SUMMARY
The development of comparative phylogenetic methods has provided a powerful toolkit for addressing adaptive hypotheses, and researchers have begun to apply these methods to test the role of pollinators in floral evolution and diversification. One approach is to reconstruct the history of both floral traits and pollination systems to determine if floral trait change is spurred by shifts in pollinators. Looking across multiple shifts, it is also possible to test for significant correlations between floral evolution and pollinators using parsimony, likelihood and Bayesian methods for discrete characters or using statistical comparative methods for continuous characters. Evolutionary shifts in pollinators and floral traits may cause changes in diversification rates, and new methods are available for simultaneously studying character evolution and diversification rates. Relatively few studies have yet applied formal comparative methods to understanding how pollinators affect floral evolution across the phylogeny, and fruitful directions for future applications are discussed.
Keywords: ancestral state reconstruction, diversification, floral evolution, PGLS, phylogenetics, pollination system, stochastic mapping, transition rate
Comparative analysis has a long history in the study of plant-pollinator interactions. By examining how floral features serve to attract pollinators and enhance reproductive success in different species of angiosperms, pollination biologists have sought to identify floral traits that are adaptations for particular modes of pollination. For example, Darwin (1877) examined how the shape, size and orientation of floral parts vary among orchid species in relation to the type of pollinators that visit them. He proposed that these differences across species, acquired since the common ancestor of orchids, are adaptations to promote cross pollination given each species’ “conditions of life.” Looking more broadly across angiosperms, pollination biologists have noted combinations of floral features (“syndromes”) that have arisen independently in distantly related lineages in association with particular modes of pollination, consistent with the idea that these syndromes reflect floral adaptation (Stebbins, 1970; Fenster et al. 2004). These comparative studies of floral morphology and pollination systems across lineages have provided the foundation for our understanding of the ecology and evolution of plant-animal interactions and have inspired numerous studies detailing the mechanisms by which pollinators have acted as agents of selection (e.g., Cresswell & Galen, 1991; Campbell et al. 1996; Meléndez -Ackerman et al, 1997).
Despite the rich history of comparing floral features across species in the context of their pollination systems, the application of formal phylogenetic approaches to the study of pollinator-mediated floral evolution has only just begun. Modern phylogenetic methods make it possible to address not only basic questions such as how many times a particular trait has arisen but also to test broader questions about how floral traits respond to pollinator shifts and how such evolutionary transitions affect the fates of lineages. To date, however, the vast majority of phylogenetic studies of floral radiations use comparative analyses only to map changes of traits on trees, and often these reconstructions are limited to floral traits or pollination modes alone. Studies that extend comparative methods to estimating evolutionary correlations among traits and pollinators or testing for associations between trait evolution and diversification rates remain rare. This gap between what is possible and what has been done may reflect the fact that many statistical comparative methods are relatively new (e.g., Pagel, 1994; Martins & Hansen, 1997, Maddison et al., 2007) and also that there are few groups in which the floral trait variation, the pollination ecology and the phylogeny have been well-studied.
This review aims to outline how phylogenetic methods and particularly statistical comparative methods can be used to test the role of pollinator shifts in floral evolution. Although we are increasingly discovering that floral traits are shaped by a multitude of biotic and abiotic forces (McCall & Irwin, 2006; Strauss & Whittall, 2006; Kessler et al., 2008; Armbruster et al., 2009), the central role played by pollinators seems undeniable (Schemske & Bradshaw, 1999; Fenster et al. 2004). I will first review how the history of floral traits and pollination systems can be reconstructed to ask whether the order of evolutionary events is consistent with floral adaptation in response to pollinator shifts. Next, I consider how one can test for correlated evolution by looking across multiple evolutionary transitions. In addition, I will briefly consider the impact of transitions in floral traits and pollination systems on the diversification of lineages, and how differential diversification of lineages might affect tests of pollinator-mediated floral evolution. Throughout the review, I focus on examples from the literature that employ independently-estimated phylogenies (not based on floral morphology) and that characterize pollination systems using field studies or observations (as opposed to inference from floral morphology). This approach minimizes the potential for circularity and allows for robust tests of the relationship between pollinators and floral trait evolution across the tree. Although my goal here is to examine how pollinators have shaped the evolution of flowers, the methods and approaches I describe could be used to test the importance of any putative selective factor on any trait.
Reconstructing the history of trait evolution
Phylogenetic methods for ancestral state reconstruction can be applied to any trait that is heritable. Although genetic material may be the first character that jumps to mind, phylogenetic analysis has been used to study the evolution of a wide range of traits, including morphological features, behavior, physiological traits, geographic range and habitat preferences. Like genetic material, descendent species tend to inherit the character state present in their ancestor. For example, an understory species is likely to give rise to other understory species, and a wind-pollinated species is likely to give rise to other wind-pollinated species (a pattern often called “niche conservatism”; Webb et al., 2002; Weins & Graham, 2005). However, in the course of evolutionary time, these character states do change, some more frequently than others. Although long-term stasis is itself an interesting evolutionary phenomenon (Levinton, 1983), we rely on shifts in character states and the resulting variation across species for testing hypotheses about the drivers of evolutionary change.
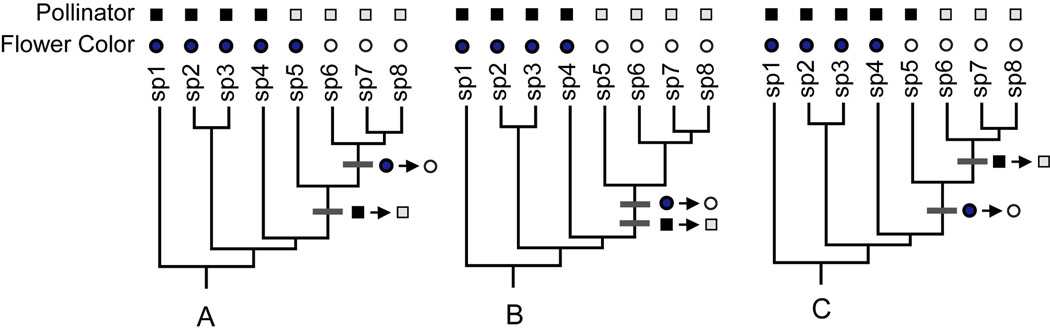
The most commonly used approach for reconstructing the history of floral traits and pollination systems is maximum parsimony, which seeks to explain the trait variation across a set of species by postulating the fewest number of character state changes. An example of parsimony reconstruction is shown in Figure 1 with a hypothetical group of 8 plant species that vary in flower color and pollination system. The three trees (Fig. 1 A–C) display different possible patterns of trait variation, and in each case, the most parsimonious reconstruction would be a single change from blue to white flowers and a single change from bee to moth pollination. It is worth noting that this parsimony reconstruction does not depend on the lengths of any of the branches, that is, all of branches are equally likely to have experienced trait shifts although they may vary markedly in their duration (as indicated by their relative length).
Fig. 1.
