Abstract
Gene amplification occurs in most solid tumors and is associated with poor prognosis. Amplification of 20q13.2 is common to several tumor types including breast cancer. The 1 Mb of sequence spanning the 20q13.2 breast cancer amplicon is one of the most exhaustively studied segments of the human genome. These studies have included amplicon mapping by comparative genomic hybridization (CGH), fluorescent in-situ hybridization (FISH), array-CGH, quantitative microsatellite analysis (QUMA), and functional genomic studies. Together these studies revealed a complex amplicon structure suggesting the presence of at least two driver genes in some tumors. One of these, ZNF217, is capable of immortalizing human mammary epithelial cells (HMEC) when overexpressed. In addition, we now report the sequencing of this region in human and mouse, and on quantitative expression studies in tumors. Amplicon localization now is straightforward and the availability of human and mouse genomic sequence facilitates their functional analysis. However, comprehensive annotation of megabase-scale regions requires integration of vast amounts of information. We present a system for integrative analysis and demonstrate its utility on 1.2 Mb of sequence spanning the 20q13.2 breast cancer amplicon and 865 kb of syntenic murine sequence. We integrate tumor genome copy number measurements with exhaustive genome landscape mapping, showing that amplicon boundaries are associated with maxima in repetitive element density and a region of evolutionary instability. This integration of comprehensive sequence annotation, quantitative expression analysis, and tumor amplicon boundaries provide evidence for an additional driver gene prefoldin 4 (PFDN4), coregulated genes, conserved noncoding regions, and associate repetitive elements with regions of genomic instability at this locus.
Genome scanning techniques such as Comparative Genomic Hybridization (CGH), Restriction Landmark Genome Scanning, and analysis of Loss of Heterozygosity (LOH) have mapped numerous regions of recurrent genome copy number abnormality in human solid tumors (Gray and Collins 2000). In breast tumors alone, >30 such regions have been identified (Kallioniemi et al. 1994) and the genomes of most other tumor types are similarly affected (Knuutila et al. 1998, 1999). Such aberrant loci are thought to encode proteins that participate in tumor progression as a result of altered gene dosage, translocations, and/or mutation. Typically, these “cancer genes” are identified by narrowly defining regions of recurrent loss or gain followed by functional assessment of candidate genes. This approach is becoming increasingly efficient with the development of high-resolution genome scanning techniques such as array CGH (Pinkel et al. 1998; Albertson et al. 2000). However, the mapping information from these techniques will be most informative only when integrated with well-annotated genomic sequence. To accomplish this, we have developed and applied a suite of software tools collectively called Genome Cryptographer (GC) to facilitate integrative analysis. GC collects genome sequence information from multiple databases and visually displays it in analysis intervals (AIs) of constant width along the genome. Displayed information includes CpG density, sequence tagged sites (STSs), expressed sequence tag (EST) clusters, locations and densities of repeated sequences (e.g., Alus, SINEs, LINEs), duplicons, similarities with syntenic murine sequences, known genes and genome copy number determined using array CGH.
We applied GC to the analysis of 1.2 Mb of 20q13.2 because it is amplified in a wide range of tumor types (Kallioniemi et al., 1994, 1998, 1999), appears to be an early event in breast cancer (Werner et al. 1999), and is associated with aggressive tumor behavior (Tanner et al. 1995), immortalization (Savelieva et al. 1997; Cuthill et al. 1999), and genome instability (Savelieva et al. 1997). The entire region is amplified in the majority of breast tumors with gain at 20q13.2 (Tanner et al. 1994, 1996). However, high-resolution fluorescent in-situ hybridization (FISH) (Collins et al. 1998), quantitative microsatellite analysis (QUMA) (Ginzinger et al. 2000), and array CGH (Albertson et al. 2000) mapping elucidated a complex amplicon structure with two regions of recurrent amplification separated by ∼600 kb (Albertson et al. 2000), one region containing ZNF217 (Collins et al. 1998) and the other CYP24 (Albertson et al. 2000). Overexpression of ZNF217 immortalizes cultured human mammary epithelial cells (HMEC) (Nonet et al. 2001) and overexpression of CYP24 has been postulated to interfere with vitamin D mediated differentiation (Albertson et al. 2000). Nevertheless, other genes in the amplicon peaks also may contribute to cancer progression. Accordingly, we sequenced and computationally analyzed the entire 1.2-Mb region to catalog all genes in the region and to attempt to identify structural features in the DNA sequence that might underlie local instability.
