaquinone-specific | 431 |
| pabB | b1812 | P05041 | aminodeoxychorismate synthase component I | 453 |
| trpE | b1264 | P00895 | anthranilate synthase component I | 520 |
| gcl | b0507 | P30146 | glyoxylate carboligase | 592 |
| ilvI | b0077 | P00893 | large subunit of acetolactase synthase III/acetohydroxybutanoate synthase III | 574 |
| poxB | b0871 | P07003 | pyruvate oxidase | 572 |
| metB | b3939 | P00935 | O-succinylhomoserine(thiol)-lyase | 386 |
| metC | b3008 | P06721 | cystathionine-beta-lyase | 395 |
| hisA | b2024 | P10371 | phosphoribosylformimino-5-amino-1-phosphoribosyl-4-imidazole carboxamide isomerase | 245 |
| hisF | b2025 | P10373 | subunit of imidazole glycerol phosphate synthase | 258 |
Sources: Gene, GenProtEC; b number, Blattner et al. 1997; accession number (primary accession no.), SWISS-PROT; enzyme, EcoCyc; length (protein length in amino acids), SWISS-PROT.
Sequence similarities between each pair of related enzymes and relationships of alignment were determined as accepted point mutation (PAM) values reported by the DARWIN analysis (Gonnet et al. 1992; http://cbrg.inf.ethz.ch/) and also by gapped BLAST (Altschul et al. 1997; http://www.ncbi.nlm.nih.gov/). Results in terms of PAM values for each of the pairs are listed in Table 3 below. PAM values ranged from 116 to 221, where 116 can be considered an unquestionably significant match and 221 is marginal. Protein domains were located with Pfam (Bateman et al. 2000; http://www.sanger.ac.uk/Software/Pfam/). Other sources of information are the databases EcoCyc (Karp et al. 2000; http://ecocyc.ai.sri.com/) for functions of the enzymes in metabolism, GenProtEC (Riley 1998; http://genprotec.mbl.edu/) for modular composition of complex proteins, and primary research literature for information on the proteins pertinent to their biochemical relationships.
Table 3.
Shared Characteristics between Sequence-Related Enzymes
| Enzymes | PAM | Mechanism1 | Substrate | Cofactor | Pathway | Sequential2 |
|---|---|---|---|---|---|---|
| EntE–EntF | 185 | + | + | − | + | − |
| PurK–PurT | 174 | + | + | − | + | − |
| GuaB–GauC | 116 | + | + | − | + | − |
| MurC–MurE | — | + | + | − | + | − |
| MurF–MurD | 196 | + | + | − | + | − |
| MurF–MurC | 193 | + | + | − | + | + |
| MurF–MurE | 185 | + | + | − | + | + |
| MurC–MurD | 173 | + | + | − | + | + |
| MurD–MurE | 158 | + | + | − | + | + |
| PurF–AsnB | — | + | + | − | − | − |
| AsnB–GlmS | 185 | + | + | − | − | − |
| GlmS–PurF | 155 | + | + | − | − | − |
| TrpE–PabB | 127 | + | + | − | − | − |
| Gcl–PoxB | 173 | + | − | + | − | − |
| PoxB–IlvI | 140 | + | − | + | − | − |
| IlvI–Gcl | 124 | + | − | + | − | − |
| MetB–MetC | 144 | + | − | + | + | + |
| MenF–PabB | 221 | − | + | − | − | − |
| MenF–TrpE | 185 | − | + | − | − | − |
| HisA–HisF | 180 | − | − | − | + | + |
Chemistry mechanism of the reaction catalyzed.
Sequential: one product is substrate for the subsequent reaction (sequential enzymes in a pathway).
“+” indicates “same,” “similar” or “yes,” and “−” indicates “different” or “no”.
Sources: Gene and PAM value, GenProtEC; mechanism of action, BRENDA and EC nomenclature; substrate, cofactor, and pathway, EcoCyc.
We assessed the nature of the phenotypic similarity between each pair of sequence-related enzymes. The reactions catalyzed by these proteins and their EC designations (Webb 1992) are listed in Table 2 with emphasis added on participation of same or similar substrates and products. (No emphasis has been applied to universal participants such as ATP and NAD because they do not discriminate at the level we are examining.) Each reaction is different; for each pair of reactions that shared one reactant, therefore, the other reactants were not the same.
Table 2.