Parsimony reconstruction of flower colors and pollination system in hypothetical group of 8 plant species. There are two possible states for pollinator, where the black squares represent bee pollination and the gray squares moth pollination. There are two flower colors, blue (dark circles) and white (open circles). Inferred changes in pollinators and flower colors are marked on the tree with bars. The scenarios in (A) and (B) are consistent with the hypothesis that white flowers evolved as an adaptation for moth pollination whereas the scenario in (C) suggests that the evolution of white flowers was not driven by the evolution of moth pollination because the flower color change occurred in bee-pollinated lineage.
Using this parsimony reconstruction, we can ask whether the order of changes is consistent with the hypothesis that pollinators served as the selective agent causing the floral trait change (Coddington, 1988; Baum & Larson, 1991). For example, if the shift from blue to white flowers followed the transition to moth-pollination (i.e., if the color change occurred in a moth-pollinated lineage), the reconstruction would support the hypothesis that the evolution of moth pollination drove the evolution of white flowers (Fig. 1A). If the shifts in color and pollination map to the same branch of the tree (Fig 1B), the pattern would be consistent with the moth adaptation hypothesis. Although it is unlikely that the changes in color and pollination system truly occurred simultaneously, a lineage possessing only one of the derived states (white flowers or moth pollination) has not survived to present day in Fig 1B. This scenario may often be the case if the selective pressure is strong, e.g., if blue-flowered plants have low fitness in the moth-pollination environment.
In contrast, Fig. 1C contradicts the hypothesis that the evolution of moth pollination drove the evolution of white flowers because the color change occurred in a lineage that was pollinated by bees instead. If white flowers nonetheless confer a fitness advantage when moths act as pollinators, white flowers are best viewed as an exaptation, a trait whose current utility does not match its function when it was originally selected (Baum & Larson, 1991; Armbruster, 1997). Understanding the evolutionary origin of white flowers in a case such as Fig. 1C would require considering additional adaptive hypotheses (e.g., fine scale shifts in pollinator fauna, changes in habitat, etc.) and also non-adaptive hypotheses (genetic drift). In addition to the three scenarios portrayed in Fig. 1, we can imagine scenarios in which there is no correspondence between the evolution of the pollination system and the flower color, suggesting that the two have evolved independently.
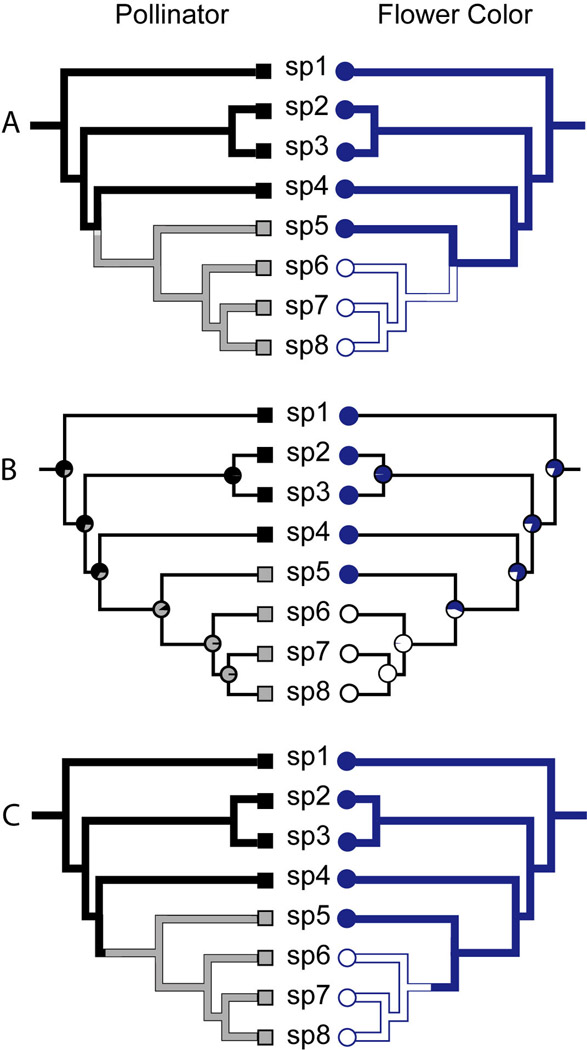
Although parsimony remains the most common approach for reconstructing evolutionary history, particularly for floral traits, comparative biologists are increasingly utilizing model-based maximum likelihood (ML) and Bayesian stochastic mapping. Both ML and Bayesian approaches offer the advantage of allowing transition rates between states to vary, estimating the statistical support for a given trait reconstruction, and using branch lengths to inform the probability of change. The latter feature derives from the expectation that change is more likely along long branches (long periods of time). Figure 2 compares parsimony mapping with ML and stochastic mapping using the data and phylogeny from Fig. 1A.
Fig. 2.
Comparison of ancestral state reconstruction methods. As in Fig. 1, there are two states for pollinator, bee pollination (black squares) and moth pollination (gray squares), and two states for flower color, blue (dark circles) and white (open circles). The characters states for each tip follow Fig. 1A. (A) Parsimony reconstruction with branches “painted” to show ancestral states. Trait changes are localized to nodes by convention. (B) Maximum likelihood reconstruction with pie charts at each node showing the relative probability of each state. (C) A single stochastic mapping realization for each character. The location of the color shift indicates the exact position of change along the branch during the realization. Note that multiple realizations of character history would be needed to obtain an estimate of the number and position of changes in each character.
All three methods provide information about the ancestral states at nodes and the timing of shifts in pollinators and flower color, but they differ in several important ways. Parsimony identifies both the number and order of the shifts, given that there is a single most parsimonious reconstruction for each character, but does not provide a measure of how confident we can be in that reconstruction (Fig 2A). In contrast, ML reconstruction (Fig. 2B) gives the relative probability of each state at each node using transition rates estimated from the tip states and the branch lengths (Pagel, 1994). Still, pinpointing the position of character changes, and thus estimating the number of shifts, remains more difficult with ML (Steele & Penny, 2000 and references therein), and software to “map” changes using ML has only recently been developed (O’Meara, 2008). Stochastic mapping, the newest of the three methods, brings together the most attractive features of parsimony and likelihood (Fig. 2C). Similar to parsimony, it directly estimates the number and direction of trait shifts, and because it is model-based like ML, it can provide statistical measures, such as the probability of ancestral states at nodes and confidence intervals for each type of character change (Huelsenbeck et al., 2003; Bollback, 2006).
To date, only a handful of studies have reconstructed the evolutionary history of both pollination system and floral traits for any group of flowering plants using an independent estimate of the phylogeny and any of the reconstruction methods described above (e.g., Baum et al., 1998; Hapeman & Inoue, 1997; Perez et al., 2006; Martén-Rodríguez et al., 2010). These studies have suggested intriguing patterns consistent with adaptive hypotheses such as the shifts in multiple floral traits (flower color and petal reflexion) coincident with the evolution of moth pollination in Schizanthus (Perez et al. 2006). However, they have also revealed several challenges with respect to addressing hypotheses of floral adaptation. A particular floral trait change may occur following or simultaneously with a pollinator shift in one part of the tree (consistent with an adaptive hypothesis) but not in another (Hapeman & Inoue, 1997). Also, uncertainty in the phylogeny and/or in the trait reconstruction can leave the order and position of shifts in traits and pollination systems ambiguous (Baum et al. 1998). In order to overcome these challenges, we must move beyond visually comparing reconstructions and apply statistical phylogenetic approaches to test for macroevolutionary patterns, such as contingent evolution and correlated evolution. In most cases, incorporating phylogenetic uncertainty into these tests is easily accomplished.
