Abstract
Protein-protein interaction (PPI) network analysis has been widely applied in the investigation of the mechanisms of diseases, especially cancer. Recent studies revealed that cancer proteins tend to interact more strongly than other categories of proteins, even essential proteins, in the human interactome. However, it remains unclear whether this observation was introduced by the bias towards more cancer studies in humans. Here, we examined this important issue by uniquely comparing network characteristics of cancer proteins with three other sets of proteins in four organisms, three of which (fly, worm, and yeast) whose interactomes are essentially not biased towards cancer or other diseases. We confirmed that cancer proteins had stronger connectivity, shorter distance, and larger betweenness centrality than non-cancer disease proteins, essential proteins, and control proteins. Our statistical evaluation indicated that such observations were overall unlikely attributed to random events. Considering the large size and high quality of the PPI data in the four organisms, the conclusion that cancer proteins interact strongly in the PPI networks is reliable and robust. This conclusion suggests that perturbation of cancer proteins might cause major changes of cellular systems and result in abnormal cell function leading to cancer.
Keywords: Cancer proteins, Cancer genes, Protein-protein interactions, Protein interaction network, Global network characteristics, Network topology
1. Introduction
Protein–protein interactions (PPIs) play fundamental roles in cellular systems such as DNA replication, transcription regulation, signal transduction, molecule transportation, and recognition and modification of foreign molecules [Xia, et al., 2010]. PPIs have been widely used in the investigation of the causal mechanisms of diseases and disease comorbidity [Goh, et al., 2007]. During the last decade, rapid progress in high-throughput experimental techniques, especially yeast two-hybrid system, has greatly accelerated the generation of PPI data [Stelzl, et al., 2005; Krogan, et al., 2006]. In parallel, a great number of computational algorithms and methods have been developed to assist investigators to predict PPIs in one or multiple organisms [von Mering, et al., 2005; Sun, et al., 2007]. With the available massive amount of PPI data, investigators further developed framework to better interpret and evaluate the data, such as construction of a comprehensive PPI network in an organism (interactome) or subnetworks [Yu, et al., 2008; Venkatesan, et al., 2009]. So far, the quantity and quality of PPIs in several model organisms, especially in humans and yeast, have enabled investigators to construct reliable interactomes that serve as references in biomedical research, especially in studying molecular mechanisms of complex diseases [Uetz, et al., 2000; Giot, et al., 2003; Li, et al., 2004; Krogan, et al., 2006]. Specifically in humans, PPI data has been widely applied to identify and prioritize disease candidate genes, understand the relationship between disease genes, explore network properties of disease genes, and reconstruct disease-specific subnetworks [Goh, et al., 2007; Chen, et al., 2009; Zanzoni, et al., 2009; Kann, 2010; Barabasi, et al., 2011; Jia, et al., 2011].
Cancer is one of the most severe human diseases. It has been widely investigated by numerous approaches, among which include examining cancer proteins’ topological features, searching dynamic modularity, and predicting novel cancer genes by subnetwork analysis [Jonsson and Bates, 2006; Platzer, et al., 2007; Taylor, et al., 2009; Sun and Zhao, 2010]. Recently, we systematically examined the global and local network characteristics of cancer proteins in the human interactome and compared their features with other proteins encoded by essential genes or control genes [Sun and Zhao, 2010]. Our analysis revealed that, relative to essential or control proteins, cancer proteins had higher connectivity and betweenness centrality values but shorter distance in the human interactome, suggesting that cancer proteins tend to interact strongly and serve as biologically important hubs [Sun, et al., 2010; Sun and Zhao, 2010]. Here, connectivity (degree) measures the number of the direct links of a protein node in the PPI network; distance measures the length of the path having the smallest number of links between a pair of selected nodes (i.e., shortest path); and betweenness centrality measures the sum of the fractions of shortest paths between all possible pairs of nodes in the network that traverse the node. However, it is well known that cancer genes and their proteins have been extensively studied due to investigators’ special interest in cancer as well as substantial funding of cancer research. As a result, the observation of strong interaction of cancer proteins might be an artifact introduced by the bias of human PPI data [Cai, et al., 2010; Sun, et al., 2010]. Whether this artifact is real or not remains unclear to us.