RESULTS AND DISCUSSION
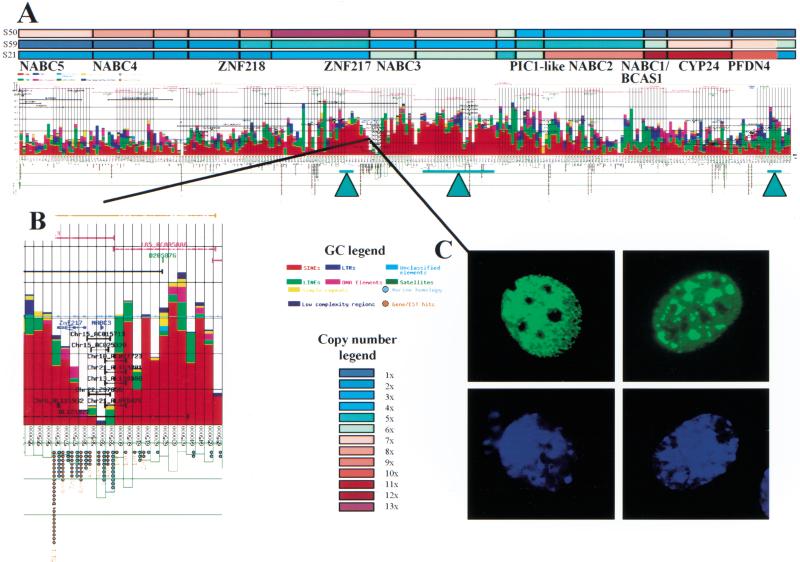
Figure 1 shows a GC analysis of a 1.2-Mb region of amplification at 20q13.2. This analysis identified six previously identified genes (Collins et al. 1998) as well as four genes (NABC3 [Novel gene Amplified in Breast Cancer], NABC4, NABC5, and prefoldin 4 [PFDN4]) that previously were not known to be present in this region (Fig. 1A) (Multiple gene prediction algorithms were used to find genes; however, these analyses failed to provide convincing evidence for additional coding sequences, and thus the data were not included.) We then manually integrated GC output and array-CGH data to map genes relative to amplicon peaks at genome sequence resolution and to identify sequence features that might play a role in the amplification process (Fig. 1A). The array-CGH mapping was performed with a contiguous set of bacterial artificial chromosome (BAC) clones spanning this amplicon (Albertson et al. 2000). Boxes indicate the genomic interval for which copy number was measured, and color corresponds to copy number with crimson representing highest copy number. The triangles point to amplicon boundaries defined as clusters of amplification breakpoints previously identified in primary tumors and breast-cancer cell lines.
Figure 1.
Integration of genome copy number and genome sequence information in a region of amplification at 20q13.2. (A) Genome Cryptographer (GC) analysis of a 1.2-Mb region of amplification. Average genome copy number values in selected tumors (S50, S59, S21) measured using array Comparative Genomic Hybridization (CGH) (Albertson et al. 2000) are shown as color-coded bars at the top of the figure. The array CGH data were obtained using a contig of BAC clones that now have been sequenced. Brick red lines represent public draft assemblies as of 2.1.01. Pink lines correspond to the exact size and position of the BAC clones used in the study. Densities and classification of repetitive elements are shown in color-coded cumulative bar chart above the X axis. CpG dinucleotide densities are plotted below the X axis as open green boxes. Sequence features such as genes are shown as horizontal lines above the X axis spanning the total extent of the sequence similarity. Exons are shown in bold lines. Genes and pseudogenes are represented by blue arrows pointing in the direction of transcription. The names of genes appear below the CGH copy number plot in black bold font. Total number of gene/EST hits and/or mouse identity regions are presented below the X axis as red or blue circles, respectively. Aquamarine triangles with bars, indicating the mapping resolution, mark the approximate positions of amplicon boundaries mapped by array CGH (Albertson et al. 2000), fluorescent in-situ hybridization (FISH) (Collins et al. 1998) and Southern hybridization (Collins et al., unpubl.). This figure can also be viewed at http://shark.ucsf.edu:8080/∼stas/GR2001/index.html. (B) Enlargement of the ZNF217-NABC3 region of 20q13.2 amplification. This panel further illustrates the ability of GC to annotate features such as public draft sequence assembly (orange), BAC template locations (pink), STSs (dark green), alignment of syntenic murine sequence (light blue line), human/murine sequence identities (light blue rectangle on line), human genes (dark blue), duplications and other identities to human genomic sequence (black). The locations of genome duplications (e.g., Chr15_AC015713) are identified above the black line indicating the chromosome 20 location of each duplicon. Ratios shown beneath EST clusters correspond to the total number of EST hits/total murine EST hits. Numbers under blue circles indicate the total number of murine sequence identities per analysis interval. (C) ZNF217-EGFP fusion proteins localize to the nucleus of HeLa cells and are excluded from the nucleoli. The top two panels show localization of ZNF217-GFP fusion and the bottom two panels show DAPI staining of cell nuclei.
The GC analysis suggests the possibility that repetitive elements are involved in amplification at 20q13.2. Figure 1 shows a markedly uneven distribution of the density and type of repetitive elements across the region. Earlier FISH- and array CGH-based studies (Collins et al. 1998; Albertson et al. 2000) mapped amplicon boundaries with a high degree of precision and revealed two classes of tumors. In one class, the copy number maximum is centered on the ZNF217-NABC3 locus (Collins et al. 1998). In the second class, a larger amplicon includes both the ZNF217-NABC3 and CYP24-PFDN4 loci (Albertson et al. 2000) with the copy number peak centered on the CYP24-PFDN4 locus. In the first class of tumors, the proximal boundary was mapped by FISH in two tumors (Collins et al. 1998) and refined by Southern blot mapping in one (C. Collins, unpubl.) to within 10 kb of the ZNF217 gene's 3∏ untranslated region (UTR). The distal boundary was mapped in three independent tumors and one cell line (Collins et al. 1998). In the second class, the boundary distal to CYP24-PFDN4 was mapped to within a single BAC in two tumors (Albertson et al. 2000). Interestingly, the average density of repetitive elements flanking amplicon boundaries is below 40%; however, each of the three amplicon boundaries fall into regions of >60% repetitive DNA content. Repetitive elements (e.g., Alu and L1) have been implicated in recombination (Moran et al. 1999), genome evolution (Brosius 1999) and disease-related aberrations (Huie et al. 1999). Thus, the association of high repetitive element density with regions of frequent chromosome breakage suggests a possible role for repetitive elements in the amplification process (e.g., as sites for recombination-driven amplification).