Detecting correlated shifts in pollinators and floral traits
The repeated evolution of a particular trait in a given environment in independent lineages is a classic source of evidence for adaptation (Pagel, 1999). For example, we might observe that red flowers tend to evolve in lineages that are hummingbird-pollinated, i.e., that the two traits, flower color and hummingbird pollination, evolve in a correlated fashion across the phylogeny. Such an observation would support the idea that red flower color is adaptive for plants pollinated by hummingbirds. However, red flowers may sometimes arise by genetic drift (non-adaptation) or due to other selective forces such as defense against herbivores. Also, some hummingbird-pollinated lineages may not have the capacity to evolve red floral pigmentation even if it would be advantageous. Thus, even if a particular trait is adaptive in a particular environment, we would not expect a perfect correspondence between trait and environment, but rather a general trend across the phylogeny. In our hummingbird example, we would predict that red flowers would evolve significantly more often in lineages with hummingbird pollination than those without.
One straightforward parsimony approach to test for correlated evolution is the concentrated changes test, or the CCT (Maddison, 1990). The histories of both the independent character (in this case, pollination) and the dependent character (a floral trait) are mapped on the phylogeny, and using either exact methods or simulations, we can determine the probability that the observed correspondence of trait shifts could have occurred by chance.
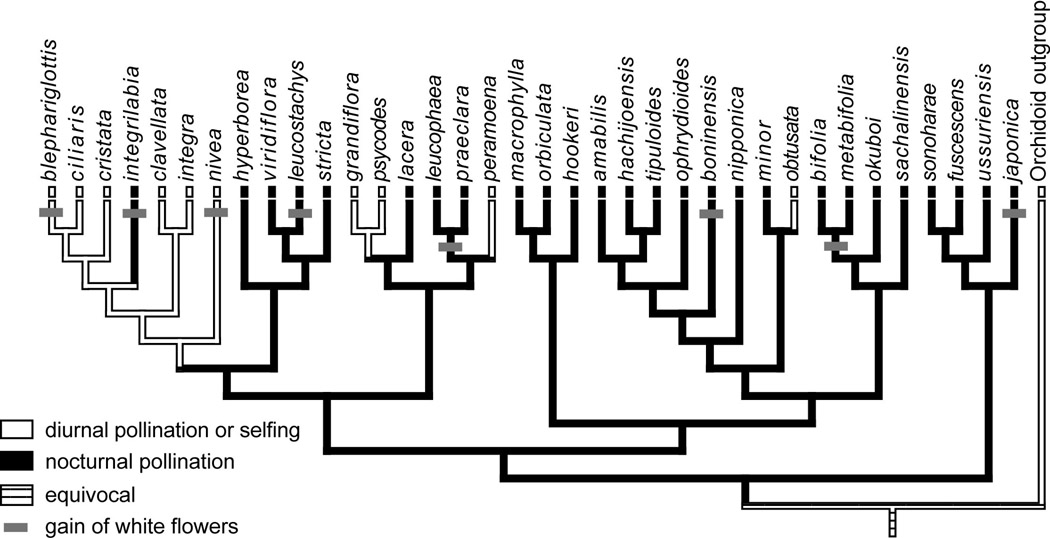
Consider the example in Fig 3, based on the Platanthera orchid dataset from Hapeman & Inoue (1997). Parsimony supports eight origins of white flowers in Platanthera. In six of these cases, the shift to white flowers occurred in lineages which rely on nocturnal moth pollinators, suggesting that the change in flower color represents an adaptive response to this pollination system. The CCT allows us to ask whether we can reject the null hypothesis that so many shifts to white flowers in nocturnally-pollinated lineages could occur by chance. Here, we cannot reject the null (P =0.39), and this is perhaps not surprising because most of the branches on the tree are reconstructed by parsimony as being nocturnally pollinated. Thus, if we were to randomly place shifts to white flowers along the tree, we would expect many to fall on nocturnally-pollinated (black) branches. Still, if all of changes (8/8 instead of 6/8) had arisen in nocturnally-pollinated lineages, we could have rejected the null hypothesis of a chance association (P=0.04). To my knowledge, the only applications of the CCT for testing associations between floral features and pollination system are Linder’s (1998) study of floral traits in wind-pollinated hamamelids and Altshuler’s (2003) study of flower color and hummingbird pollination, which both found significant support for correlated evolution.
Fig. 3.
Multiple origins of white flowers in Platanthera orchids. Parsimony reconstruction of diurnal and nocturnal pollination is shown on the ITS phylogeny from Hapeman & Inoue (1997) and gains of white flowers are indicated with gray bars. Relationships in the P. blephariglotis-P. ciliaris-P. cristata clade were randomly resolved before mapping.
