Abstract
Disruptions of interregional correlations in the blood oxygenation level dependent fMRI signal have been reported in multiple diseases, including Alzheimer’s disease and mild cognitive impairment. “Default network” regions that overlap with areas of earliest amyloid deposition have been highlighted by these reports, and abnormal default network activity is also observed in unimpaired elderly subjects with high amyloid burden. However, one limitation of current methods for analysis of interregional correlations is that they rely on transformation of functional data to an atlas volume (e.g., Talairach-Tournoux or Montreal Neurological Institute atlases) and may not adequately account for anatomic variation between subjects, particularly in the presence of atrophy. We assessed the utility of the FreeSurfer cortical parcellation to analyze default network functional correlations on the native surfaces of individual subjects. Group-level quantitative analysis was accomplished by comparing correlations between equivalent structures in different subjects. The method was applied to resting-state fMRI data from young, healthy subjects; preliminary results were also obtained from cognitively unimpaired elderly subjects and patients with Alzheimer’s disease, Parkinson’s disease, Parkinson’s disease dementia, and dementia with Lewy bodies.
Keywords: resting state, default network, fMRI, functional connectivity, dementia, FreeSurfer, atlas, registration
1 Introduction
Characteristic patterns of low-frequency correlations have been repeatedly identified in the blood oxygenation level dependent (BOLD) fMRI signal when subjects are asked to simply lie still in the scanner (Biswal et al., 1995; Greicius et al., 2003; Buckner et al., 2008). The relative consistency of these patterns across studies and analysis methods, as well as the simplicity of the instructions, has led to considerable interest in their potential application as a biomarker in disease (Fox and Raichle, 2007; Rogers et al., 2007; Greicius, 2008; Auer, 2008; van den Heuvel and Hulshoff Pol, 2010). Particular attention has been paid to a collection of regions called the default network1 and the disruption of correlations across these regions in Alzheimer’s disease (Greicius et al., 2004; Wang et al., 2006; Allen et al., 2007; Supekar et al., 2008; Koch et al., 2010; Zhang et al., 2010; for review, see Greicius, 2008; Sorg et al., 2009). Disruptions in so-called functional connectivity in the default network have also been reported in conditions believed to precede onset of Alzheimer’s disease, including patients with amnestic mild cognitive impairment (Sorg et al., 2007; Pihlajamäki et al., 2009; Bai et al., 2009) and cognitively unimpaired subjects with high amyloid burden (Hedden et al., 2009; Sheline et al., 2010).
Anatomical variability across subjects gives rise to two notable challenges in the analysis of spontaneous BOLD correlations within the default network. First, if analyses are to be extended beyond qualitative assessment in individual subjects, a method of comparing results across subjects is critical. Second, the network of interest has to be identified in each subject. In seed-correlation analyses, this is typically accomplished by choosing an a priori seed region known to lie within the network (e.g., Hedden et al., 2009; Sheline et al., 2010). For independent component analyses (ICA), a template is used to identify the component that best matches the default network (e.g., Greicius et al., 2004; Seeley et al., 2009).
Currently, both of these challenges are addressed by performing analyses in atlas-volume space. Anatomical and functional images from each subject are transformed, or warped, to match a canonical brain (e.g., Talairach-Tournoux or Montreal Neurological Institute template). Once in a standardized, or atlas, volume, seed regions and templates from the literature or other data sets can be applied to the spatially transformed data to identify the default network. The process of transforming data to an atlas volume also permits direct comparison of analysis results across subjects and studies.
Unfortunately, atlas-space results are only valid to the extent that the warping process is valid, a point of particular concern in conditions where participants’ brains differ considerably from the atlas due to disease. Functional correlation analyses are subject to concerns similar to some known issues with voxel-based morphometry, a method for structural MRI analysis which also depends heavily on accurate registration to a template. Improper registration can lead to misleading results in both cases because, for example, a given coordinate represents gray matter in the template but lies in cerebrospinal fluid in a patient’s warped brain. Voxel-based morphometry gives varied results depending on the particular warping algorithm used (Senjem et al., 2005), and even algorithms identified as “optimized,” which include multiple steps to improve normalization, are still prone to errors when atrophy causes gross changes in brain structure (Bookstein, 2001; Ashburner and Friston, 2001; Senjem et al., 2005). Despite the crucial role warping plays in functional correlation analyses and the known pitfalls of common methods in the face of structural brain pathology, accuracy of individual transformations are rarely, if ever, reported or displayed.