Reactions Catalyzed by Sequence-Related Enzymes
| Gene | EC | Reaction | Mechanism |
|---|---|---|---|
| entE | 6.3.2.- | ATP + 2,3-dihydroxybenzoate ⇒ pyrophosphate + 2,3-Dihydroxybenzoyl-AMP | Adenylation |
| entF | — | ATP + L-serine → pyrophosphate + L-Seryl-AMP | Adenylation |
| purK | 4.1.1.21 | CO2 + H2O + AIR ⇔ ADP + phosphate + AICAR | 1 carbon incorporation |
| purT | 2.1.2.- | HCOOH + ATP + GAR ⇔ ADP + phosphate + FGAR | 1 carbon incorporation |
| guaB | 1.1.1.205 | XMP + NADH ⇔ IMP + NAD + H2O | Redox reaction |
| guaC | 1.6.6.8 | GMP + NADPH ⇔ IMP + NADP + NH3 | Redox reaction |
| murC | 6.3.2.8 | ATP + UDP-N-acetylmuramate + L-alanine ⇔ X + phosphate + ADP | Amino acid ligation |
| murD | 6.3.2.9 | ATP + X + D-glutamate ⇔ XY + phosphate + ADP | Amino acid ligation |
| murE | 6.3.2.13 | ATP + XY + meso-diaminopimelate ⇔ XYZ + phosphate + ADP | Amino acid ligation |
| murF | 6.3.2.15 | ATP + XYZ + D-alanyl-D-alanine ⇔ XYZW + phosphate + ADP | Amino acid ligation |
| asnB | 6.3.5.4 | L-glutamine + L-aspartate + ATP + H2O ⇔ L-glutamate + L-asparagine + pyrophosphate + AMP | Amide group transfer |
| glmS | 2.6.1.16 | L-glutamine + fructose-6-phosphate ⇔ D-glucosamine-6-phosphate + L-glutamate | Amide group transfer |
| purF | 2.4.2.14 | L-glutamine + PRPP + H2O ⇔ 5-phosphoribosylamine + pyrophosphate + L-glutamate | Amide group transfer |
| menF | 5.4.99.6 | chorismate ⇒ isochorismate | Mutase |
| pabB | 4.1.3.- | chorismate + NH3 ⇔ 4-amino-4-deoxychorismate | Oxo-acid lyase |
| trpE | 4.1.3.27 | chorismate + NH3 ⇔ anthranilate + pyruvate | Oxo-acid lyase |
| gcl | 4.1.1.47 | 2 glyoxylate ⇔ CO2 + tartronate semialdehyde | Decarboxylation |
| ilvI | 4.1.3.18 | 2 pyruvate ⇔ CO2 + 2-aceto-lactate (valine biosynthesis); pyruvate + 2-oxobutanoate ⇔ CO2 + 2-aceto-2-hydroxy-butyrate (isoleucine biosynthesis) | Decarboxylation |
| poxB | 1.2.2.2 | pyruvate + ferricytochrome-b1 + H2O ⇔ CO2 + acetate + ferrocytochrome-b1 | Decarboxylation |
| metB | 4.2.99.9 | L-cysteine + o-succinyl-L-homoserine ⇔ succinate + cystathionine | Carbon-oxygen lyase |
| metC | 4.4.1.8 | cystathionine + H2O ⇔ pyruvate + NH3 + homocysteine | Carbon-sulfur lyase |
| hisA | 5.3.1.16 | phosphoribosyl-formimino-AICAR-P ⇔ PRFAR | Isomerization |
| hisF | 2.4.2.- | PRFAR + L-glutamine ⇔ D-erythro-imidazole-glycerol-phosphate + AICAR + L-glutamate | Amidation |
Sources: Gene, GenProtEC; EC (Enzyme Commission number), ExPASy–ENZYME; reaction, modified from EcoCyc; mechanism, BRENDA and EC nomenclature
In reactions catalyzed by murCDEF: X = UDP-N-acetylmuramoyl-L-alanine; XY = UDP-N-acetylmuramoyl-L-alanyl-D-glutamate; XYZ = UDP-N-acetylmuramoyl-L-alanyl-D-glutamyl-meso-2, 6-diaminoheptanedioate; XYZW = UDP-N-acetylmuramoyl-L-alanyl-D-glutamyl-meso-2, 6-diaminoheptanedioate-D-alanyl-D-alanine.
Some of the sequence-related enzymes grouped in Table 2 appear to have similar binding sites for similar or identical reactants, whereas at the same time they differ for other reactants. This is the case for the pairs of enzymes EntE–EntF, PurK–PurT, and GuaB–GuaC, which function in the same pathway, and for the triplets AsnB–GlmS–PurF and TrpE–PabB–MenF, which are in different pathways.
Requiring more explanation are the pairs in which the product of the reaction catalyzed by one of the pair is the substrate of the reaction catalyzed by the other. This is the case for the Mur enzymes (MurC–D–E–F), for the MetB–MetC pair, and the HisA–HisF pair. Although they form a chain in which the product of one reaction is the substrate for another, the Mur enzymes carry out similar ligation reactions. The same holds true for the reactions of MetB and MetC, which are both lyases, although of a different type.
Not related by pathway are the two pairs MenF–PabB and MenF–TrpE. They use the same reactant, chorismate, but the reaction of MenF differs from that of the other two.
Another group not related by pathway is the Gcl–IlvI–PoxB group. For the IlvI–PoxB pair, one of the substrates is the same, pyruvate, and when one includes the related Gcl, one sees that all three enzymes modify α-keto acids. Although the reactions appear at first sight to be different, they have common features to be discussed below.
These relationships between the sequence-related enzymes are summarized in Table 3.