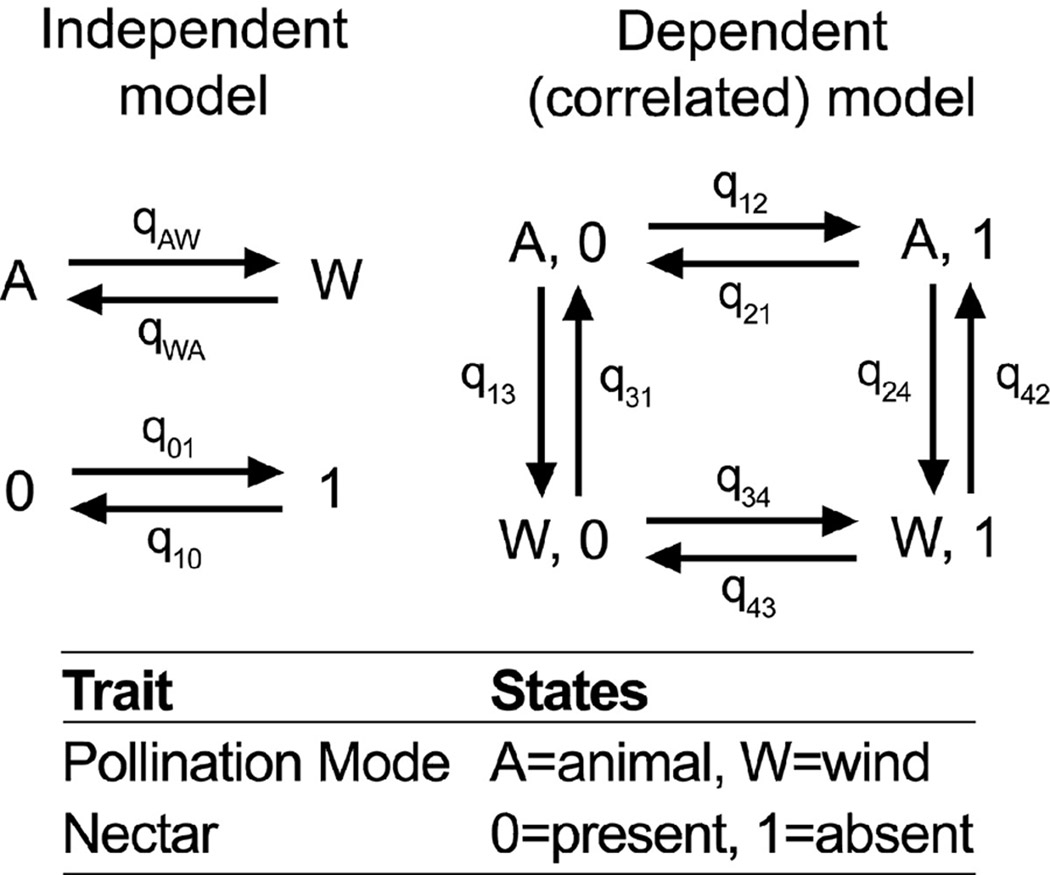
Similar questions can be addressed by applying the ML transition rate models (Pagel, 1994) described earlier in the context of ancestral state reconstruction (Fig. 2B). A useful example of this approach is provided by Friedman & Barrett (2008) who examined changes in floral traits associated with wind pollination. In this test for correlated evolution, two transition rate models are compared: one in which each trait evolves independently and one in which the change in one character depends on the state of the other character (as shown with wind pollination and nectar in Fig. 4). The fit of the two models is compared with a likelihood ratio test. Friedman and Barrett (2008) found that the dependent or correlated model was a significantly better fit (P<0.0001, df=4), indicating that shifts between wind and animal pollination are strongly correlated with the presence of nectar across angiosperms. Notice that in comparison to the parsimony-based CCT, this conclusion does not depend on particular reconstruction of the character states, only on how well the transition-rate models fit the data. Thus, if we were interested in knowing where on the tree these changes happened, we would have to undertake additional analyses such as those shown in Fig. 2.
Fig. 4.
Independent and correlated transition rate models for wind pollination and nectar adapted from Friedman & Barrett (2008). The independent model has four rate (q) parameters, and the correlated model has eight.
By constraining various parameters of the dependent model, a wide range of additional analyses can be undertaken to determine the nature of the correlated evolution. For example, one could test whether changes in nectar production are contingent on the shift to wind-pollination (see Friedman & Barrett, 2008). Also, one could examine whether any of the transitions, say from animal-pollinated without nectar to animal pollinated with nectar, are directional or, in the extreme, irreversible (Pagel, 1994). Despite this powerful framework for phylogenetic hypothesis testing, Friedman and Barrett’s (2008) paper appears to be the only study to apply ML transition rate models to test the relationship between pollination system and floral trait evolution. It would be particularly interesting to see tests of correlated evolution for other pollination systems (insect versus bird or nocturnal versus diurnal) and other floral traits (symmetry, color, flower size and shape, etc.).
Stochastic mapping offers a Bayesian alternative for testing correlated evolution. Both traits are mapped as shown in Fig. 2C, and from the mapping, we can determine the amount of time (i.e., the amount of branch length across the tree) spent in each character state. Following the wind-pollination example from Friedman and Barrett (2008), we might observe that across the tree, 86% of the total branch length was animal-pollinated and 14% was wind-pollinated. Put another way, at any particular point in the tree, the probability of being in state “animal-pollinated” is 0.86 and “wind-pollinated” is 0.14. We repeat this for the second character, nectar, and find the probability of having nectar at any point along the tree (state 0) is 0.62 and having no nectar (state 1) is 0.38. Assuming the characters evolve independently, we can compute the probability of any combination by multiplying their individual probabilities (Table 1). For example, the probability of being wind-pollinated and having nectar is 0.14 × 0.62 = 0.09. This is also the portion of the total branch length over which that we expect to find this character combination if the characters evolve independently.
Table 1.
Stochastic mapping test for correlation using the wind pollination and nectar example. Pollination mode has two states, animal-pollinated (A) and wind-pollinated (W), and nectar also has two states, present (0) or absent (1). Hypothetical observed and expected probabilities of finding each character state and state combination (e.g. A, 0) are given. The difference (d) is the observed minus the expected for each cell and D is the sum of the absolute value of the individual differences.
| Observed | Expected | ||||||||
| Pollination | Pollination | Difference (d) | |||||||
| Nectar | A | W | Nectar | A | W | ||||
| 0 | 0.60 | 0.02 | 0.62 | 0 | 0.53 | 0.09 | 0.62 | 0.07 | −0.07 |
| 1 | 0.26 | 0.12 | 0.38 | 1 | 0.33 | 0.05 | 0.38 | −0.07 | 0.07 |
| 0.86 | 0.14 | 0.86 | 0.14 | D = 0.28 | |||||
The comparison of the observed and expected associations of the character states in multiple stochastic mapping realizations can then be used to determine the strength of the association (Huelsenbeck et al., 2003). In the hypothetical example, we find that wind-pollinated lineages lack nectar more often than expected if the two traits evolved independently, and conversely, animal-pollinated lineages produce nectar more often than expected. We can compute the difference between the observed and expected (Table 1), and determine if this difference is significant using the straightforward simulation approach laid out in Huelsenbeck et al. (2003) and implemented by Bollback (2006). Martén-Rodríguez et al. (2010) recently applied this approach to test for correlations in Caribbean Gesnerieae and found that several pollination systems (e.g., hummingbird, bat and generalist) were significantly associated with particular flower shapes, colors and phenologies.
The ML transition rate and stochastic mapping approaches for testing correlated trait evolution both offer the advantage of easily incorporating phylogenetic uncertainty by using a sample of Bayesian trees. Because more probable trees will be visited more frequently in Bayesian analysis, these trees will be more common in the sample and have proportionally greater influence on the results. Incorporation of phylogenetic uncertainty, whether using a Bayesian sample of trees or another approach (e.g. bootstrapping), is relatively common in ancestral state reconstruction (Kay et al. 2005, Wilson et al. 2007), but unfortunately remains rare in studies applying the tests of correlated evolution of discrete traits described here (but see Leschen & Buckley, 2007).
Up to this point, I have considered floral traits and pollination systems that can be coded as discrete binary characters. The concentrated changes test can only be applied to binary characters, but stochastic mapping (Bollback, 2006), and transition rate models (as implemented in Brownie; O’Meara et al., 2006) can accommodate characters with multiple states. Still, the requirement that the traits be coded as discrete units may not be biologically realistic for many floral traits and pollination systems. Luckily, a parallel set of questions can be addressed with phylogenetic statistical methods suited for continuously valued variables.