Here, we examined this important issue by comparing network characteristics of cancer proteins with other proteins in three other organisms (fly, worm, and yeast), whose interactomes are largely not biased toward cancer or other diseases. We first identified four sets of human genes: cancer genes, non-cancer essential genes, non-cancer disease genes, and control genes. Then, we found their homologous genes in three organisms: fly (Drosophila melanogaster), worm (Caenorhabditis elegans), and yeast (Saccharomyces cerevisiae). Finally, we examined their network properties by three common measurements: degree, distance and betweenness centrality. Our results confirmed that cancer proteins interact strongly not only in humans but also in all the three non-human organisms, suggesting that perturbation of cancer proteins might cause major changes of cellular systems and result in abnormal cell function leading to cancer.
2. Materials and methods
2.1 Protein-protein interaction data
We downloaded the most recent version of the PPI data from the Protein Interaction Network Analysis (PINA) platform (http://csbi.ltdk.helsinki.fi/pina/, March 4, 2010) [Wu, et al., 2008]. We retrieved PPI data in four organisms: human (Homo sapiens), fly (Drosophila melanogaster), worm (Caenorhabditis elegans), and yeast (Saccharomyces cerevisiae), because these four organisms have largest amount of available annotated PPI data. Only the experimentally verified PPI data was extracted for analysis. The protein identifications were then mapped to the official gene symbols in the National Center for Biotechnology Information Gene database (NCBI, http://www.ncbi.nlm.nih.gov/gene/). After removing redundancy and self-interactions, we used the PPIs with matched gene symbols to construct protein interaction networks for the four organisms. Table 1 summarizes the number of proteins and their interactions in the four organisms.
Table 1.
Summary of genes (proteins) and their protein-protein interactions in four organisms
| Organism | No. of interactions | No. of proteins | Proportion of genome (%)a | No. of genes (proteins)b |
|||
|---|---|---|---|---|---|---|---|
| Cancer genes | Non-cancer disease genes | Essential genes | Control genes | ||||
| Human | 50,100 | 10,324 | 43.70 | 370 (420) | 3132c (4666) | 1832c (2253) | 5857 (5857) |
| Fly | 25,026 | 7474 | 54.06 | 94 (123) | 834 (1205) | 596 (796) | 1412 (1908) |
| Worm | 6876 | 3885 | 19.25 | 34 (74) | 331 (911) | 249 (584) | 574 (1469) |
| Yeast | 54,376 | 5433 | 92.37 | 35 (35) | 355 (362) | 213 (217) | 652 (669) |
Proportion of the proteins in the interactome over the total number of proteins encoded by the protein-coding genes in the genome.
Number of genes whose proteins could be found in the protein-protein interaction network. The numbers in parentheses are all the available genes categorized by the corresponding annotations in each organism (i.e., some genes whose proteins could not be found in the PPI dataset).
Specifically in the human genome, there are 867 genes overlapped between non-cancer disease genes and essential genes.
2.2 Classification of human protein-coding genes
First, we retrieved 427 human cancer genes and their annotations from the Cancer Gene Census database (CGC, http://www.sanger.ac.uk/genetics/CGP/Census/, 2010-03-30 version) [Futreal, et al., 2004]. Among them, 420 cancer genes had official gene symbols in the NCBI Gene database. We considered these genes as cancer genes. Second, we obtained human disease genes from two public resources: Online Mendelian Inheritance in Man (OMIM, http://www.ncbi.nlm.nih.gov/omim/, downloaded on October 18, 2010) [Hamosh, et al., 2005] and Genetic Association Database (GAD, http://geneticassociationdb.nih.gov/, downloaded on October 26, 2010) [Blekhman, et al., 2008]. We mapped the disease genes in these two databases to the genes with official gene symbols in NCBI. Then, we excluded the cancer genes retrieved from the CGC database. This step resulted in a total of 4666 genes; we considered them as non-cancer disease gene set. Third, we obtained a human essential gene set from our previous work based on mouse lethality phenotype [Sun and Zhao, 2010]. After we excluded the cancer genes, we had 2253 non-cancer essential genes. Finally, we used all the protein-coding genes included in the human interactome as a representative set of well-characterized human genes. Those genes that did not appear in any of the above three gene sets were considered as control genes. This process resulted in a total of 5857 control genes. In the next step, we identified genes in these four sets that could be mapped to the human interactome. We had 370 cancer genes, 3132 non-cancer disease genes, 1832 essential genes, and 5857 control genes whose proteins were included in the human interactome; we defined them as four protein sets accordingly (Table 1).