Sequence-Related Enzymes Sharing Reaction Chemistry and Substrate Specificity
Although the pairs and groups of sequence-related enzymes were chosen as likely to be examples of evolutionary divergence, when examined closely the basis of the sequence relatedness became clear. The following pairs and groups of enzymes share reaction chemistry and substrate specificity: EntE–EntF, PurK–PurT, GuaB–GuaC, MurC–D–E–F, AsnB–GlmS–PurF, and TrpE–PabB as shown in Table 2. These enzymes have similar specificity for reactants, either a pair of substrates or substrates and products of the reactions. All enzyme pairs bind the same substrates, or the product of one reaction is the substrate of the other, or they produce a common product. The pairs and groups of enzymes that do not share reaction chemistry, but do relate in one way or another in small molecule specificity are Gcl–IlvI–PoxB, MetB–MetC, MenF–PabB, MenF–TrpE, and HisA–HisF.
There are four sequence-related groups of enzymes sharing reaction chemistry and substrate specificity (Tables 1–3). EntE and EntF are of unequal length. However for this pair, sequence alignment shows that the active parts of the polypeptides are similar in sequence. EntF is multimodular, and it is the C-terminal part of EntF that pairs with EntE. EntE and EntF are polypeptides that are components of the EntB–EntE–EntF multienzyme complex enterobactin synthase (Gehring et al. 1998). Although the catalytic activities seem different because EntE is a ligase and EntF is an activation enzyme (Table 1), both proteins have an AMP-binding domain in the C-terminal portion and both catalyze the adenosinylation of their substrates (2,3-dihydroxybenzoate and L-serine, respectively), using ATP as AMP donor. Therefore, they are similar in reaction chemistry.
The enzymes PurK and PurT are both enzymes of purine biosynthesis. Their EC numbers are completely different (Table 2), and the enzyme activities seem unlike (Table 1). PurK can be viewed as a carboxylase or a carboxy-lyase depending on reaction direction, whereas PurT is a formyltransferase. These enzymes share the ATP-grasp domain, which is present in several ATP-dependent carboxylate-amine ligases (Pfam). There are fundamental similarities of reactions. The ribosyl phosphate moieties of the substrates and products of both reactions are similar and both reactions incorporate one-carbon moieties into the substrates with the cleavage of ATP. PurK incorporates CO2 into the substrate and PurT incorporates HCOOH. Therefore, although EC designations differ, the two enzymes share both reaction chemistry and substrate and product similarities (Table 3). A possible evolutionary relationship between these two enzymes has been suggested before (Marolewski et al. 1994).
The enzymes GuaB and GuaC are a dehydrogenase and a reductase, respectively, in guanosine nucleotide metabolism. Although EC numbers differ because of the different directions in which the reactions are viewed, the reactions are both reversible redox reactions. By rewriting one reaction direction, the two enzymes can be seen as catalyzing similar reactions yielding IMP as product with either NADH or NADPH as cofactor (Table 2). GuaB and GuaC are members of the common nucleoside diphosphate-binding-site TIM barrel family (Zhang et al. 1999).
MurC, MurD, MurE, and MurF are enzymes that catalyze consecutive steps in peptidoglycan biosynthesis. The consecutive steps have a great deal of similarity. Unlike most of the other sequence-related groups examined, these bear similar EC designations (EC 6.3.2.-), because they are all acid-amino-acid ligases (peptide synthases) forming carbon–nitrogen bonds (Eveland et al. 1997). They all catalyze ATP-dependent ligation reactions and act on substrates that share the UDP-N-acetylmuramoyl moiety. The ATP-binding consensus sequence GXXGKT/S and seven other amino acids are invariants among Mur enzymes (Bouhss et al. 1997, 1999). The Mur enzymes may also be seen as transferases transferring different groups as follows: L-alanine (MurC), D-glutamate (MurD), diaminopimelic acid (MurE), and D-alanyl alanine (MurF). For each pair of enzymes, there is a shared compound because the product of one reaction corresponds to the substrate of the subsequent reaction. Therefore, even though it is unusual for four enzymes that are consecutive in a metabolic pathway to catalyze similar reactions, the four Mur proteins are related both by reaction chemistry and by substrate specificity. That they function in the same pathway is a consequence of the process of building peptidoglycans by sequential additions.
AsnB, GlmS, and PurF function in different pathways: asparagine biosynthesis and degradation (AsnB), hexosamine biosynthesis (GlmS), and de novo purine biosynthesis (PurF). According to EC Nomenclature, AsnB is a carbon nitrogen ligase with glutamine as amido-N-donor (EC 6.3.5.-), GlmS is a transaminase transferring a nitrogenous group (EC 2.6.1.-), and PurF is a pentosyltransferase (glycosyltransferase) (EC 2.4.2.-; Table 1). However, the classification refers to the holoenzyme, not to the subunits AsnB, GlmS, and PurF. These three polypeptides all exhibit the same activity, amido group transfer forming a carbon–nitrogen bond. They are amidotransferases that use L-glutamine as amido donor and aspartate, fructose-6-phosphate, or PRPP as acceptors, respectively (Table 2). They share a glutamine amidotransferase (GATase) domain in the N-terminal part of the polypeptides, shown by structural data as well as the sequence similarity (Kim et al. 1996).