Modeling pollination systems and floral traits as continuous characters
The degree of specialization varies widely across angiosperms with some species reliant on a single pollinator species and others utilizing a wide array of pollinators (Waser et al., 1996). Thus, whereas we may infer shifts between naturally discrete pollination modes in some clades (e.g., fragrance-collecting male bees and resin-collecting female bees, Armbruster, 1993), we are often likely to see shifts along a continuum, with changes occurring for the most part in the relative contribution of different pollinators to effective pollination (e.g., Smith et al. 2008a). In the latter scenario, pollination systems are best treated as a continuously varying trait and studied using phylogenetic approaches appropriate for continuous data.
Phylogenetic comparative methods (PCMs) for continuous characters encompass many of the techniques used in non-phylogenetic statistics, such as correlation, regression, and principal component analysis (PCA). However, because species are not independent due to their shared evolutionary history, PCMs must explicitly account for the effect of phylogeny and incorporate a model of how the traits evolve along the tree. The two most popular PCMs are phylogenetic independent contrasts (PICs, Felsenstein, 1985) and phylogenetic generalized least squares (PGLS, Grafen, 1989). The PICs method assumes an underlying Brownian motion model of trait evolution and transforms raw data for N taxa into N-1 independent contrasts, which can then be used in standard correlation and regression analyses. PICs is a special case of PGLS, which can be used to incorporate a variety of models of trait evolution in addition to Brownian motion (Martins & Hansen, 1997; Garland & Ives, 2000). PGLS can also be used to perform phylogenetic ANOVAs and ANCOVAs where the residual variation among species is correlated due to their shared history (Lavin et al., 2008). In additions to PICs and PGLS, there are many other PCMs, such as the phylogenetic mixed model (Housworth et al., 2004) and generalized estimating equations (Paradis & Claude, 2002), which accommodate different models of trait evolution and different types of comparative data.
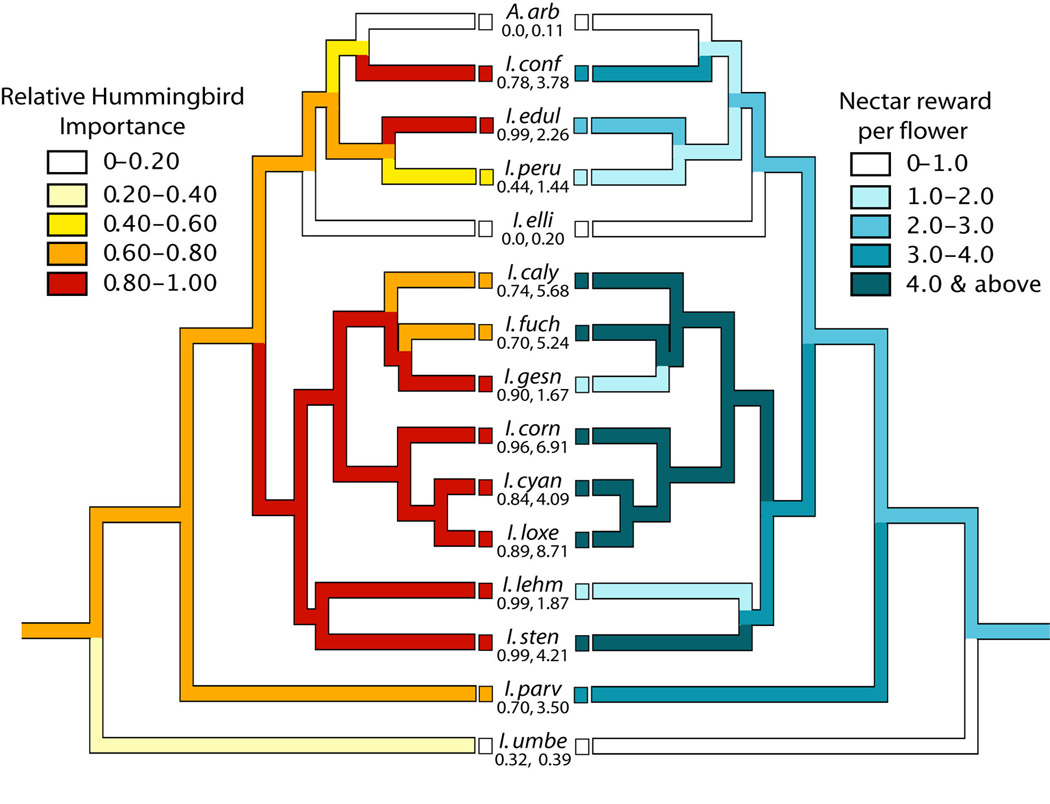
These statistical comparative methods have only rarely been applied to testing the relationship between floral trait evolution and pollination systems. For example, Armbruster (1996) examined the relationship between bract color and pollination mode in Dalechampia (Euphorbiaceae). After accounting for phylogenetic relatedness, he found no significant relationships and concluded that the color variation is likely due to indirect selection for pigmentation in vegetative tissue. Also, Smith et al. (2008a) used PGLS to test for correlations between pollinator importance and a suite of floral traits in Iochroma (Solanaceae). This study found that shifts in the relative importance of hummingbird and insect pollinators were significantly associated with changes in nectar reward and display size but not with changes in flower color or length. For example, Iochroma species that offered higher nectar rewards had significantly greater hummingbird pollination (Fig. 5). This finding is in accord with the idea that hummingbirds have higher energetic demands than most insect pollinators and thus select for larger nectar rewards (Heinrich, 1975; Stiles, 1978). Although both Armbruster (1996) and Smith et al. (2008a) grouped pollinators before analyses, it would also be possible to perform principal components analyses on pollinator visitation and/or effectiveness data (Wilson et al., 2004) and to use these PC scores for each species as continuous measures of pollination system (analogous to the study of piscivory of Collar et al., 2009).
Fig. 5.
The evolution of hummingbird pollination and nectar in 15 species of Iochrominae. I. is Iochroma and A. is Acnistus, a monotypic genus nested in Iochroma. Data are from Smith et al. (2008a). Branch colors indicate inferred state using squared change parsimony. For both characters, darker shades equal higher values. The values for each continuous character were grouped into 5 bins, and the raw values for each species are given beneath the name. Specific epithets for each species were abbreviated to the first four letters.
An added benefit of these comparative methods (e.g., PICs and PGLS) is the potential for simultaneously analyzing multiple variables. For instance, Smith et al. (2008a) conducted stepwise multiple correlation analyses to determine which sets of floral traits best explain the variation in pollinator importance across Iochroma and to estimate partial correlations between pollinator and floral traits. Multivariate analyses are also possible with discrete characters, and the software for implementing such analyses has recently been developed as part of the Brownie program (O’Meara et al. 2006).
The effects of plant-pollinator interactions on diversification rates
An outstanding question in the evolution of flowering plants is the extent to which shifts in pollination system affect diversification rate (Kay & Sargent, 2009). For example, we might hypothesize that an evolutionary transition from a generalized pollination system to a specialized system will restrict subsequent diversification if dependence on a small group of pollinators makes a species more prone to extinction (Futuyma & Moreno, 1988). Alternately, extreme specialization may result in rapid speciation of both the plants and their pollinators (Kiester et al., 1984), as is perhaps the case with figs and fig wasps (Rønsted et al., 2005). We could also imagine that other kinds of shifts in pollination systems that affect the type of pollinator but not necessarily the amount of specialization might affect subsequent diversification.