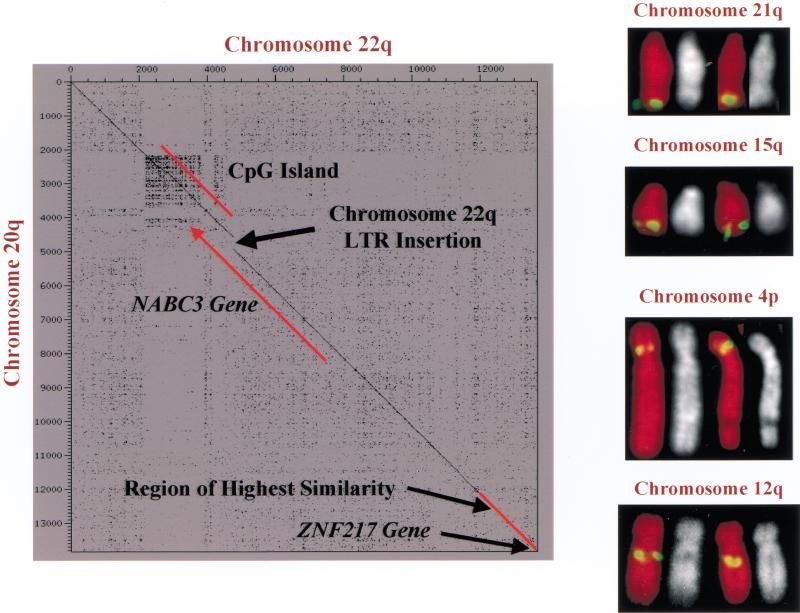
GC analysis also revealed a 14-Kb duplicon (Eichler 1998) 167 bp distal to ZNF217. This is significant because duplicons have been associated with evolutionarily unstable chromosomal loci in primates. Homologous recombination between duplicons has been implicated in the formation of duplications, deletions, inversions, translocations, and formation of supernumerary marker chromosomes (Ji et al. 2000), some of which are disease-related (Eichler 1998; Christian et al. 1999; Peoples et al. 2000). Thus, this element may play a role in amplification of the ZNF217-NABC3 locus in cancer. The duplicon includes NABC3 and a CpG island and is ∼97% identical to elements found on the long arms of chromosomes 15q and 22q (Fig. 2). Hybridization of probes spanning the duplicon to the CalTech D BAC library resulted in identification of 16 BAC clones. These were FISH-mapped to chromosomes 4p, 12q, 15q, 21q (Fig. 2), 20q, and 22q. In addition, some of the BAC clones decorated the pericentromeric regions of multiple chromosomes (data not shown). Although we do not know if each mapped BAC contains a complete element, we do know from GC analysis that chromosome arms 20q, 22q, and 15q do in fact have complete elements, and that chromosomes 10, 21, and 13 harbor fragments of the duplicon.
Figure 2.
Dotter (http://www.cgr.ki.se/cgr/groups/sonnhammer/Dotter.html) analysis of 14 kb 20q:22q duplication showing very high primary structure conservation. This plot corresponds to coordinates ∼580,000–594,000 in (B) and is an alignment between Chr22_Z97056 and sequence at 20q13.2. The positions of a CpG island, the NABC3 gene interrupted by insertion of a LTR on chromosome 22, and the start of the ZNF217 gene are annotated. Results of fluorescence in situ hybridization (FISH) mapping of four bacterial artificial chromosome (BAC) clones isolated by screening the Caltech D human BAC library with duplicon-specific probes. The FISH mapping confirms chromosome duplications shown in Figures 1A, 1C, 3A, and in this figure.
The degree of sequence conservation and pattern of chromosomal distribution provides compelling evidence that this element is indeed a duplicon (Eichler 1998). A retroviral LTR inserted in the chromosome 22 element disrupts the paralogous NABC3 gene (Fig. 2). Comparative analysis of human and syntenic mouse sequence identified an orthologous NABC3 gene at mouse chromosome 2H3 (syntenic to human chromosome 20q13.2). In addition, the position, size, and presumably function of the 1.8-Kb CpG island also is conserved (Fig. 3A). FISH mapping indicates that in mouse the NABC3 gene is single copy (data not shown). This finding is consistent with the current view that duplicons do not occur outside of primates (Eichler et al. 1999). Thus, duplicon's pangenomic migration most likely occurred after the primate-mouse divergence with 20q13.2 being the ancestral element. The finding of a duplicon within 20q13.2 amplicon is intriguing however, in the absence of data regarding the presence of duplicons in other amplicons, its role in mediating amplification remains unclear.