MenF, PabB, and TrpE also form a group of sequence-related polypeptide components of multimeric enzymes. PabB is the main subunit of the enzyme that catalyzes the first step in the biosynthesis of p-aminobenzoate (PABA) in the pathway of folate biosynthesis. p-Aminobenzoic acid synthase is an enzyme complex containing two nonidentical polypeptide chains. Component I (PabB) contains the binding site for chorismate and catalyzes the formation of 4-amino-4-deoxychorismate using ammonia rather than glutamine. (PabA provides the glutamine amidotransferase function and is component II of the holoenzyme.) TrpE in the pathway of tryptophan synthesis is a similar case. Anthranilate synthase also contains two nonidentical polypeptide chains. The N-terminal module of TrpE is anthranilate synthase component I (EC 4.1.3.27). It contains the binding site for chorismate and catalyzes the formation of anthranilate using ammonia rather than glutamine. (TrpD provides the glutamine amidotransferase function and is component II of the holoenzyme.) The third sequence-related protein MenF, however, is different. It is a mutase and does not seem to share reaction chemistry with PabB and TrpE (Dahm et al. 1998). The pairs of MenF with PabB or TrpE were therefore placed into the fourth section of Table 3 because the reaction chemistries for these pairs are not similar. However, the three enzymes are related in that all have a binding site for chorismate.
Sequence-Related Enzymes Sharing Reaction Chemistry and Cofactor Binding
At first sight, the Gcl–IlvI–PoxB group does not seem to share features of reactivity (Table 1). Gcl is part of glyoxylate catabolism. IlvI corresponds to the large subunit of acetolactate synthase III/acetohydroxybutanoate synthase III enzyme, which catalyzes the first of a set of shared reactions in valine, isoleucine, and leucine biosynthesis. PoxB oxidizes and decarboxylates pyruvate. The corresponding EC numbers are different, characterizing the respective reactions as carbon–carbon ligation (Gcl and IlvI) or pyruvate oxidation (PoxB; Table 2). However, inspection of the reactions showed that another aspect of the reaction is shared that is not reflected in the EC numbers, that of decarboxylation using thiamine diphosphate (ThDP) as cofactor (Table 3). They all contain a thiamine diphosphate-binding domain (Pfam). All also have FAD-binding sites. The PoxB enzyme is a classic two-electron flavin dehydrogenase in contrast with the IlvI and Gcl enzymes, which are considered anomalous flavoproteins, because they have an absolute requirement for flavin but do not catalyze a redox reaction (Chang and Cronan 1988). The three enzymes are therefore related not by substrate, but rather by bound cofactor and redox prosthetic group, and by the same reaction chemistry, decarboxylation using ThDP cofactor.
Enzymes Functioning in the Same Pathway
The pairs and groups of enzymes EntE–EntF, PurK–PurT, GuaB–GuaC, and MurC–D–E–F each belongs to a particular biochemical pathway or area of metabolism (Table 3). The Mur enzymes are all in the pathway of peptidoglycan synthesis, EntE and EntF are part of an enzyme complex in enterobactin synthesis, PurK and PurT are part of purine biosynthesis, and GuaA and GuaB are part of purine nucleotide metabolism. Two other pairs of enzymes we considered are together in pathways: MetB and MetC in methionine biosynthesis and HisA and HisF in histidine biosynthesis. Both cases have been well studied in the past.
The enzymes MetB and MetC catalyze, respectively, the second and third committed steps in methionine synthesis. The MetB enzyme is a synthase for cystathionine; the MetC enzyme cleaves cystathionine to form homocysteine (Table 2). The sequence similarity between these two enzymes is well known (Belfaiza et al. 1986). MetB and MetC share a cys/met-metabolism PLP-dependent enzymes domain (PFAM). The similarity in sequence seems to reflect many relationships: reaction chemistry, cofactor, and substrate/product relationships. Both enzymes are lyases having pyridoxal 5′-phosphate as prosthetic group. Studies of amino acid sequence and structural alignments revealed that MetB and MetC are very similar, but critical differences in the substrate-binding characteristics determine the different reactions catalyzed by these enzymes (Clausen et al. 1996, 1998). Not only to each other but in a larger context, MetB and MetC are both related to other pyridoxal 5′-phosphate-dependent enzymes (Alexander et al. 1994; Mehta and Christen 2000).
The enzymes of histidine biosynthesis also have been well studied, and the similarity between the HisA and HisF proteins has been analyzed (Fani et al. 1995, 1998). HisA is an isomerase that catalyzes conversion of phosphoribosylformimino-AICAR-P to phosphoribulosylformimino-AICAR-P (PRFAR); HisF is one subunit of the HisFH dimer that constitutes imidazole glycerol phosphate synthase (Table 2). Alone, HisF can catalyze a multistep, ammonia-dependent reaction converting PRFAR and ammonia to AICAR and IGP. (When complexed with HisH, glutamine serves as the source of the amino group.) The product of the HisA reaction is the substrate of the HisF reaction in histidine biosynthesis; these proteins therefore seem to be related by substrate/product rather than by catalytic mechanism. Common ancestry has been proposed (Fani et al. 1997, 1998).