Analysis on a subject’s native surface offers potential advantages over atlas-volume methods. First, possible ambiguity about precise anatomic locations is reduced. Measuring functional correlations on native surfaces also facilitates accounting for anatomic effects of disease and age. Moreover, by preserving inter-individual anatomic variability, longitudinal patient studies can better avoid confounds due to disease-related structural changes that affect an individual patient’s brain over time. Comparison of functional measures to other individual markers is also straightforward on native surfaces, in particular cross-modal imaging markers such as amyloid imaging results and regional cortical thickness. There is also a clinical appeal to obtaining and displaying functional imaging results on the brain surface of an individual patient. Prior studies have pointed to the potential of functional correlations to provide meaningful results in individual patients (Greicius et al., 2004; Buckner and Vincent, 2007; Koch et al., 2010); analyzing functional data on native surfaces may be an important step toward that aim.
We assessed the utility of the FreeSurfer (http://surfer.nmr.mgh.harvard.edu) cortical parcellation to analyze functional correlations on the native surfaces of individual subjects. Automated processes are employed to anatomically parcellate each subject’s cortical surface into distinct regions of cortex (subcortical gray matter structures are included after a similar automated volume segmentation). One cortical region, the isthmus cingulate, is proposed as a suitable native-surface seed for identification and analysis of the default network. Parcellation and segmentation regions are then used for group-level analyses by comparing interregional correlations between equivalent regions in different subjects. Additionally, registration of native sulcal and gyral patterns to an average surface allows display of group-level results after quantitative parcellation analysis on native surfaces.
Here we present results from the application of this method to BOLD data from young, healthy subjects as a proof of concept. The primary findings were reproduced in preliminary data from multiple disease populations, including Alzheimer’s disease (AD), Parkinson’s disease dementia (PDD), Dementia with Lewy bodies (DLB), and cognitively unimpaired elderly controls.
2 Methods
2.1 Subjects
Subject demographics are provided in Table 1. Patients designated ‘Alzheimer’s disease’ had a clinical diagnosis of probable AD based on the NINCDS/ADRDA criteria (McKhann et al., 1984); diagnoses for dementia with Lewy bodies and Parkinson’s disease dementia were based on the criteria established by the Movement Disorders Society Task Force (Geser et al., 2005; McKeith, 2007). Diagnosis for all patients was made by consensus of two or more neurologists in the UCSD Shiley-Marcos Alzheimer’s Disease Research Center Clinical Core. Elderly controls with no cognitive impairment had a mini mental status exam score of at least 27 and a CDR score of zero.
Table 1.