2.3 Human homologous genes in other organisms
The homologs of human cancer genes, essential genes, non-cancer disease genes, and control genes in the other three organisms (i.e. fly, worm and yeast) were retrieved from the NCBI HomoloGene database (http://www.ncbi.nlm.nih.gov/sites/entrez?db=homologene, release 64). For comparison purposes, those genes were considered as the corresponding cancer genes, non-cancer disease genes, essential genes, and control genes in the corresponding organisms.
The total number of protein-coding genes in each of the four genomes was retrieved from the GENE_INFO file downloaded from ftp://ftp.ncbi.nlm.nih.gov/gene/DATA/GENE_INFO (downloaded on October 18, 2010).
2.4 Network topological measures and statistical analysis
In a PPI network, a node denotes a protein encoded by a gene and an edge denotes an interaction between two proteins. We used three common network topological measures, i.e., degree, distance and betweenness centrality, to examine network topological properties of four sets of proteins in the PPI network. We briefly describe the measurement here; more details were provided in our previous study [Sun and Zhao, 2010]. For a node in a PPI network, degree (also connectivity) measures the number of links for the node to other nodes; distance measures the number of links of the shortest path traveling from the node to another node; and betweenness centrality measures the sum of the fractions that the node is a member of the set of shortest paths that connect all the pairs of nodes in the network [Freeman, 1977]. We used an R package, igraph (http://igraph.sourceforge.net/), to calculate these network topological measures.
To evaluate the significance of the network properties of each protein set, we applied an empirical re-sampling approach. First, for each protein set of interest that had n proteins, we randomly selected n proteins from all the available proteins (i.e., random protein set) and calculated the network properties (degree, distance and betweenness centrality). We repeated this re-sampling process 1000 times. Next, to estimate the significance of average degree or betweenness centrality observed in a protein set of interest, we counted the number of random protein sets whose average degree (Nd) or betweenness centrality (Nb) was higher than the observed average degree or betweenness centrality, respectively. Similarly, for the distance, we counted the number of random protein sets whose average distance (Ns) was shorter than the observed distance. Finally, we calculated their empirical P value by Nd/1000, Nb/1000, and Ns/1000, respectively, for these three corresponding network topological measures.
3. Results and discussion
3.1 Data summary for cancer protein analysis
We reconstructed the whole human interaction network based on all the available experimentally verified PPIs. It contained 10,324 nodes (corresponding to 10,324 unique genes or proteins) and 50,100 edges (corresponding to 50,100 unique PPIs). Similarly, there were 7474 nodes and 25,026 PPIs in the whole fly network, 3885 nodes and 6876 PPIs in the whole worm network, and 5433 nodes and 54,376 PPIs in the whole yeast network (Table 1). Those proteins in the interactomes accounted for 43.70%, 54.06%, 19.25% and 92.37% of the total proteins encoded by the protein-coding genes in the human, fly, worm and yeast genomes, respectively. The amount of data indicated it is overall sufficient for a comparative PPI study for cancer and other proteins. In humans, we had 420, 4666, 2253 and 5857 cancer proteins, non-cancer disease proteins, essential proteins and control proteins, among which, 370, 3132, 1832 and 5857 could be mapped onto the whole human PPI network, respectively. Note that the essential protein set is not mutually exclusive from the non-cancer disease protein set – there were 867 proteins shared. For these four sets of proteins, their homologous genes (proteins) were identified in fly, worm, and yeast and were summarized in Table 1. Except for the worm data, the majority of proteins in each protein set could be mapped to the corresponding network. For example, among the 362 homologous non-cancer disease proteins in yeast, 355 could be mapped to the whole yeast network (Table 1).