DISCUSSION
Generation of diversity in protein evolution is thought to depend on recruitment of a protein to take on a new role. Specifically, the process of duplication and divergence during evolution is believed to have generated groups of proteins of similar sequence that share features of binding site specificity and/or reaction chemistry but carry out new reactions (Alexander et al. 1994; Babbitt and Gerlt 1997; Galperin and Koonin 1997; Gerlt and Babbitt 1998). We have tested this hypothesis by close examination of pairs or groups of sequence-related enzyme proteins in E. coli that seemed on the surface to be unrelated. We found that in spite of appearance to the contrary, relationships of either reaction chemistry or binding specificity, sometimes both, existed in all cases examined.
Of the 20 pairs, 17 pairs shared reaction chemistry and shared small molecules as substrates, products, or cofactors; 11 bore EC numbers different in either the first or second positions; and six were related as components of multimeric enzymes. Altogether 11 were in the same pathway, nine were not. The relationships for each pair are shown in Table 3. The most common relationship was of reaction chemistry, followed closely by recognition of same small molecules, and 13 pairs shared both kinds of similarity. Reaction chemistry and ligand recognition were therefore, as seen by this study, used repeatedly in the recruitment of new enzymes for different metabolic functions from existing enzymes.
It may seem a contradiction for some of the sequence-related enzymes with grossly different EC numbers, that the reactions catalyzed have close similarity (Table 2). In these cases, the EC numbers do not reflect the similarity of the particular aspect of the reactions that seems to have been conserved in evolution. Any reaction can be characterized from several points of view. When sequence-related pairs or groups of enzymes have different EC numbers, the EC numbers do not always reflect a known similarity between the reactions catalyzed. Reactions are cataloged in the EC numeric system by the chemical composition of reactants and products, not by features of the reaction chemistry itself (Webb 1992). Evolution seems not always to have produced variant proteins by the features used by the Enzyme Nomenclature system to categorize biochemical reactions.
Multimeric enzymes are examples of the problem. They can make classification difficult. The glutamine amidotransferases reported in E. coli include GlmS, PurF, AsnB, PabA, and TrpG proteins (Riley and Serres 2000). The first two are homomultimers whose reactions could have been classified as amidotransferases (EC 2.6.99.1). However, GlmS was classified as an aminotransferase rather than an amidotransferase. GlmS and PurF were classified for other aspects of their reactions than the amido group transfer. AsnB, PabA, and TrpG are all subunits of heteromultimer enzymes. EC number assignments focused on the overall reactions catalyzed by the holoenzymes, not referring to the particular activities of each of the subunit polypeptides.
Finally, looking at similar enzymes within single metabolic pathways is relevant to the idea of retrograde evolution, the generation of neighboring enzymes in a pathway by duplication and divergence (Horowitz 1945, 1965; Roy 1999). Although few examples have been found supporting this view, among the enzyme pairs collected here the four Mur enzymes in a row in the pathway to peptidoglycan synthesis; the two enzymes in methionine synthesis, MetB and MetC; and the two enzymes in histidine biosynthesis, HisA and HisF, would fit with this hypothesis. However, these are the only instances we found in the pathways of intermediary metabolism in E. coli.
We can ask if there are reasonable chemical mechanisms to account for divergence of different catalytic characteristics from a common ancestor. Some of the cases presented in Table 2 appear to involve no more than conservation of one binding site (for similar reactants) with differences in specificity toward other reactants. In other more complex cases the product of the reaction catalyzed by one of the pair is the substrate of the reaction catalyzed by the other. In one such case, MetB and MetC, the structures of the enzymes have been analyzed in detail (Clausen et al. 1998). Both the MetB and MetC enzymes are homotetramers and both are lyases that use pyridoxal-5-phosphate as cofactor. The structures of the two monomers have similar folds that map together very closely except at the ends of the chains. In the main body of the proteins there are substitutions at a few critical residues that affect the shape of the active site channel and the hydrophobicity of the substrate-binding site. Substitution of a few residues has completely changed the reaction catalyzed. Thus relatively minor divergence has given rise to two enzymes that carry out quite different reactions. Finally, a set of enzymes whose relationships may not be immediately obvious is Gcl–IlvI–PoxB. The reactions catalyzed, identified by EC number, are written in the Enzyme Commission database as:
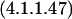 |
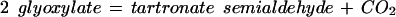 |
 |
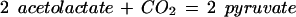 |
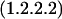 |
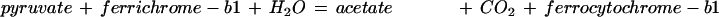 |
It is not immediately obvious what the relationship of the third reaction is to the other two. However, there is a common mechanism. All three enzymes use thiamine diphosphate cofactor. Viewing the second reaction in reverse direction (Table 2) positions all three reactions as decarboxylation of α-keto acids mediated by thiamine diphosphate. The first two are nonoxidative, the third oxidative. All proceed through carbanion intermediates (Silverman 2000).
In the case of the 4.1.3.18 reaction, deprotonation of the cofactor gives an -ylide form of thiamine diphosphate, which adds to the α carbon of one of the pyruvate molecules, destabilizing the carbonyl group, causing decarboxylation. Addition of this complex intermediate to the second pyruvate molecule is followed by elimination of the thiamine diphosphate, producing acetolactate. The 4.1.1.47 reaction is entirely analogous. Finally, the oxidative decarboxylation of the 1.2.2.2 reaction also passes through the thiamine diphosphate addition product, following which the carbanion intermediate is oxidized. All three enzymes are flavoproteins, although only the pyruvate oxidoreductase uses the flavin group to pass electrons (Chang et al. 1993). The shared features of the three enzymes that suggest a common ancestor are (1) the enzymatic promotion of the addition of thiamine diphosphate to the α carbon of a keto acid, producing a carbanion intermediate, which then loses the destabilized carbonyl group; and (2) a flavin prosthetic group, which is active in one case, not in the other two. The proposed ancestor would have been an α-keto acid decarboxylase flavoprotein that used a cofactor similar to thiamine diphosphate and had flexible substrate specificity.