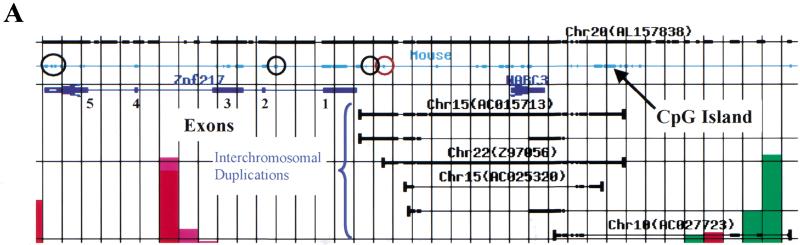
Figure 3.
(A) A high-resolution Genome Cryptographer (GC) analysis showing human/mouse sequence alignment. GC analysis was carried out in an analysis interval of 1 kb. This figure shows a chromosome 20 PAC (AL157838) in black. The extent of syntenic mouse sequence is indicated by a thin blue line with sequence identities shown as heavy lines. Human genes ZNF217 and NABC3 appear as dark blue arrows pointing in the direction of transcription. Bracketed lines show interchromosomal duplications. Their extent is shown as thin black lines with actual sequence identities indicated by heavy black lines (e.g., Chr15, AC015713). (B) Sequence alignment of noncoding conserved human and mouse sequence (circled in red on the GC analysis in A).
The 1.2-Mb encodes 10 genes, three CpG islands, and two pseudogenes. The ZNF217, NABC3, CYP24, and PFDN4 genes are of particular interest because they are located at amplification maxima. ZNF217 has been shown to immortalize HMEC and thus has properties consistent with it being a bona fide oncogene. Structurally, ZNF217 resembles a transcription factor having eight C2H2 motifs, a nuclear localization signal, and a proline-rich domain (Collins et al. 1998). The NABC3 cDNA has a poly-A tail, lacks an open reading frame, does not share identity with any known genes, lacks introns, and is expressed in a wide range of tissues (data not shown). Analysis of the predicted RNA secondary structure using MFOLD (http://bioweb.pasteur.fr/seqanal/interfaces/mfold.html) shows that it is unusually stable. These features suggest that NABC3 may encode an RNA gene rather than a processed pseudogene. Its possible role in cancer remains unclear. PFDN4 is a subunit of the heterohexameric chaperone protein prefoldin family (Vainberg et al. 1998). It captures unfolded actin and tubulin for delivery of cytosolic chaperone (CTT) (Vainberg et al. 1998; Hansen et al. 1999). PFDN4 may function as a transcription factor or cofactor in cell-cycle regulation (Iijima et al. 1996).
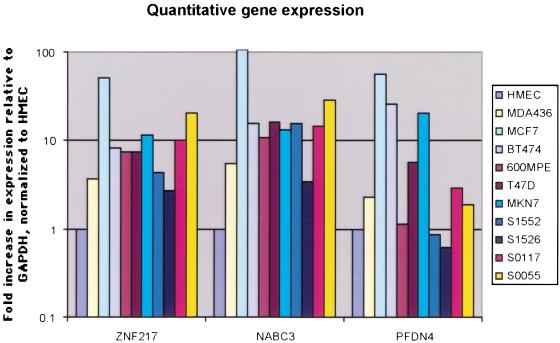
Expression levels of ZNF217, NABC3, and PFDN4 were analyzed in normal cultured human breast epithelial cells, breast-cancer cell lines, and primary tumors (Fig. 4) using quantitative reverse transcriptase-polymerase chain reaction (RT-PCR). Expression of NABC3 was strikingly similar to that of ZNF217, including high-level expression in the cell lines 600MPE and T47D in which they are not amplified. The coordinate expression of ZNF217 and NABC3 suggests utilization of common regulatory elements. To identify putative regulatory elements, we aligned syntenic mouse sequence spanning the ZNF217-NABC3 locus (Fig. 3). This alignment and a percent identity plot (PIP) analysis (http://nog.cse.psu.edu/pipmaker/) identified several conserved noncoding elements in and around the region encoding ZNF217-NABC3. In Figure 3A, these regions of conserved noncoding DNA in and flanking ZNF217 are circled. A cluster of such motifs occurs in and proximal to the 3′ untranslated region, in the first intron, and distal to the first exon. An example of an actual sequence alignment for one of the elements circled in red is shown in Figure 3B. These candidate regulatory elements now can be assessed for activating mutations and epigenetic modifications in 600MPE, T47D, and primary breast tumors in which ZNF217 and NABC3 are overexpressed in the absence of amplification. PFDN4 was overexpressed in cell lines in which it was amplified. Thus, both NABC3 and PFDN4 remain viable candidate oncogenes requiring further biological assessment. It will be important to determine if a synergistic relationship exists between these genes and ZNF217.
Figure 4.
RNA expression levels of ZNF217, NABC3, and PFDN4 in six cell lines and four mammary tumors. Transcript levels are calculated as 2 -ΔN (Albertson et al. 2000) with GAPDH as a reference gene and relative to the expression levels as measured in the human mammary epithelial cells (HMECs). As a control, expression levels were measured with GUS as a reference gene, which also showed nearly identical expression profiles for ZNF217 and NABC3 (not shown). Cultured HMECs, cell lines MCF7, MDA436, BT474, 600MPE, T47D, and MKN7, primary tumors S1552, S1526, S0117, and S0055 were used as a source of template mRNA for this experiment.