Subject demographics
| Subject group | n | Age range | Field strength |
|---|---|---|---|
| Young adults | 15 | 22–28 | 3.0 Tesla |
| Elderly controls | 7 | 69–90 | 1.5 Tesla |
| Alzheimer’s disease | 4 | 61–94 | 1.5 Tesla |
| Parkinson’s disease dementia | 8 | 65–86 | 1.5 Tesla |
| Dementia with Lewy bodies | 7 | 61–75 | 1.5 Tesla |
2.2 MRI Acquisition
Functional imaging of each subject consisted of two T2*-weighted sequences of approximately seven minutes each on a General Electric Signa Excite HDx using an eight-channel phased-array head coil (General Electric Healthcare, Waukesha, WI). Data for young subjects were acquired on a 3.0 Tesla system (TE: 30 ms; TR: 2124 ms; flip angle: 90°; matrix: 64 × 64; voxel size: 3.75 mm × 3.75 mm × 4 mm; 36 adjacent sagittal slices; 205 samples per series); data for elderly subjects, including patients, were acquired on a 1.5 Tesla system (TE: 45 ms; TR: 2624 ms; flip angle: 90°; matrix: 64 × 64; voxel size: 3.75 mm × 3.75 mm × 5 mm; 32 adjacent sagittal slices; 155 samples per series). The initial five samples from each functional run were excluded to allow for T1-equilibration. Immediately prior to each functional series, a spin-echo volume was acquired with opposite phase-encoding polarity for field inhomogeneity correction (Holland et al., 2010). Instructions for the young subjects were to rest motionless with eyes closed. Instructions for the elderly subjects were to rest motionless with eyes open; this modification was adopted after it was suggested that functional correlations in the default network are more robust with eyes open (Yan et al., 2009; Van Dijk et al., 2009). In addition to the functional volumes, a high-resolution, three-dimensional, T1-weighted volume was acquired for each subject during the same session (TE: 2.8 ms/3.8 ms; TR: 6.5 ms/8.5 ms; TI: 600 ms/500 ms; flip angle: 8°/10°; matrix: 256 × 256; voxel size: 0.9375 mm × 0.9375 mm × 1.2000 mm; values separated by ‘/’ are for 3.0 T data/1.5 T data). Respiratory effort and heart rate were monitored with a pressure transducer (BioPac Systems Inc., Goleta, CA) and a pulse oximeter (BioPac Systems and InVivo, Orlando, FL), respectively.
2.3 Structural MRI Processing
A model of each subject’s cortical surface was reconstructed from the T1-weighted MRI volume (Dale et al., 1999; Fischl et al., 1999a). To ensure accuracy, the automatically generated boundaries were overlaid on the original T1-weighted volume as thin colored lines to aid visual confirmation of the tissue boundaries on each slice—yellow for the boundary between white and gray matter, and red for the boundary between gray matter and cerebrospinal fluid. Where these automatically generated lines deviated from the visually identified boundaries, manual control points were created, and the automated algorithms were applied again. Final surfaces were visually inspected to search for gross errors; none were found in the present data set.
The surface model was then anatomically parcellated using the Desikan-Killiany atlas and standard FreeSurfer tools (Fischl et al., 2004; Desikan et al., 2006). This process assigns each point (vertex) on the native surface to the most probable anatomical label (e.g., inferior parietal, precentral, parahippocampal, etc.) based on registration to a probabilistic atlas of surface folding patterns and on the observed surface geometry at that location of the native surface (Fischl et al., 2004). Subcortical structures were similarly identified by volume segmentation (Fischl et al., 2002). Automated parcellation by this method has been shown to be comparable to manual labeling (Fischl et al., 2004). Additionally, the parcellation for each subject was visually inspected to search for gross errors; none were found in the present data set.
For direct comparison with prevailing methods, the T1-weighted volume from an Alzheimer’s patient exhibiting atrophy was submitted to common registration algorithms to warp the atrophied brain to the Montreal Neurological Institute (MNI) 152 T1 reference brain provided in standard software packages. Affine transformation with 12 degrees of freedom was performed using 3dWarpDrive in AFNI (http://afni.nimh.nih.gov/afni). Nonlinear transformations were performed using FNIRT in FSL (http://www.fmrib.ox.ac.uk/fsl/index.html), the “normalise” process in SPM2 (http://www.fil.ion.ucl.ac.uk/spm/), and the DARTEL process in SPM8. An older version of SPM (SPM2) was included because it appears to be among the more common packages used in the relevant literature (e.g., Wang et al., 2007, 2010; Supekar et al., 2008; Buckner et al., 2009; Van Dijk et al., 2009). All registration procedures followed the configurations and parameters recommended in the documentation provided with the corresponding software. Simple alignment of the original volume was also performed using manually defined markers in AFNI in order to display the original images in a similar orientation to the registered images.