In summary, our data indicate that the similarity found in sequence-related pairs and groups of enzymes in E. coli is in some cases related to the chemistry of the reaction catalyzed and in other cases to binding-site specificity. Often both aspects are used in the related protein. Neither EC numbers nor enzyme names can be relied upon to reflect such evolutionary connections. Grouping by distant sequence relatedness allows us to collect together proteins that differ, but whose molecular specificity, binding sites, and/or reaction chemistries are similar, revealing commonalities that probably reflect common ancestry. As we continue analysis of examples of sequence-related proteins in which divergence is ongoing today, we will be contributing to an understanding of the mechanisms of protein evolution.
Finally, this information has relevance to the arena of functional genomics in which sequence similarity between known and unknown proteins is used to ascribe function to the unknown protein. In such functional annotation we must be aware that weak but significant sequence similarity may reflect conservation of substrate, substrate/product, cofactor, or reaction chemistry. To understand which features are conserved and to make specific attributions of function, additional information is needed. In the absence of additional information, we suggest that attributions should be conservative, perhaps simply stating similarity to the known protein or to the class of enzyme, regulator, or transporter, rather than conferring a function without disclaimer.
METHODS
Selection of Examples of Divergence from Paralogous Groups
Proteins in E. coli K12 were grouped into sequence-related families using DARWIN (Data Analysis and Retrieval With Indexed Nucleotide/Peptide Sequences) programs (Gonnet et al. 1992; http://cbrg.inf.ethz.ch/) as described elsewhere (Riley and Labedan 1997; P. Liang, B. Labedan, and M. Riley, in prep.).
The potential examples of divergence were selected by the following criteria. We examined all paralogous groups in E. coli that had at least one partner with a different EC number in the first or second place plus paralogs that were in the same pathway. We relied more on EC numbers and same pathway than on gene and protein names because names are extremely variable.
Information about the paralogous groups was extracted from GenProtEC, a database of the genome and proteome of E. coli K-12 chromosomal genes (Liang et al. 2000). Members of these groups are sequence-related pairs with alignments of 100 amino acids or more and PAM values of 250 or less. (PAM value corresponds to the number of accepted point mutations per 100 residues separating two sequences.) GenProtEC can be accessed directly on the World Wide Web (http://genprotec.mbl.edu/).
Distribution of Similarity throughout Proteins
Protein sequences in the FASTA format were collected from SWISS-PROT protein sequence database (release 38) (Bairoch and Apweiler 2000; http://www.expasy.ch/sprot/).
Database searches for sequence similarity were performed with the DARWIN system (Gonnet et al. 1992) and gapped BLAST version 2.0 (Altschul et al. 1997; http://www.ncbi.nlm.nih.gov/). A minimum alignment of 100 amino acids was required. Results for significance of pairwise alignments are expressed as PAM values, which were calculated by DARWIN from the amino acid substitution tables appropriate to the distance between each pair. With the BLASTP program, we have searched the SWISS-PROT database, the BLOSUM62 matrix, and an Expect value (E) cutoff of 0.001.
Search for the Domain Similarities
We performed a Hidden Markov Model (HMM) search using PFAM protein domain database (Bateman et al. 2000) as provided by the HMMER2 package (HMMER 2.1.1; http://hmmer.wustl.edu/). The E-value cutoff level of 1.0 was adopted in this analysis.
Search for Functional Data
The Enzyme Commission (EC) numbers were collected from the ENZYME database (Bairoch 2000) in the ExPASy (Expert Protein Analysis System) proteomics server. This database is primarily based on the recommendations of the Nomenclature Committee of the International Union of Biochemistry and Molecular Biology (Webb 1992).
The information relative to the metabolic pathways, reactions, cofactors, and prosthetic groups was extracted from EcoCyc (Encyclopedia of E. coli Genes and Metabolism) (Karp et al. 2000; http://ecocyc.doubletwist.com/).
Acknowledgments
L.A.N. was supported by FAPESP (Fundação de Amparo à Pesquisa do Estado de São Paulo, Brazil) and M.R. by subcontract from NIH R01 RR07861 and the Marine Biological Laboratory Astrobiology Institute. We thank Alida Pellegrini-Toole for assistance with EcoCyc and Ping Liang with GenProtEC. Thanks also to Margrethe Serres and Thomas McCormack for assistance with revisions.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL mriley@mbl.edu; FAX (508) 289-7388.
Article and publication are at http://www.genome.org/cgi/doi/10.1101/gr.180901.