Next we extended this functional genomic analysis to the protein level. As one of the first steps of the systematic functional annotation of all proteins identified in the amplicon, we sought to determine their subcellular localization. Because ZNF217 maps to a narrow tumor amplicon, is overexpressed in all tumors in which it is amplified and some in which it is not, and can immortalize HMECs upon ectopic expression, we sought to determine its subcellular localization first. To this end we constructed a vector expressing a ZNF217–green fluorescent protein (GFP) fusion and microinjected this construct into HeLa cells. As shown in Figure 1C, the ZNF217-GFP fusion localizes to the nucleus in a punctate pattern. These data are consistent with the presence of nuclear localization signals in ZNF217 identified by PSORT (http://psort.nibb.ac.jp/). Of the two genes mapped to distal amplicon peak, CYP24 has been localized to the mitochondria in previous studies (Beckman and DeLuca 1997) and a manuscript is in preparation with detailed analysis of PFDN4 including its subcellular localization.
Finally, we note that the 1.2 Mb of assembled sequence reported here is consistent with the NCBI draft assembly (NT_011484 and NT_019675) and bridges the gap between these two sequence contigs (Fig. 1A). However, GC analysis did reveal a number of annotation errors and sequencing artifacts present in the public database. We found 11 regions of putative sequence identity between this sequence and chromosome 5 averaging ∼300 bp. Exhaustive characterization of these regions including RH mapping, and PCR on individual BAC clones from chromosomes 5 and 20, showed conclusively that the identities are, in fact, chromosome 20 sequences contaminating that of chromosome 5 BACs. In addition, we identified two BACs (AC026267, AC021970) annotated as chromosomes 4 and 16, respectively. These BACs share >99% sequence identity in entirety and contain only chromosome 20 STSs. Thus, we conclude that these represent annotation errors. The graphical representation of the 1.2 Mb sequence immediately revealed the presence and extent of both types of artifacts and facilitated the design of experiments to distinguish artifacts from real paralogous sequences.
Conclusion
This is the first tumor amplicon to be completely sequenced and biologically annotated. GC analysis provides a comprehensive view of the genomic landscape including distribution of genes, repetitive elements, duplications, cross-species homologies, and amplicon structure and suggests the possibility that NABC3 and PFDN4 may play a role in cancer progression. These results also suggest that repeated sequences and/or duplications may be involved in aberration formation and indicate specific genomic sequences that can be interrogated to test this hypothesis. Integration of high-resolution array CGH data with genomic sequence in other recurrent amplicons will provide an important test of the overall importance of repeat sequences and duplicons in gene amplification in humans.
METHODS
Genome Sequence
A BAC and P1 contig was assembled between D20S902 and D20S609 as described by (Collins et al. 1998) and a minimum tiling path of clones was selected for genomic sequencing (Fig. 1A). Sequencing was performed at the Department of Energy's Joint Genome Institute (http://www.jgi.doe.gov) and resulted in the assembly of ∼1.2 Mb. In addition, 865 kb of murine draft sequence spanning the gene ZNF217 was generated. Genomic and comparative sequence analyses were performed using Sequin (ftp://ncbi.nlm.nih.gov/sequin/), enhanced with a suite of programs for automation of data entry, PIP (http://bio.cse.psu.edu/pipmaker/), and Genome Cryptographer.
Accession Numbers
Human BAC and P1 clone accession numbers are as follows: BAC109: AC004499, P141: AC004505, P130: AC004504, BAC185: AC005808, BAC189: AC005914, P12: AC006076, P128: AC004762, BAC99: AC005220, BAC121: AC004501, H119: AF312913, P139: AF312912, H79/H117: AF312915, H143: AF312914. Mouse BAC clone accession numbers are as follows. M1: AC023610, M10: AC073667, and M12: AC073727. All accession numbers are from GenBank.
Sequence and Copy Number Annotation
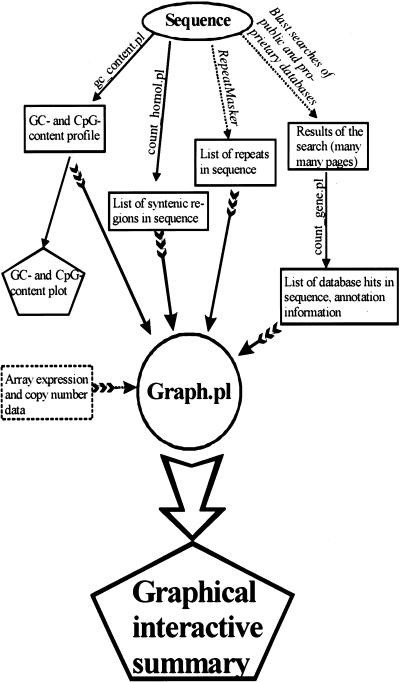
We have developed Genome Cryptographer (GC), which is a suite of Perl programs to facilitate megabase-scale analysis of genomic sequence (Fig. 5). This suite is built of separate modules that exchange information via intermediate text files. Data in intermediate files are written in a consistent format: sequence name, sequence length, window size, appropriate data for a given window (the number of these “data” lines equals the number of windows that are contained per sequence and, optionally, after a blank line, annotation data.