3.2 Strong cancer protein interaction in four organisms
In this section, we compared four sets of proteins in four organisms by three network topological measurements: degree, distance, and betweenness centrality. Table 2 summarizes the network properties of proteins encoded by cancer, non-cancer disease, essential and control genes.
Table 2.
Network topology properties of four sets of proteins in four organisms
| Organism | Cancer proteins | Non-cancer disease proteins | Essential proteins | Control proteinsa |
|---|---|---|---|---|
| Degree | ||||
| Human | 24.48 (0)b | 10.70 (0) | 16.17 (0) | 7.14 (1) |
| Fly | 12.36 (0) | 7.26 (0.023) | 8.54 (0) | 7.24 |
| Worm | 8.50 (0) | 4.82 (0) | 5.30 (0) | 4.23 |
| Yeast | 38.29 (0.017) | 28.13 (0.001) | 32.80 (0.001) | 31.08 |
| Distance | ||||
| Human | 3.60 (0) | 3.94 (0.836) | 3.77 (0) | 3.98 (1) |
| Fly | 4.13 (0.011) | 4.24 (0.023) | 4.20 (0.001) | 4.24 |
| Worm | 4.30 (0.010) | 4.60 (0.446) | 4.49 (0.005) | 4.58 |
| Yeast | 2.77 (0.003) | 2.83 (0) | 2.80 (0) | 2.82 |
| Betweenness centrality | ||||
| Human | 5.34×104 (0) | 1.79×104 (0) | 2.95×104 (0) | 0.84×104 (1) |
| Fly | 3.08×104 (0) | 1.40×104 (0.023) | 1.74×104 (0) | 1.32×104 |
| Worm | 2.01×104 (0.007) | 1.04×104 (0) | 1.06×104 (0.001) | 0.83×104 |
| Yeast | 0.72×104 (0.102) | 0.70×104 (0.105) | 0.82×104 (0.090) | 0.85×104 |
The P-values of control proteins in fly, worm and yeast were not included because proteins homologous to human control proteins did not account for a large proportion of the proteins in the interactome of the three organisms.
The P-values based on randomization analysis are included in parentheses.
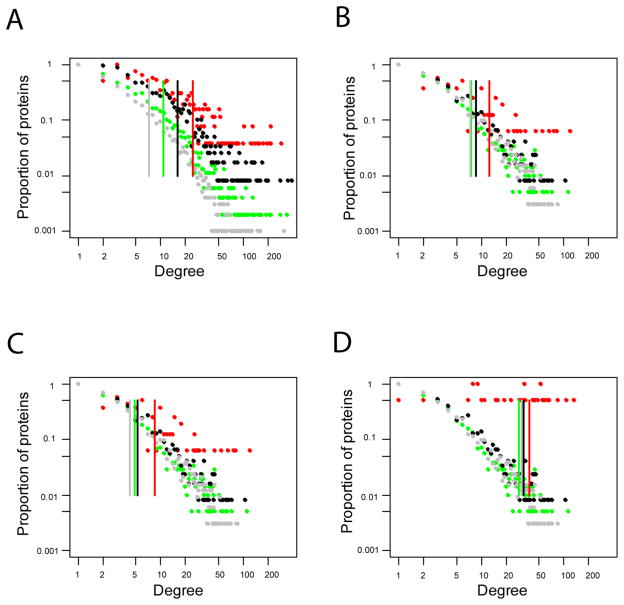
Degree is the most elementary characteristic in a network. In humans, the average degree of cancer proteins was 24.48, which was more than two times that of non-cancer disease proteins (10.70), approximately one and a half times that of essential proteins (16.17), and more than three times that of control proteins (7.14). This observation confirmed our previous report of stronger interaction of cancer proteins than other proteins in the whole human network [Sun and Zhao, 2010]. We then examined this features in the other three organisms whose PPI data was not biased towards cancer or general disease studies. As shown in Table 2, cancer proteins had much stronger interactions than any other proteins, regardless of whether the other proteins were categorized as non-cancer proteins, essential proteins, or control proteins. Specifically, cancer proteins had approximately 1.7, 1.8 and 1.4 times as many interaction partners as non-cancer disease proteins in fly, worm and yeast, respectively. For a more detailed view of the degree characteristic, we separated the nodes by their degree value and examined their degree distribution (Fig. 1). For human proteins, we found that the cancer proteins tend to skew toward a higher degree than the non-cancer disease, essential or control proteins (Fig. 1A). This feature was subsequently confirmed in the fly, worm and yeast data, though the extent was different (Fig. 1B, 1C, 1D). We noted that the difference in distributions between cancer proteins and any other proteins is always strong in the human, fly and worm; this difference was seen in the yeast too. However, compared to human PPI distribution, the difference among the non-cancer disease, essential or control proteins was not as prominent in fly, worm, or yeast (Fig. 1). It is worth noting that yeast exhibits a flatter trend than human, fly and worm (Fig. 1). This observation might be caused by the small number of homologs of human genes used in the analysis of the yeast interactome (Table 1).