2.4 fMRI Data Pre-analysis Processing
All fMRI pre-analysis processing was performed using custom software written in MATLAB (Mathworks, Natick, MA), except where noted. Functional images were first corrected for distortion due to inhomogeneity in the static magnetic field (Holland et al., 2010). Effects of respiratory fluctuations were modeled and removed from time series using RETROICOR (Glover et al., 2000). Similar removal of cardiac fluctuations did not have a meaningful impact on the results in any group, and this step was not included in final analyses for the sake of consistency across all subjects (pulse recordings were sporadically lost in approximately 20% of scans in elderly and disease subjects due to technical problems). After interpolation for slice acquisition timing, rigid body volume registration was performed using AFNI (Cox and Jesmanowicz, 1999), followed by voxel-wise regression of six head motion parameters and a cubic polynomial baseline from each functional series. Functional data were next projected onto the subject’s cortical surface model using FreeSurfer, and a bandpass filter of 0.01–0.08 Hz was applied to the time series from each vertex on the surface. BOLD correlation analyses typically include a smoothing step with a Gaussian kernel to account for functional and anatomic variation across subjects, but this step is not necessary for native-surface parcellation analysis.
2.5 fMRI Correlation Analysis
We designed an fMRI correlation analysis that takes advantage of the FreeSurfer surface generation and parcellation tools and avoids transforming functional data to an atlas volume. All steps were performed using custom software written in MATLAB, except where noted. Functional time series were averaged across surface vertices within the left isthmus cingulate region to serve as the seed time series for correlation analyses. Average time series were also calculated from each of the other 30 cortical surface parcellation regions in the Desikan-Killiany atlas not adjacent to the seed (see Figure 4), as well as from five volume segmentation regions from the left hemisphere (hippocampus, caudate, pallidum, putamen, and amygdala). A Pearson’s correlation coefficient was calculated for the correlation between the seed time series and each region’s average time series, and Fisher’s z-transform was applied to these coefficients. The same process was repeated for right hemisphere regions, with the right isthmus cingulate region as the seed. Region time series were obtained by loading both the subject’s functional data and the parcellated native surface (which contains a region code at each vertex location) in MATLAB; time series at each vertex could then be classified by the region code at the corresponding location in the parcellated surface.
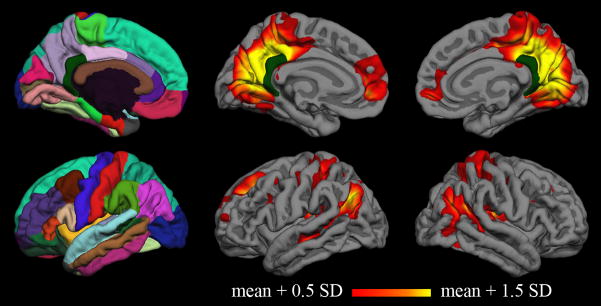
Figure 4. Group correlation map for young subjects.
Vertex-wise average correlation map across fifteen young subjects after surface-based registration to the FreeSurfer “fsaverage” subject. The Desikan-Killiany cortical parcellation atlas is shown in the first column, and the isthmus cingulate seed is shown in dark green on all medial surfaces.
Results from native-surface parcellation analysis were summarized in two ways. First, the z-transformed correlation coefficients from each region were averaged across subjects. Second, as it is possible that relative changes in correlation coefficient may also be informative, all 35 regions per hemisphere were ranked in order of highest to lowest coefficient for a given subject. These ranks were summarized by calculating the median rank across subjects for each region.
A power analysis was performed to give an estimate of the number of subjects needed to detect a difference between two groups. For each region, the standard deviation (across subjects) was calculated for the z-transformed correlation coefficient with the isthmus cingulate seed. This standard deviation was included in Cohen’s equation sample size for a population difference (Cohen, 1988; Dawson and Trapp, 2004). The expected effect size (i.e., population difference) was assumed to be 0.2; the actual value is unknown and specific to the populations studied, but available published values suggest this value is conservative for comparing unimpaired elderly with high risk for Alzheimer’s disease to age-matched controls (Fleisher et al., 2009; Hedden et al., 2009; Koch et al., 2010). All calculations also assumed 80% power and an alpha value of 0.05. The final result of the power calculations was an estimated sample size for each region, corresponding to the number of subjects necessary to detect a population difference of 0.2 in the correlation coefficient.