Figure 5.
Genome Cryptographer (GC) flowchart. The names of programs are given above solid arrows lacking feathers. Programs from the public domain are shown in italics. The final graphics output is presented in pentagrams. Intermediate data are shown in rectangles. Input of information into the graphics module (graph.pl) is shown by feathered arrows. A module for integrating expression and copy number array data is under development. GC and GC tutorial are available at http://kinase.ucsf.edu/gc.
Analysis of the sequence is done in the following stages:
Using script gc_plot.pl, we generate the plot of the GC-content and number of CpG dinucleotides per AI. The CpG dinucleotide density is weighted by adding 0.25 to the dinucleotide count for each CpG dinucleotide that is found within 20 bp of another. This makes CpG islands more apparent as peaks in CpG dinucleotide density plots. The script also produces the graphic plot of the GC- and CpG-content and, if available, can annotate the plot with features from the output of the count_gene.pl script (making it easier to correlate changes in GC and CpG content with sequence features).
The sequence is analyzed for repeats using publicly available RepeatMasker program (Smit and Green, http://repeatmasker.genome.washington.edu/cgi-bin/RM2_req.pl). RepeatMasker output files are saved. Masked sequence is used for searches of public and proprietary databases. Currently, GC employs the NCBI version of BLAST (ftp://ncbi.nlm.nih.gov/blast/). Sequence is compared to nonredundant, HTGS, dbSTS, and dbEST divisions of GenBank. Sequence similarity criteria are set to reduce the probability of identifying ESTs from members of closely related gene families (cutoff of expect score 10–20).
Optionally, masked sequence is searched against a database containing syntenic sequences of model organisms (in our case, mouse sequence from syntenic region of mouse chromosome 2).
count_gene.pl and count_homol.pl are used to analyze output of the blast searches, creating a list of the number of relevant hits per AI. count_gene.pl also generates a first draft of sequence annotation data, by capturing all the database hits that exceed in length, a user-selectable threshold. If desired, this annotation can be extended and updated by the user manually. We capture the exact coordinates of regions of identity of database hits used for annotation. This information proved to be invaluable for analysis of the gene relationships, because the alignment of cDNA sequence to genomic sequence automatically yields intron-exon organization of the corresponding gene.
Finally, graph.pl is used to gather information produced by gc_plot.pl (CpG distribution data) RepeatMasker (repeat distribution data), count_gene.pl (annotation and distribution of database hits) and count_homol.pl (distribution of conserved regions) and produce a graphical summary. Currently we are working on the extension of graph.pl capabilities (to make output interactive and to add capability to include gene expression and copy number data from array-based experiments). The first version of the Genome Cryptographer software is accessible at http://kinase.ucsf.edu/gc.
FISH Mapping
FISH mapping was performed as described in Kallioniemi et al. (1992) and Stokke et al. (1995). Briefly, BAC DNA was extracted from overnight cultures and labeled with digoxigenin-11-dUTP by nick translation. Hybridization to metaphase chromosomes was carried out in the presence of human Cot1 DNA overnight and hybridized signal detected using anti-digoxigenin conjugated with FITC. Chromosomes were counterstained with DAPI to localize the hybridization signal.
Microinjection and Fluorescence Microscopy
ZNF217-EGFP (Clontech) cellular targeting was monitored after microinjection of 10 ng/mL recombinant plasmid into HeLa cells grown on glass coverslips in 10% fetal calf serum in Dulbecco's modified Eagle's medium as previously described (Tominaga et al. 2000). Two hours after microinjection, cells were fixed and stained with Hoechst 33258 to visualize DNA and fluorescent images were captured with a SPOT CCD camera mounted on a Leica microscope equipped with a 100X oil-immersion objective.
Quantitative PCR
Quantitative PCR (Taqman) was performed as described previously (Albertson et al. 2000). PCR primer and probe sequences are as follows:
ZNF217: Forward TTTTTCCGTTCAAATTATTACCTCAA, Reverse GCAGCATATTCACAAAATTCACATT, and the TaqMan probe: FAM-CATCTCAGAACGCATACAGGTGAAAAAC CATAC-TAMRA.
NABC3: Forward CTACGCTGTAGGACACACAGTGG, Reverse TAAATGGCGGTTGCAGTGGT, and the TaqMan probe: FAM-CAATAATACAGGACCCCCAAACTGGCCA TAMRA.
PFDN4: Forward TTGGTGATGTCTTCATTAGCCATT, Reverse TTCCACTCTGGATTCTAAGGCG, and the TaqMan probe: FAM-AAGAAACGCAAGAAATGTTAGAAGAAG CAAAGAAAAAT.
Acknowledgments
We thank Dr. Vivienne Watson for critical review of this manuscript and Tim Andriese for coordinating sequencing of the mouse BACs. We thank Dr. Ung Jin Kim for screening the CalTech BAC library and Dr. Genevieve Nonet for providing the ZNF217-EGFP plasmid. This work was supported by Breast Cancer SPORE Grant CA 58207 and performed in part under the auspices of the US Department of Energy by the Joint Genome Institute under contracts DE-AC-03–76SF00098, W-7405-ENG-48, W-7405-ENG-36, and Vysis.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL collins@cc.ucsf.edu; FAX (415) 476-8218.