Fig. 1.
Degree distribution and average degrees of four sets of proteins (cancer, non-cancer disease, essential, and control proteins) in four organisms. (A) Human. (B) Fly. (C) Worm. (D) Yeast. Y-axis represents the proportion of proteins having a specific degree. Note that the scale of both the X-axis and Y-axis is not linear. The average degree of each protein set is labeled in vertical line. Red dots denote cancer proteins, green dots denote non-cancer disease proteins, black dots denote essential proteins, and grey dots denote control proteins.
Furthermore, the cancer proteins, non-cancer proteins, and essential proteins had significantly more direct interactions than randomly selected proteins (empirical P values were 0, 0 and 0, respectively; here P=0 means there was no randomly selected protein set having a higher average degree than the observed value of cancer proteins, non-cancer disease proteins or essential proteins), while control proteins had the opposite characteristic (empirical P=1; all randomly selected protein sets had a higher average degree than the observed values of the control proteins) (Table 2). Similar to the human data, we found the empirical P values for the observed degree of the cancer proteins, non-cancer disease proteins, and essential proteins in the other three organisms were all less than 0.05, indicating that the features were not random. For example, the empirical P values were 0, 0.023, and 0 for fly cancer proteins, non-cancer disease proteins, and essential proteins, respectively (Table 2).
We next examined distance. In a network, distance measures how many nodes need to pass through from one node to another [Barabasi and Oltvai, 2004]. The average distance for the cancer proteins (3.60) was shorter than that of non-cancer disease proteins (3.94), essential proteins (3.77), or control proteins (3.98) in humans. Moreover, cancer proteins and essential proteins had significantly shorter distance than randomly selected proteins (both empirical P values were 0), while non-cancer disease proteins and control proteins had the opposite characteristic (empirical P values were 0.836 and 1, respectively). This is not surprising because non-cancer disease proteins (3132) and control proteins (5857) accounted for the majority of proteins in the human protein network (Table 1). Importantly, we confirmed that cancer proteins had shorter distance than other proteins in fly, worm, and yeast (Table 2). For example, in fly, the average distance of cancer proteins was 4.13, shorter than that of the non-cancer disease proteins (4.24), essential proteins (4.20), or control proteins (4.24).
Finally, we examined betweenness centrality of the four sets of proteins. Betweenness centrality measures how many shortest paths go through a particular node in a specific network. It can be used to reflect how many signals might have paths through the node in a network. In humans, the average betweenness centrality of the cancer proteins (5.34 × 104) was substantially greater than that of the non-cancer disease proteins (1.79 × 104), essential proteins (2.95 × 104), or control proteins (0.84 × 104), respectively. This pattern was confirmed in fly and worm, e.g., the average betweenness centrality of cancer homologous proteins had the highest values among the four protein sets. For example, the average betweenness centrality of cancer proteins was 3.08 × 104, greater than that of non-cancer disease proteins (1.40 × 104) or essential proteins (1.74 × 104) in fly. The empirical P values of proteins encoded by cancer, non-cancer disease and essential genes were 0, 0.023 and 0, respectively. These results suggested that the cancer proteins tended to occupy network positions that are of global importance in communications between protein pairs compared to the non-cancer disease proteins, essential proteins or control proteins. However, compared to humans, fly, and worm, betweenness centrality values among four sets of proteins were similar in yeast: 0.72 × 104 (cancer proteins), 0.70 × 104 (non-cancer disease proteins), 0.82 × 104 (essential proteins), and 0.85 × 104 (control proteins). This might be partially due to the small sample size of yeast cancer proteins relative to the yeast interactome size. Further investigation of this feature is needed in future.