Vertex-wise correlation analysis was performed in addition to the parcellation analysis, allowing visualization of entire hemispheres at finer resolution. Individual maps were produced by calculating the Fisher’s z-transformed correlation coefficient for the average seed region time series and the time series of each vertex on the surface. Individual native surfaces were registered to the FreeSurfer fsaverage surface using a spherical-based algorithm in FreeSurfer (Fischl et al., 1999b). That registration was used to transform the individual maps to the fsaverage surface, also using FreeSurfer tools. Group maps were created by loading the fsaverage versions of the individual maps in MATLAB and taking the average across subjects. A surface-based smoothing process was applied using FreeSurfer for display in the figures (28 iterative steps, approximately equivalent to a 6 mm full-width half-maximum Gaussian kernel in two dimensions). Group maps were calculated from unsmoothed individual maps so that the smoothing process was only applied once. The minimum threshold for both group and individual hemisphere maps was set as the mean coefficient across all vertices on the surface plus 0.5 times the standard deviation; the maximum threshold was set as the mean coefficient plus 1.5 times the standard deviation.
2.6 Comparative Analysis: Volume Atlas vs. Native Surface
To assess the degree to which warping to a volume atlas affects functional correlation results, all functional data from this study were also analyzed after nonlinear transformation to the MNI152 volume atlas. All pre-analysis processing and correlation procedures for the volume-atlas analysis were performed as in the native-surface analysis up to the point of projection of time series data to the surface. Instead of directly projecting processed functional time series to each subject’s native surface, all data were transformed to volume-atlas space using standard tools in a software package cited in several resting correlation studies (SPM2). The transformation to volume-atlas space was calculated for each subject using the individual high-resolution T1-weighted volumes. The time series in MNI space were then projected onto the fsaverage surface using the standard transformation provided with FreeSurfer. Subsequent bandpass filtering and correlation analysis (using the fsaverage surface parcellation) were performed as in the native-surface analysis.
Any inaccuracies in registration to the MNI volume atlas could change the definition of the seed and therefore affect correlation measures across the entire brain even if the rest of the brain was perfectly registered to MNI. To isolate the effect of whole-brain registration to a volume atlas, the time series from each subject’s native-surface isthmus cingulate was used as the seed in both the native-surface and volume-atlas analyses. Additionally, the correlation coefficient between the native-surface isthmus cingulate time series and the MNI-transformed isthmus cingulate time series was calculated for each subject to quantify the effect of MNI transformation on the seed. Note that, in order to maintain intuitive interpretation of the values, these seed-to-seed correlation coefficients were not Fisher-transformed.
MNI volume-atlas correlation coefficients for each region on the fsaverage surface were compared to the corresponding region on the native surfaces. As volume-atlas registration inaccuracies lead to a heterogeneous mix of both increases and decreases in correlation values, the magnitude of the differences was used for comparing the methods. The mean (across-subjects) difference between MNI-transformed and native-surface results was calculated for each region. Paired t-tests were applied to evaluate the statistical significance of any regional differences between the two methods.
Vertex-by-vertex comparison of the MNI volume-atlas results (projected on the fsaverage surface) to the native-surface results was achieved using the native-surface maps that were registered to the fsaverage surface (see section 2.5). Difference maps were calculated by taking the root-mean-square (RMS) average, at each vertex, of the difference between MNI-transformed and native-surface results.