Article published on-line before print: Genome Res., 10.1101/gr.174301.
Article and publication are at www.genome.org/cgi/doi/10.1101/gr.174301.
REFERENCES
- Albertson DG, Ylstra B, Segraves R, Collins C, Dairkee SH, Kowbel D, Kuo WL, Gray JW, Pinkel D. Quantitative mapping of amplicon structure by array CGH identifies CYP24 as a candidate oncogene. Nat Genet. 2000;25:144–146. doi: 10.1038/75985. [DOI] [PubMed] [Google Scholar]
- Beckman MJ, DeLuca HF. Assay of 25-hydroxyvitamin D 1 alpha-hydroxylase and 24-hydroxylase. Methods Enzymol. 1997;282:200–213. doi: 10.1016/s0076-6879(97)82108-8. [DOI] [PubMed] [Google Scholar]
- Brosius J. RNAs from all categories generate retrosequences that may be exapted as novel genes or regulatory elements. Gene. 1999;238:115–134. doi: 10.1016/s0378-1119(99)00227-9. [DOI] [PubMed] [Google Scholar]
- Christian SL, Fantes JA, Mewborn SK, Huang B, Ledbetter DH. Large genomic duplicons map to sites of instability in the Prader- Willi/Angelman syndrome chromosome region (15q11-q13) Hum Mol Genet. 1999;8:1025–1037. doi: 10.1093/hmg/8.6.1025. [DOI] [PubMed] [Google Scholar]
- Collins C, Rommens JM, Kowbel D, Godfrey T, Tanner M, Hwang SI, Polikoff D, Nonet G, Cochran J, Myambo K, et al. Positional cloning of ZNF217 and NABC1: Genes amplified at 20q13.2 and overexpressed in breast carcinoma. Proc Natl Acad Sci. 1998;95:8703–8708. doi: 10.1073/pnas.95.15.8703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins FS, Patrinos A, Jordan E, Chakravarti A, Gesteland R, Walters L. New goals for the U.S. Human Genome Project: 1998–2003. Science. 1998;282:682–689. doi: 10.1126/science.282.5389.682. [DOI] [PubMed] [Google Scholar]
- Cuthill S, Agarwal P, Sarkar S, Savelieva E, Reznikoff CA. Dominant genetic alterations in immortalization: Role for 20q gain. Genes Chromosomes Cancer. 1999;26:304–311. [PubMed] [Google Scholar]
- Eichler EE. Masquerading repeats: Paralogous pitfalls of the human genome [published erratum appears in Genome Res. 10: 1095] Genome Res. 1998;8:758–762. doi: 10.1101/gr.8.8.758. [DOI] [PubMed] [Google Scholar]
- Eichler EE, Archidiacono N, Rocchi M. CAGGG Repeats and the Pericentromeric Duplication of the Hominoid Genome. Genome Res. 1999;9:1048–1058. doi: 10.1101/gr.9.11.1048. [DOI] [PubMed] [Google Scholar]
- Ginzinger DG, Godfrey TE, Nigro J, Moore DH, Suzuki S, Pallavicini MG, Gray JW, Jensen RH. Measurement of DNA copy number at microsatellite loci using quantitative PCR analysis. Cancer Res. 2000;60:5405–5409. [PubMed] [Google Scholar]
- Gray JW, Collins C. Genome changes and gene expression in human solid tumors. Carcinogenesis. 2000;21:443–452. doi: 10.1093/carcin/21.3.443. [DOI] [PubMed] [Google Scholar]
- Hansen WJ, Cowan NJ, Welch WJ. Prefoldin-nascent chain complexes in the folding of cytoskeletal proteins. J Cell Biol. 1999;145:265–277. doi: 10.1083/jcb.145.2.265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huie ML, Shanske AL, Kasper JS, Marion RW, Hirschhorn R. A large Alu-mediated deletion, identified by PCR, as the molecular basis for glycogen storage disease type II (GSDII) Hum Genet. 1999;104:94–98. doi: 10.1007/s004390050916. [DOI] [PubMed] [Google Scholar]
- Iijima M, Kano Y, Nohno T, Namba M. Cloning of cDNA with possible transcription factor activity at the G1-S phase transition in human fibroblast cell lines. Acta Med Okayama. 1996;50:73–77. doi: 10.18926/AMO/30489. [DOI] [PubMed] [Google Scholar]
- Ji Y, Eichler EE, Schwartz S, Nicholls RD. Structure of chromosomal duplicons and their role in mediating human genomic disorders. Genome Res. 2000;10:597–610. doi: 10.1101/gr.10.5.597. [DOI] [PubMed] [Google Scholar]