In this study, we excluded self-interactions of proteins in the measurement of network topological properties. Examination of self-interactions might shed additional light on the molecular mechanisms of cancer, as protein duplication usually stabilizes the PPI network but cancer proteins tend to have strong perturbation in the network. We examined protein dimers (self-interacting proteins) in the four organisms. There were 1069, 69, 3, and 183 protein dimers in the human, fly, worm, and yeast interactomes, respectively. Among them, we found 56, 4, 0, and 2 dimers belonged to cancer proteins in these four organisms. While the number of cancer protein dimers is small and consistently smaller than that in other three categories of protein sets (e.g., 2, 23, 10, 148 dimers in the cancer proteins, non-cancer disease proteins, essential proteins, and control proteins in the yeast interactome), the proportion of dimers in the cancer protein set is not always smaller than that in other three categories of protein sets (e.g. the proportion is 0.057, 0.065, 0.047, and 0.227 in yeast cancer, non-cancer disease, essential, and control protein sets, respectively). Considering the likely biases in the human data and smaller number of protein dimers in other three organisms, this analysis of self-interactions could not provide much insight on network stabilization reduction of cancer proteins, and thus, this is a preliminary analysis.
4. Conclusion
In this study, we examined whether the previous report of stronger cancer protein interactions and its unique network topological characteristics is due to the bias of the protein interaction data generated by prevalent cancer research in humans. We addressed this issue by uniquely comparing the network topological characteristics of cancer proteins with other proteins in four organisms, three of which (fly, worm, and yeast) did not have biased PPI data towards cancer. We confirmed that cancer proteins have stronger connectivity, shorter distance, and larger betweenness centrality than non-cancer disease proteins, essential proteins, and control proteins in all four of these organisms. Our statistical evaluation indicated that such observations were overall unlikely due to the randomness of the data. Although the PPI data is still incomplete in any organisms, considering the large size and high quality of the PPI data in the four organisms used in this study, the conclusion that cancer proteins interact strongly in the PPI networks should be reliable and robust. This conclusion has important implications for cancer proteins’ biological function in cellular system, especially at the interactome level. It also helps cancer investigators to better interpret cancer risk genes and design effective drug targets.
Acknowledgments
This work was partially supported by the National Institutes of Health Grant Nos. R03LM009598-02 and R03LM009598-02W1. Z.Z. was supported by Vanderbilt’s Specialized Program of Research Excellence in GI Cancer grant P50CA95103 and the VICC Cancer Center Core grant P30CA68485.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Barabasi AL, Gulbahce N, Loscalzo J. Network medicine: a network-based approach to human disease. Nat Rev Genet. 2011;12:56–68. doi: 10.1038/nrg2918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barabasi AL, Oltvai ZN. Network biology: understanding the cell’s functional organization. Nat Rev Genet. 2004;5:101–113. doi: 10.1038/nrg1272. [DOI] [PubMed] [Google Scholar]
- Blekhman R, et al. Natural selection on genes that underlie human disease susceptibility. Curr Biol. 2008;18:883–889. doi: 10.1016/j.cub.2008.04.074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai J, Borenstein E, Petrov D. Broker genes in human disease. Genome Biol Evol. 2010;2:815–825. doi: 10.1093/gbe/evq064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J, Aronow B, Jegga A. Disease candidate gene identification and prioritization using protein interaction networks. BMC Bioinformatics. 2009;10:73. doi: 10.1186/1471-2105-10-73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freeman LC. Set of Measures of Centrality Based on Betweenness. Sociometry. 1977;40:35–41. [Google Scholar]