Thus far, a handful of studies have examined how diversification varies among lineages with different pollination systems (reviewed in Kay & Sargent, 2009). Several authors find support for greater diversification in lineages with biotic pollination than abiotic pollination (Dodd et al., 1999; Kay et al., 2006). Schiestl & Schluter (2009) found that increased specialization was associated with increased species richness in orchids, but Smith et al. (2008b) found no such pattern in Yucca (Agavaceae). Overall these studies suggest that changes in pollination systems can affect diversification rates and that specialized pollination is not likely a dead-end (see also Tripp & Manos, 2008); however they all apply methods which suffer from several statistical problems and biases (Vamosi & Vamosi, 2005; Maddison et al., 2007; Goldberg & Igic, 2008). Perhaps the most problematic issue is the use of the phylogeny to test the effect of a character on diversification when the character itself has shaped the tree.
The recently described binary state-dependent speciation and extinction (BiSSE) method (Maddison et al., 2007) offers a solution to this problem by simultaneously estimating rates of character change along with speciation and extinction rates in each character state. An added benefit of this method is that it can be applied even when the phylogeny of the group of interest is incomplete (FitzJohn et al., 2009). Although BiSSE has not yet been applied to test the effects of differences in pollination system on diversification rates, it has been used in two studies of floral character evolution. Armbruster et al. (2009) examined the evolution of defensive floral traits Dalechampia (Euphorbiaceae) and found no effect of these traits on diversification rates. In contrast, Smith et al. (2010) found that lineages lacking floral anthocyanins speciated at a rate roughly three times lower than that of pigmented lineages in the Quamoclit clade of Ipomoea (Convolvulaceae). As with any such analysis, the caveat must be added that the observed effect in Quamoclit could also be due to another character (such as pollination system) which evolves in a correlated fashion with floral anthocyanins.
Because floral traits and pollination systems have the potential to affect diversification rates and thus to cause biases in inferences about evolutionary history, BiSSE analyses or other methods incorporating state-dependent speciation and extinction are likely to become more prevalent in comparative studies. Methods for reconstructing ancestral states (as in Fig. 2) are already available in a BiSSE framework (Goldberg & Igic, 2008). Other extensions, such as the application to continuous characters and tests for correlated evolution, are currently being developed (R. E. FitzJohn, pers. comm.). Overall, this approach holds great promise for understanding how shifts in floral traits and pollination systems affect the diversification of plant lineages.
Conclusions
With the wealth of comparative phylogenetic methods that has developed over the past three decades, plant evolutionary biologists and ecologists are well positioned to test the role of pollinator shifts in floral diversification at fine and broad scales. By bringing together comparative pollination ecology with measures of floral variation in a phylogenetic context, we can determine whether the wide array of traits classically considered as floral adaptations for particular pollinators do indeed show evidence of correlated evolution across the phylogeny. Although a perfect correspondence is not expected, a floral trait that is adaptive for a particular pollination system should arise significantly more frequently in lineages with that pollination system than in lineages without, resulting in an overall pattern of correlated shifts across the tree. Looking across floral traits, some traits may more predictably follow pollinator shifts (i.e., be more tightly correlated) while others may only evolve in certain contexts (e.g., in particular geographic areas or in the presence of other traits). Cases in which a hypothesized floral adaptation does not correlate with pollinator shifts are themselves quite interesting because they may lead to the investigation of alternative hypotheses that may involve pollinators (e.g., variation due to selection for enhanced pollinator constancy) or may not (e.g., variation due to selection for defense against herbivores).
Indeed, macroevolutionary studies are best viewed as complementary to other studies of pollinator-mediated selection. While comparative approaches can reveal general patterns (correlations, directional trends, diversification effects), ecological, behavioral and genetic investigations at finer scales are needed to elucidate the evolutionary processes that give rise to these patterns. For example, the selective mechanisms underlying correlated shifts in floral traits and pollination systems across the phylogeny can be examined by studying how trait variation segregating within populations is related to pollinator visitation and effectiveness. In cases where floral transitions appear directional or, in the extreme, irreversible, evolutionary genetic studies may reveal that the genetic basis for the phenotypic change constrains reverse evolution (Zufall & Rausher, 2004). Similarly, evolutionary shifts in pollination ecology may be constrained if the nature of selection is directional, for example, if birds can impose selection on flowers adapted to bee pollination but not vice versa (Thomson & Wilson, 2008). When the evolution of particular traits (e.g., floral asymmetry, nectar spurs) consistently results in increased diversification rates, the mechanisms thought to underlie these associations (e.g., enhanced reproductive isolation due to more precise pollen placement or greater pollinator specificity) can be examined in the field using populations or sister species that exhibit variation in the trait (Hileman et al., 2003). This effort to connect macroevolutionary patterns with microevolutionary studies will ultimately provide a more complete understanding of nature of floral diversification and the importance of pollinators in this process.
Acknowledgements
The author is grateful for helpful comments from H. Kong, N. Waser, C. Wessinger, P. Wilson and an anonymous reviewer on an earlier draft. This work was supported by an NIH Ruth L. Kirschstein NRSA fellowship and the Center for Evolutionary Genomics at Duke University and the Duke Institute for Genome Sciences and Policy.