REFERENCES
- Alexander FW, Sandmeier E, Mehta PK, Christen P. Evolutionary relationships among pyridoxal-5′-phosphate-dependent enzymes. Regio-specific α, β and γ families. Eur J Biochem. 1994;219:953–960. doi: 10.1111/j.1432-1033.1994.tb18577.x. [DOI] [PubMed] [Google Scholar]
- Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babbitt PC, Gerlt JA. Understanding enzyme superfamilies. Chemistry as the fundamental determinant in the evolution of new catalytic activities. J Biol Chem. 1997;272:30591–30594. doi: 10.1074/jbc.272.49.30591. [DOI] [PubMed] [Google Scholar]
- Bairoch A. The ENZYME database in 2000. Nucleic Acids Res. 2000;28:304–305. doi: 10.1093/nar/28.1.304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bairoch A, Apweiler R. The SWISS-PROT protein sequence database and its supplement TrEMBL in 2000. Nucleic Acids Res. 2000;28:45–48. doi: 10.1093/nar/28.1.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bateman A, Birney E, Durbin R, Eddy SR, Howe KL, Sonnhammer EL. The Pfam protein families database. Nucleic Acids Res. 2000;28:263–266. doi: 10.1093/nar/28.1.263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belfaiza J, Parsot C, Martel A, de la Tour CB, Margarita D, Cohen GN, Saint-Girons I. Evolution in biosynthetic pathways: Two enzymes catalyzing consecutive steps in methionine biosynthesis originate from a common ancestor and possess a similar regulatory region. Proc Natl Acad Sci. 1986;83:867–871. doi: 10.1073/pnas.83.4.867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouhss A, Mengin-Lecreulx D, Blanot D, van Heijenoort J, Parquet C. Invariant amino acids in the Mur peptide synthetases of bacterial peptidoglycan synthesis and their modification by site-directed mutagenesis in the UDP-MurNAc:l-alanine ligase from Escherichia coli. Biochemistry. 1997;36:11556–11563. doi: 10.1021/bi970797f. [DOI] [PubMed] [Google Scholar]
- Bouhss A, Dementin S, Parquet C, Mengin-Lecreulx D, Bertrand JA, Le Beller D, Dideberg O, van Heijenoort J, Blanot D. Role of the ortholog and paralog amino acid invariants in the active site of the UDP-MurNAc-l-alanine:d-glutamate ligase (MurD) Biochemistry. 1999;38:12240–12247. doi: 10.1021/bi990517r. [DOI] [PubMed] [Google Scholar]
- Chang YY, Cronan JE., Jr Common ancestry of Escherichia coli pyruvate oxidase and the acetohydroxy acid synthases of the branched-chain amino acid biosynthetic pathway. J Bacteriol. 1988;170:3937–3945. doi: 10.1128/jb.170.9.3937-3945.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang YY, Wang AY, Cronan JE., Jr Molecular cloning, DNA sequencing, and biochemical analyses of Escherichia coli glyoxylate carboligase. An enzyme of the acetohydroxy acid synthase–pyruvate oxidase family. J Biol Chem. 1993;268:3911–3919. [PubMed] [Google Scholar]
- Clausen T, Huber R, Laber B, Pohlenz HD, Messerschmidt A. Crystal structure of the pyridoxal-5′-phosphate dependent cystathionine β-lyase from Escherichia coli at 1.83 A. J Mol Biol. 1996;262:202–224. doi: 10.1006/jmbi.1996.0508. [DOI] [PubMed] [Google Scholar]
- Clausen T, Huber R, Prade L, Wahl MC, Messerschmidt A. Crystal structure of Escherichia coli cystathionine γ-synthase at 1.5 A resolution. EMBO J. 1998;17:6827–6838. doi: 10.1093/emboj/17.23.6827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dahm C, Muller R, Schulte G, Schmidt K, Leistner E. The role of isochorismate hydroxymutase genes entC and menF in enterobactin and menaquinone biosynthesis in Escherichia coli. Biochim Biophys Acta. 1998;1425:377–386. doi: 10.1016/s0304-4165(98)00089-0. [DOI] [PubMed] [Google Scholar]
- Eveland SS, Pompliano DL, Anderson MS. Conditionally lethal Escherichia coli murein mutants contain point defects that map to regions conserved among murein and folyl poly-γ-glutamate ligases: Identification of a ligase superfamily. Biochemistry. 1997;36:6223–6229. doi: 10.1021/bi9701078. [DOI] [PubMed] [Google Scholar]
- Fani R, Lio P, Lazcano A. Molecular evolution of the histidine biosynthetic pathway. J Mol Evol. 1995;41:760–774. doi: 10.1007/BF00173156. [DOI] [PubMed] [Google Scholar]
- Fani R, Tamburini E, Mori E, Lazcano A, Lio P, Barberio C, Casalone E, Cavalieri D, Perito B, Polsinelli M. Paralogous histidine biosynthetic genes: Evolutionary analysis of the Saccharomyces cerevisiae HIS6 and HIS7 genes. Gene. 1997;197:9–17. doi: 10.1016/s0378-1119(97)00146-7. [DOI] [PubMed] [Google Scholar]