- Futreal P, et al. A census of human cancer genes. Nat Rev Cancer. 2004;4:177–183. doi: 10.1038/nrc1299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giot L, et al. A protein interaction map of Drosophila melanogaster. Science. 2003;302:1727–1736. doi: 10.1126/science.1090289. [DOI] [PubMed] [Google Scholar]
- Goh K, et al. The human disease network. Proc Natl Acad Sci USA. 2007;104:8685–8690. doi: 10.1073/pnas.0701361104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamosh A, et al. Online Mendelian Inheritance in Man (OMIM), a knowledgebase of human genes and genetic disorders. Nucleic Acids Res. 2005;33:D514–D517. doi: 10.1093/nar/gki033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia P, et al. dmGWAS: dense module searching for genome-wide association studies in protein-protein interaction networks. Bioinformatics. 2011;27:95–102. doi: 10.1093/bioinformatics/btq615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jonsson P, Bates P. Global topological features of cancer proteins in the human interactome. Bioinformatics. 2006;22:2291–2297. doi: 10.1093/bioinformatics/btl390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kann M. Advances in translational bioinformatics: computational approaches for the hunting of disease genes. Brief Bioinf. 2010;11:96–110. doi: 10.1093/bib/bbp048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krogan NJ, et al. Global landscape of protein complexes in the yeast Saccharomyces cerevisiae. Nature. 2006;440:637–643. doi: 10.1038/nature04670. [DOI] [PubMed] [Google Scholar]
- Li S, et al. A map of the interactome network of the metazoan C. elegans. Science. 2004;303:540–543. doi: 10.1126/science.1091403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Platzer A, Perco P, Lukas A, Mayer B. Characterization of protein-interaction networks in tumors. BMC Bioinformatics. 2007;8:224. doi: 10.1186/1471-2105-8-224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stelzl U, et al. A human protein-protein interaction network: a resource for annotating the proteome. Cell. 2005;122:957–968. doi: 10.1016/j.cell.2005.08.029. [DOI] [PubMed] [Google Scholar]
- Sun J, et al. Schizophrenia Gene Networks and Pathways and Their Applications for Novel Candidate Gene Selection. PloS One. 2010;5:e11351. doi: 10.1371/journal.pone.0011351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun J, et al. InPrePPI: an integrated evaluation method based on genomic context for predicting protein-protein interactions in prokaryotic genomes. BMC Bioinformatics. 2007;8:414. doi: 10.1186/1471-2105-8-414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun J, Zhao Z. A comparative study of cancer proteins in the human protein-protein interaction network. BMC Genomics. 2010;11(Suppl 3):S5. doi: 10.1186/1471-2164-11-S3-S5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor I, et al. Dynamic modularity in protein interaction networks predicts breast cancer outcome. Nat Biotechnol. 2009;27:199–204. doi: 10.1038/nbt.1522. [DOI] [PubMed] [Google Scholar]
- Uetz P, et al. A comprehensive analysis of protein-protein interactions in Saccharomyces cerevisiae. Nature. 2000;403:623–627. doi: 10.1038/35001009. [DOI] [PubMed] [Google Scholar]
- Venkatesan K, et al. An empirical framework for binary interactome mapping. Nat Methods. 2009;6:83–90. doi: 10.1038/nmeth.1280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Mering C, et al. STRING: known and predicted protein-protein associations, integrated and transferred across organisms. Nucleic Acids Res. 2005;33:D433–437. doi: 10.1093/nar/gki005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J, et al. Integrated network analysis platform for protein-protein interactions. Nat Methods. 2008;6:75–77. doi: 10.1038/nmeth.1282. [DOI] [PubMed] [Google Scholar]
- Xia J, Han K, Huang D. Sequence-based prediction of protein-protein interactions by means of rotation forest and autocorrelation descriptor. Protein Pept Lett. 2010;17:137–145. doi: 10.2174/092986610789909403. [DOI] [PubMed] [Google Scholar]
- Yu H, et al. High-quality binary protein interaction map of the yeast interactome network. Science. 2008;322:104–110. doi: 10.1126/science.1158684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zanzoni A, Soler-Lopez M, Aloy P. A network medicine approach to human disease. FEBS Lett. 2009;583:1759–1765. doi: 10.1016/j.febslet.2009.03.001. [DOI] [PubMed] [Google Scholar]