References
- Altshuler DL. Flower color, hummingbird pollination and habitat irradiance in four Neotropical forests. Biotropica. 2003;35:344–355. [Google Scholar]
- Armbruster WS. Evolution of plant pollination systems: hypotheses and tests with the neotropical vine Dalechampia. Evolution. 1993;47:1480–1505. doi: 10.1111/j.1558-5646.1993.tb02170.x. [DOI] [PubMed] [Google Scholar]
- Armbruster WS. Evolution of floral morphology and function: an integrated approach to adaptation, constraint, and compromise in Dalechampia (Euphorbiaceae) In: Lloyd D, Barrett S, editors. Floral biology. New York: Chapman & Hall; 1996. pp. 241–272. [Google Scholar]
- Armbruster WS. Exaptations link evolution of plant-herbivore and plant-pollinator interactions: A phylogenetic inquiry. Ecology. 1997;78:1661–1672. [Google Scholar]
- Armbruster WS, Lee J, Baldwin BG. Macroevolutionary patterns of defense and pollination in Dalechampia vines: Adaptation, exaptation, and evolutionary novelty. Proceedings of the National Academy of Sciences USA. 2009;106:18085–18090. doi: 10.1073/pnas.0907051106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baum DA, Larson A. Adaptation reviewed: A phylogenetic methodology for studying character macroevolution. Systematic Zoology. 1991;40:1–18. [Google Scholar]
- Baum DA, Small RL, Wendel JF. Biogeography and floral evolution of baobabs (Adansonia, Bombacaceae) as inferred from multiple data sets. Systematic Biology. 1998;47:181–207. doi: 10.1080/106351598260879. [DOI] [PubMed] [Google Scholar]
- Bollback JP. SIMMAP: Stochastic character mapping of discrete traits on phylogenies. BMC Bioinformatics. 2006;7:88. doi: 10.1186/1471-2105-7-88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell DR, Waser NM, Price MV. Mechanisms of hummingbird-mediated selection for flower width in Ipomopsis aggregata. Ecology. 1996;77:1463–1472. [Google Scholar]
- Coddington JA. Cladistic tests of adaptational hypotheses. Cladistics. 1988;4:3–22. doi: 10.1111/j.1096-0031.1988.tb00465.x. [DOI] [PubMed] [Google Scholar]
- Collar DC, O’Meara BC, Wainwright PC, Near TJ. Piscivory limits diversification of feeding morphology in Centrarchid fishes. Evolution. 2009;63:1557–1573. doi: 10.1111/j.1558-5646.2009.00626.x. [DOI] [PubMed] [Google Scholar]
- Cresswell JE, Galen C. Frequency-dependent selection and adaptive surfaces for floral character combinations—the pollination of Polemonium viscosum. American Naturalist. 1991;138:1342–1353. [Google Scholar]
- Darwin C. On the Various Contrivances by Which British and Foreign Orchids Are Fertilized. Chicago: University of Chicago Press; 1877. Republished 1984. [Google Scholar]
- Dodd ME, Silvertown J, Chase MW. Phylogenetic analysis of trait evolution and species diversity variation among angiosperm families. Evolution. 1999;53:732–744. doi: 10.1111/j.1558-5646.1999.tb05367.x. [DOI] [PubMed] [Google Scholar]
- Garland T, Ives AR. Using the past to predict the present: Confidence intervals for regression equations in phylogenetic comparative methods. American Naturalist. 2000;155:346–364. doi: 10.1086/303327. [DOI] [PubMed] [Google Scholar]
- Goldberg EE, Igic B. On phylogenetic tests of irreversible evolution. Evolution. 2008;62:2727–2741. doi: 10.1111/j.1558-5646.2008.00505.x. [DOI] [PubMed] [Google Scholar]
- Grafen A. The phylogenetic regression. Philosophical Transactions of the Royal Society of London, Series B. 1989;326:119–157. doi: 10.1098/rstb.1989.0106. [DOI] [PubMed] [Google Scholar]
- Hapeman JR, Inoue K. Plant-pollinator interactions and floral radiation in Platanthera (Orchidaceae) In: Givnish TJ, Sytsma KJ, editors. Molecular Evolution and Adaptive Radiation. New York: Cambridge University Press; 1997. pp. 432–454. [Google Scholar]
- Housworth EA, Martins EP, Lynch M. The phylogenetic mixed model. American Naturalist. 2004;163:84–96. doi: 10.1086/380570. [DOI] [PubMed] [Google Scholar]
- Huelsenbeck JP, Nielsen R, Bollback JP. Stochastic mapping of morphological characters. Systematic Biology. 2003;52:131–158. doi: 10.1080/10635150390192780. [DOI] [PubMed] [Google Scholar]
- Felsenstein J. Phylogenies and the comparative method. American Naturalist. 1985;125:1–15. doi: 10.1086/703055. [DOI] [PubMed] [Google Scholar]
- Fenster CB, Armbruster WS, Wilson P, Dudash MR, Thomson JD. Pollination syndromes and floral specialization. Annual Review of Ecology and Systematics. 2004;35:375–403. [Google Scholar]
- FitzJohn RG, Maddison WP, Otto SP. Estimating trait-dependent speciation and extinction rates from incompletely resolved phylogenies. Systematic Biology. 2009;58:595–611. doi: 10.1093/sysbio/syp067. [DOI] [PubMed] [Google Scholar]
- Friedman J, Barrett SCH. A phylogenetic analysis of the evolution of wind pollination in the angiosperms. International Journal of Plant Sciences. 2008;169:49–58. [Google Scholar]
- Futuyma DJ, Moreno G. The evolution of ecological specialization. Annual Review of Ecology and Systematics. 1988;19:207–233. [Google Scholar]
- Heinrich B. Energetics of pollination. Annual Review of Ecology and Systematics. 1975;6:139–170. [Google Scholar]
- Hileman LC, Kramer EM, Baum DA. Differential regulation of symmetry genes and the evolution of floral morphologies. Proceedings of the National Academy of Sciences USA. 2003;100:12814–12819. doi: 10.1073/pnas.1835725100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kay KM, Reeves P, Olmstead R, Schemske DW. Rapid speciation and the evolution of hummingbird pollination in Neotropical Costus subgenus Costus (Costaceae): Evidence from nrDNA ITS and ETS sequences. American Journal of Botany. 2005;92:1899–1910. doi: 10.3732/ajb.92.11.1899. [DOI] [PubMed] [Google Scholar]
- Kay KM, Voelckel C, Yang JY, Hufford KM, Kaska DD, Hodges SA. Floral characters and species diversification. In: Harder LD, Barrett SCH, editors. Ecology and Evolution of Flowers. New York: Oxford University Press; 2006. pp. 311–325. [Google Scholar]
- Kay KM, Sargent RD. The role of animal pollination in plant speciation: Integrating ecology, geography,and genetics. Annual Review of Ecology and Systematics. 2009;40:637–656. [Google Scholar]
- Kiester AR, Lande R, Schemske DW. Models of coevolution and speciation in plants and their pollinators. American Naturalist. 1984;124:220–243. [Google Scholar]
- Kessler D, Gase K, Baldwin IT. Field experiments with transformed plants reveal the sense of floral scents. Science. 2008;321:1200–1202. doi: 10.1126/science.1160072. [DOI] [PubMed] [Google Scholar]
- Lavin SR, Karasov WH, Ives AR, Middleton KM, Garland T., Jr Morphometrics of the avian small intestine compared with that of non-flying mammals: a phylogenetic approach. Physiological and Biochemical Zoology. 2008;81:526–550. doi: 10.1086/590395. [DOI] [PubMed] [Google Scholar]
- Leschen RAB, Buckley TR. Multistate characters and diet shifts: evolution of Erotylidae (Coleoptera) Systematic Biology. 2007;56:97–112. doi: 10.1080/10635150701211844. [DOI] [PubMed] [Google Scholar]