3 Results
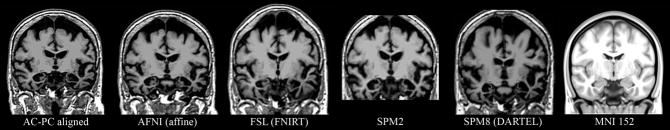
Both linear and nonlinear algorithms successfully aligned the atrophied brain to the MNI template (Figure 1). The nonlinear methods (FSL, SPM2, SPM8) appear to have reduced ventricular spaces and stretched the brain tissue to fill portions of the adjacent CSF space (distortions to the skull and other tissues in the nonlinear examples should be ignored, as the methods are optimized for registration of the brain, not extraparenchymal tissues). None of the transformations, however, fully accounted for the bulk atrophy in the superior brain or the enlarged sulci evident throughout the cortex. Many MNI coordinates that lie within cortical regions in the template correspond to cerebrospinal fluid in the transformed brains for this patient.
Figure 1. Warping to an atlas volume.
T1-weighted volume from a patient with atrophy transformed to an atlas volume using popular software packages. AC-PC aligned: original image, after rotation and cropping for comparison with transformed images (rotation in AFNI after manual landmark identification). AFNI: after affine (12 degrees of freedom) transformed with 3dWarpDrive. FSL: after nonlinear warping with FNIRT. SPM2: after nonlinear warping with “Normalise” tool. SPM8: after nonlinear warping with the “DARTEL” process. MNI 152: standard T1 atlas volume used for all transformations presented here. None of these methods adequately accounted for the structural effects of atrophy in this patient. One consequence of the poor alignment is some cortical MNI coordinates correspond to cerebrospinal fluid in the patient’s transformed brain.
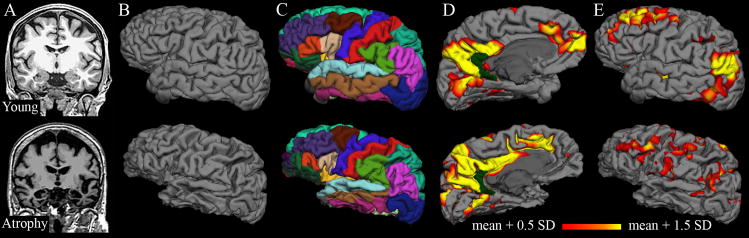
The T1-weighted volume and reconstructed cortical surface model are shown in Figure 2A and 2B, respectively, for the brain of a young subject and the atrophied brain from Figure 1. Anatomic features of each individual brain were preserved by the surface reconstructions. The depression of the superior aspect of the atrophied brain is reflected in the relatively flat superior aspect of the surface. Similarly, the patient’s enlarged sulci are readily observable in the corresponding surface. Neither of these abnormalities prevented successful automated parcellation of the cortical surface (Figure 2C). The isthmus cingulate seed region is identified for each subject within the parcellation (Figure 2D, dark green), allowing calculation of correlation coefficients for the rest of the cortex on the subject’s native surface (Figure 2D, 2E).
Figure 2. Analysis on native surfaces.
The top row shows structural and functional analysis for a young subject; the bottom row shows corresponding images from a patient with atrophy (see Figure 1). A: Original T1-weighted volumes after AC-PC alignment. B: Cortical surface models generated from T1-weighted volumes; anatomical features from individual subjects, including effects of atrophy, are preserved. C: Automated anatomical parcellation of cortical surface for each subject. D–E: Individual correlation maps consisting of coefficients (z-transformed) from correlation of each surface vertex with the average time series of the isthmus cingulate seed (dark green). The isthmus cingulate region was defined on the native surface for each subject during the automated anatomical parcellation. Thresholds (for display only) were determined separately for each subject from the mean and standard deviation of correlation coefficients for the vertices on the individual surface.
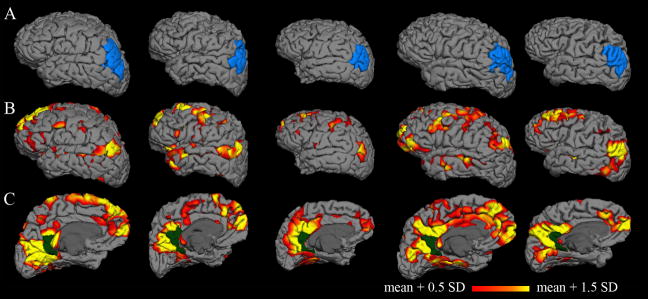
Cortical surface models reconstructed from five individual young subjects are shown in Figure 3A (all subjects are included in supplementary material). As expected, substantial variability in anatomy is observed between individuals; brain size, gyral patterns, and structural landmarks are all unique for each surface. The inferior parietal region identified by the FreeSurfer parcellation is also shown for each subject.