- Kallioniemi A, Kallioniemi OP, Piper J, Tanner M, Stokke T, Chen L, Smith HS, Pinkel D, Gray JW, Waldman FM. Detection and mapping of amplified DNA sequences in breast cancer by comparative genomic hybridization. Proc Natl Acad Sci. 1994;91:2156–2160. doi: 10.1073/pnas.91.6.2156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kallioniemi OP, Kallioniemi A, Kurisu W, Thor A, Chen LC, Smith HS, Waldman FM, Pinkel D, Gray JW. ERBB2 amplification in breast cancer analyzed by fluorescence in situ hybridization. Proc Natl Acad Sci. 1992;89:5321–5325. doi: 10.1073/pnas.89.12.5321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knuutila S, Aalto Y, Autio K, Bjorkqvist AM, El-Rifai W, Hemmer S, Huhta T, Kettunen E, Kiuru-Kuhlefelt S, Larramendy ML, et al. DNA copy number losses in human neoplasms. Am J Pathol. 1999;155:683–694. doi: 10.1016/S0002-9440(10)65166-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knuutila S, Bjorkqvist AM, Autio K, Tarkkanen M, Wolf M, Monni O, Szymanska J, Larramendy M L, Tapper J, Pere H, et al. DNA copy number amplifications in human neoplasms: review of comparative genomic hybridization studies. Am J Pathol. 1998;152:1107–1123. [PMC free article] [PubMed] [Google Scholar]
- Moran JV, DeBerardinis RJ, Kazazian HH., Jr Exon shuffling by L1 retrotransposition. Science. 1999;283:1530–1534. doi: 10.1126/science.283.5407.1530. [DOI] [PubMed] [Google Scholar]
- Nonet GH, Stampher MR, Chin K, Gray JW, Collins CC, Yaswen P. The ZNF217 gene amplified in breast cancers promotes immortalization of human mammary epithelial cells. Cancer Res. 2001;61:1250–1254. [PubMed] [Google Scholar]
- Peoples R, Franke Y, Wang YK, Perez-Jurado L, Paperna T, Cisco M, Francke U. A physical map, including a BAC/PAC clone contig, of the williams- beuren syndrome-deletion region at 7q11.23. Am J Hum Genet. 2000;66:47–68. doi: 10.1086/302722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinkel D, Segraves R, Sudar D, Clark S, Poole I, Kowbel D, Collins C, Kuo W L, Chen C, Zhai Y, et al. High resolution analysis of DNA copy number variation using comparative genomic hybridization to microarrays. Nat Genet. 1998;20:207–211. doi: 10.1038/2524. [DOI] [PubMed] [Google Scholar]
- Savelieva E, Belair CD, Newton MA, DeVries S, Gray JW, Waldman F, Reznikoff CA. 20q gain associates with immortalization: 20q13.2 amplification correlates with genome instability in human papillomavirus 16 E7 transformed human uroepithelial cells. Oncogene. 1997;14:551–560. doi: 10.1038/sj.onc.1200868. [DOI] [PubMed] [Google Scholar]
- Stokke T, Collins C, Kuo WL, Kowbel D, Shadravan F, Tanner M, Kallioniemi A, Kallioniemi OP, Pinkel D, Deaven L, et al. A physical map of chromosome 20 established using fluorescence in situ hybridization and digital image analysis. Genomics. 1995;26:134–137. doi: 10.1016/0888-7543(95)80092-z. [DOI] [PubMed] [Google Scholar]
- Tanner MM, Tirkkonen M, Kallioniemi A, Collins C, Stokke T, Karhu R, Kowbel D, Shadravan F, Hintz M, Kuo WL, et al. Increased copy number at 20q13 in breast cancer: Defining the critical region and exclusion of candidate genes. Cancer Res. 1994;54:4257–4260. [PubMed] [Google Scholar]
- Tanner MM, Tirkkonen M, Kallioniemi A, Holli K, Collins C, Kowbel D, Gray JW, Kallioniemi OP, Isola J. Amplification of chromosomal region 20q13 in invasive breast cancer: prognostic implications. Clin Cancer Res. 1995;1:1455–1461. [PubMed] [Google Scholar]
- Tanner MM, Tirkkonen M, Kallioniemi A, Isola J, Kuukasjarvi T, Collins C, Kowbel D, Guan XY, Trent J, Gray JW, Meltzer P, Kallioniemi OP. Independent amplification and frequent co-amplification of three nonsyntenic regions on the long arm of chromosome 20 in human breast cancer. Cancer Res. 1996;56:3441–3445. [PubMed] [Google Scholar]
- Tominaga T, Sahai E, Chardin P, McCormick F, Courtneidge SA, Alberts AS. Diaphanous-related formins bridge Rho GTPase and Src tyrosine kinase signaling. Mol Cell. 2000;5:13–25. doi: 10.1016/s1097-2765(00)80399-8. [DOI] [PubMed] [Google Scholar]
- Vainberg IE, Lewis SA, Rommelaere H, Ampe C, Vandekerckhove J, Klein HL, Cowan NJ. Prefoldin, a chaperone that delivers unfolded proteins to cytosolic chaperonin. Cell. 1998;93:863–873. doi: 10.1016/s0092-8674(00)81446-4. [DOI] [PubMed] [Google Scholar]
- Werner M, Mattis A, Aubele M, Cummings M, Zitzelsberger H, Hutzler P, Hofler H. 20q13.2 amplification in intraductal hyperplasia adjacent to in situ and invasive ductal carcinoma of the breast. Virchows Arch. 1999;435:469–472. doi: 10.1007/s004280050429. [DOI] [PubMed] [Google Scholar]