- Levinton JS. Stasis in progress: the empirical basis of macroevolution. Annual Review of Ecology and Systematics. 1983;14:103–137. [Google Scholar]
- Linder HP. Morphology and the evolution of wind pollination. In: Owens SJ, Rudall PJ, editors. Reproductive biology. Kew, UK: Royal Botanic Gardens; 1998. pp. 123–135. [Google Scholar]
- Maddison WP. A method for testing the correlated evolution of two binary characters: Are gains or losses concentrated on certain branches of a phylogenetic tree? Evolution. 1990;44:539–557. doi: 10.1111/j.1558-5646.1990.tb05937.x. [DOI] [PubMed] [Google Scholar]
- Maddison WP, Midford PE, Otto SP. Estimating a binary character’s effect on speciation and extinction. Systematic Biology. 2007;56:701–710. doi: 10.1080/10635150701607033. [DOI] [PubMed] [Google Scholar]
- Martén-Rodríguez S, Fenster CB, Agnarsson I, Skog LE, Zimmer EA. Evolutionary breakdown of pollinator specialization in a Caribbean plant radiation. New Phytologist. 2010 doi: 10.1111/j.1469-8137.2010.03330.x. in review. [DOI] [PubMed] [Google Scholar]
- Martins EP, Hansen TF. Phylogenies and the comparative method: A general approach to incorporating phylogenetic information into the analysis of interspecific data. American Naturalist. 1997;149:646–667. [Google Scholar]
- McCall AC, Irwin RE. Florivory: the intersection of pollination and herbivory. Ecology Letters. 2006;9:1351–1365. doi: 10.1111/j.1461-0248.2006.00975.x. [DOI] [PubMed] [Google Scholar]
- Meléndez-Ackerman E, Campbell DR, Waser NM. Hummingbird behavior and mechanisms of selection on flower color in Ipomopsis. Ecology. 1997;78:2532–2541. [Google Scholar]
- O'Meara BC, Ané C, Sanderson MJ, Wainwright PC. Testing for different rates of continuous trait evolution using likelihood. Evolution. 2006;60:922–933. [PubMed] [Google Scholar]
- O’Meara BC. Using trees: Myrmecocystus phylogeny, character evolution new methods for investigating trait evolution and species delimitation. Nature Precedings. 2008 http://dx.doi.org/10.1038/npre.2008.2261.1. [Google Scholar]
- Pagel M. Detecting correlated evolution on phylogenies: a general method for the comparative analysis of discrete characters. Proceedings of the Royal Society B. 1994;255:37–45. [Google Scholar]
- Pagel M. Inferring the historical patterns of biological evolution. Nature. 1999;401:877–884. doi: 10.1038/44766. [DOI] [PubMed] [Google Scholar]
- Paradis E, Claude J. Analysis of comparative data using generalized estimating equations. Journal of Theoretical Biology. 2002;218:175–185. doi: 10.1006/jtbi.2002.3066. [DOI] [PubMed] [Google Scholar]
- Perez F, Arroyo MTK, Medel R, Hershkovitz MA. Ancestral reconstruction of flower morphology and pollination systems in Schizanthus (Solanaceae) American Journal of Botany. 2006;93:1029–1038. doi: 10.3732/ajb.93.7.1029. [DOI] [PubMed] [Google Scholar]
- Rønsted N, Weiblen GD, Cook JM, Salamin N, Machado CA, Savolainen V. 60 million years of co-divergence in the fig-wasp symbiosis. Proceedings of the Royal Society of London Series B. 2005;272:2593–2599. doi: 10.1098/rspb.2005.3249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schemske DW, Bradshaw HD., Jr Pollinator preference and the evolution of floral traits in monkeyflowers (Mimulus) Proceedings of the National Academy of Sciences USA. 1999;96:11910–11915. doi: 10.1073/pnas.96.21.11910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schiestl FP, Schluter PM. Floral isolation, specialized pollination, and pollinator behavior in orchids. Annual Review of Entomology. 2009;54:425–446. doi: 10.1146/annurev.ento.54.110807.090603. [DOI] [PubMed] [Google Scholar]
- Smith CI, Pellmyr O, Althoff DM, Balcazar-Lara M, Leebens-Mack J, Segraves KA. Pattern and timing of diversification in Yucca (Agavaceae): specialized pollination does not escalate rates of diversification. Proceedings of the Royal Society B. 2008b;275:249–258. doi: 10.1098/rspb.2007.1405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith SD, Ane C, Baum DA. The role of pollinator shifts in the floral diversification of Iochroma (Solanaceae) Evolution. 2008a;62:793–806. doi: 10.1111/j.1558-5646.2008.00327.x. [DOI] [PubMed] [Google Scholar]
- Smith SD, Miller RE, Otto SP, FitzJohn RG, Rausher MD. The effects of flower color transitions on diversification rates in morning glories (Ipomoea subg. Quamoclit, Convolvulaceae). In: Long M, editor. Darwin's Heritage Today: Proceedings of the Darwin 200 Beijing International Conference; Beijing: Higher Education Press; 2010. in press. [Google Scholar]
- Stebbins GL. Adaptive radiation of reproductive characteristics in angiosperms. I: Pollination mechanisms. Annual Review of Ecology and Systematics. 1970;1:307–326. [Google Scholar]
- Steele M, Penny D. Parsimony, likelihood, and the role of models in molecular phylogenetics. Molecular Biology and Evolution. 2000;17:839–850. doi: 10.1093/oxfordjournals.molbev.a026364. [DOI] [PubMed] [Google Scholar]
- Stiles GF. Ecological and evolutionary implications of bird pollination. American Zoologist. 1978;18:715–727. [Google Scholar]
- Strauss SY, Whittall JB. Non-pollinator agents of selection on floral traits. In: Harder LD, Barrett SCH, editors. The ecology and evolution of flowers. Oxford, UK: Oxford University Press; 2006. pp. 120–138. [Google Scholar]
- Thomson JD, Wilson P. Explaining evolutionary shifts between bee and hummingbird pollination: Convergence, divergence, and directionality. International Journal of Plant Sciences. 2008;169:23–38. [Google Scholar]
- Tripp EA, Manos PS. Is floral specialization an evolutionary dead-end? Pollination system transitions in Ruellia (Acanthaceae) Evolution. 2008;62:1712–1736. doi: 10.1111/j.1558-5646.2008.00398.x. [DOI] [PubMed] [Google Scholar]
- Vamosi SM, Vamosi JC. Endless tests: guidelines for analyzing non-nested sister-group comparisons. Evolutionary Ecology Research. 2005;7:567–579. [Google Scholar]
- Waser NM, Chittka L, Price MV, Williams N, Ollerton J. Generalization in pollination systems, and why it matters. Ecology. 1996;77:1043–1060. [Google Scholar]
- Webb CO, Ackerly DD, McPeek MA, Donoghue MJ. Phylogenies and community ecology. Annual Review of Ecology and Systematics. 2002;33:475–505. [Google Scholar]
- Wiens JJ, Graham CH. Niche conservatism: Integrating evolution, ecology, and conservation biology. Annual Review of Ecology and Systematics. 2005;36:519–539. [Google Scholar]
- Wilson P, Castellanos MC, Hogue JN, Thomson JD, Armbruster WS. A multivariate search for pollination syndromes among penstemons. Oikos. 2004;104:345–361. [Google Scholar]
- Wilson P, Wolfe AD, Armbruster WS, Thomson JD. Constrained lability in floral evolution: Counting convergent origins of hummingbird pollination in Penstemon and Keckiella. New Phytologist. 2007;176:883–890. doi: 10.1111/j.1469-8137.2007.02219.x. [DOI] [PubMed] [Google Scholar]
- Zufall RA, Rausher MD. Genetic changes associated with floral adaptation restrict future evolutionary potential. Nature. 2004;428:847–850. doi: 10.1038/nature02489. [DOI] [PubMed] [Google Scholar]