- Fani R, Mori E, Tamburini E, Lazcano A. Evolution of the structure and chromosomal distribution of histidine biosynthetic genes. Orig Life Evol Biosph. 1998;28:555–570. doi: 10.1023/a:1006531526299. [DOI] [PubMed] [Google Scholar]
- Fitch WM. Distinguishing homologous from analogous proteins. Syst Zool. 1970;19:99–113. [PubMed] [Google Scholar]
- Galperin MY, Koonin EV. A diverse superfamily of enzymes with ATP-dependent carboxylate-amine/thiol ligase activity. Protein Sci. 1997;6:2639–2643. doi: 10.1002/pro.5560061218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gehring AM, Mori I, Walsh CT. Reconstitution and characterization of the Escherichia coli enterobactin synthetase from EntB, EntE, and EntF. Biochemistry. 1998;37:2648–2659. doi: 10.1021/bi9726584. [DOI] [PubMed] [Google Scholar]
- Gerlt JA, Babbitt PC. Mechanistically diverse enzyme superfamilies: The importance of chemistry in the evolution of catalysis. Curr Opin Chem Biol. 1998;2:607–612. doi: 10.1016/s1367-5931(98)80091-4. [DOI] [PubMed] [Google Scholar]
- Gonnet GH, Cohen MA, Benner SA. Exhaustive matching of the entire protein sequence database. Science. 1992;256:1443–1445. doi: 10.1126/science.1604319. [DOI] [PubMed] [Google Scholar]
- Horowitz NH. On the evolution of biochemical syntheses. Proc Natl Acad Sci USA. 1945;31:153–157. doi: 10.1073/pnas.31.6.153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ————— . The evolution of biochemical syntheses—Retrospect and prospect. In: Bryson V, Vogel HJ, editors. Evolving genes and proteins. New York: Academic Press; 1965. pp. 15–23. [DOI] [PubMed] [Google Scholar]
- Karp PD, Riley M, Saier M, Paulsen IT, Paley SM, Pellegrini-Toole A. The EcoCyc and MetaCyc databases. Nucleic Acids Res. 2000;28:56–59. doi: 10.1093/nar/28.1.56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JH, Krahn JM, Tomchick DR, Smith JL, Zalkin H. Structure and function of the glutamine phosphoribosylpyrophosphate amidotransferase glutamine site and communication with the phosphoribosylpyrophosphate site. J Biol Chem. 1996;271:15549–15557. doi: 10.1074/jbc.271.26.15549. [DOI] [PubMed] [Google Scholar]
- Labedan B, Riley M. Gene products of Escherichia coli: Sequence comparisons and common ancestries. Mol Biol Evol. 1995;12:980–987. doi: 10.1093/oxfordjournals.molbev.a040291. [DOI] [PubMed] [Google Scholar]
- ————— . Genetic inventory: Escherichia coli as a window on ancestral proteins. In: Charlebois RL, editor. Organization of the prokaryotic genome. Washington, DC.: ASM Press; 1999. pp. 311–329. [Google Scholar]
- Marolewski A, Smith JM, Benkovic SJ. Cloning and characterization of a new purine biosynthetic enzyme: A non-folate glycinamide ribonucleotide transformylase from E. coli. Biochemistry. 1994;33:2531–2537. doi: 10.1021/bi00175a023. [DOI] [PubMed] [Google Scholar]
- Mehta PK, Christen P. The molecular evolution of pyridoxal-5′-phosphate-dependent enzymes. Adv Enzymol Relat Areas Mol Biol. 2000;74:129–184. doi: 10.1002/9780470123201.ch4. [DOI] [PubMed] [Google Scholar]
- Riley M. Genes and proteins of Escherichia coli K-12 (GenProt EC) Nucl Acids Res. 1998;26:50–53. doi: 10.1093/nar/26.1.54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riley M, Labedan B. Protein evolution viewed through Escherichia coli protein sequences: Introducing the notion of a structural segment of homology, the module. J Mol Biol. 1997;268:857–868. doi: 10.1006/jmbi.1997.1003. [DOI] [PubMed] [Google Scholar]
- Riley M, Serres MH. Interim report on genomics of Escherichia coli. Annu Rev Microbiol. 2000;54:341–411. doi: 10.1146/annurev.micro.54.1.341. [DOI] [PubMed] [Google Scholar]
- Roy S. Multifunctional enzymes and evolution of biosynthetic pathways: Retro-evolution by jumps. Proteins. 1999;37:303–309. doi: 10.1002/(sici)1097-0134(19991101)37:2<303::aid-prot15>3.0.co;2-6. [DOI] [PubMed] [Google Scholar]
- Silverman RB. The organic chemistry of enzyme-catalyzed reactions. 1st ed. San Diego, CA: Academic Press; 2000. pp. 335–339. [Google Scholar]
- Webb EC, editor. Enzyme Nomenclature 1992. Recommendations of the Nomenclature Committee of the International Union of Biochemistry and Molecular Biology on the nomenclature and classification of enzymes. New York: Academic Press; 1992. [Google Scholar]
- Zhang R, Evans G, Rotella FJ, Westbrook EM, Beno D, Huberman E, Joachimiak A, Collart FR. Characteristics and crystal structure of bacterial inosine-5′-monophosphate dehydrogenase. Biochemistry. 1999;38:4691–4700. doi: 10.1021/bi982858v. [DOI] [PubMed] [Google Scholar]