Figure 3. Individual correlation maps.
Columns represent individual subjects. A: Cortical surface models with a single parcellation region (inferior parietal) highlighted in blue. The substantial anatomic variability across subjects is captured by the individual surfaces. B–C: Individual correlation maps for five young subjects, consisting of coefficients (z-transformed) from correlation of each vertex on the surface with the average time series of the isthmus cingulate seed (dark green). The isthmus cingulate region was defined on the native surface for each subject during the automated anatomical parcellation. Individual correlation maps for all fifteen young subjects are included in the supplementary material.
Functional correlation maps are displayed on the native surfaces for the same five individual young subjects in Figure 3B. Many individual subject maps resemble the well-known default network pattern typically reported in group averages. As with the anatomy, though, the functional maps demonstrate considerable variability from one individual to another.
Native-surface regions most consistently correlated with the seed region in each hemisphere are shown in Table 2. All of the top five regions in each hemisphere are among those frequently included in the default network (dorsolateral prefrontal, medial prefrontal, inferior parietal, and medial temporal). Both the mean correlation coefficient and median rank measures identify default network regions as the most strongly correlated with the isthmus cingulate seed.
Table 2. Native-space correlation analysis in young subjects.
Regions most consistently correlated with the seed region in each hemisphere. A median rank of 2 for the superior frontal region in the left hemisphere indicates that this region is among the top 2 regions most correlated with the seed in at least 50% of subjects (interquartile ranges give analogous results for the 25th and 75th percentiles). Mean z and SE indicate the population mean z-transformed correlation coefficient and standard error, respectively. Sample size indicates the estimated sample size to detect a difference in mean z of 0.2 with 80% power and alpha value set to 0.05.
| Left Hemisphere | |||||
|---|---|---|---|---|---|
| region ranks | correlation coefficients | ||||
| Region name | median | quartiles | meanz | SE | sample size |
| Superior Frontal | 2 | (1–2.75) | 1.06 | 0.06 | 21 |
| Inferior Parietal | 3 | (2–6.25) | 1.02 | 0.07 | 33 |
| Medial Orbitofrontal | 3 | (2–7.75) | 1.02 | 0.06 | 21 |
| Hippocampus | 6 | (5–10.5) | 0.90 | 0.05 | 16 |
| Parahippocampal | 7 | (4–13.25) | 0.89 | 0.06 | 18 |
| Right Hemisphere | |||||
| region ranks | correlation coefficients | ||||
| Region name | median | quartiles | meanz | SE | sample size |
| Superior Frontal | 4 | (2–7.75) | 0.91 | 0.06 | 22 |
| Inferior Parietal | 5 | (1.25–8.75) | 0.94 | 0.08 | 37 |
| Hippocampus | 6 | (3–10.75) | 0.89 | 0.07 | 28 |
| Parahippocampal | 7 | (4.25–18.75) | 0.80 | 0.08 | 37 |
| Medial Orbitofrontal | 8 | (2.25–13.75) | 0.87 | 0.05 | 15 |
Group-average functional correlation maps for the young subjects are displayed in Figure 4. While interindividual variability is lost in the group average, the pattern at the group level confirms that typical default network patterns are identified using the isthmus cingulate seed.
Power analysis estimated the sample size required for a difference in population mean for the regions in Table 2. Sample sizes for the regions in Table 2 had a median of 21.5 subjects. Among these regions, the right inferior parietal and right parahippocampal regions had the greatest estimated sample sizes at 37 subjects each. Right medial orbitofrontal had the smallest estimated sample size at 15 subjects.
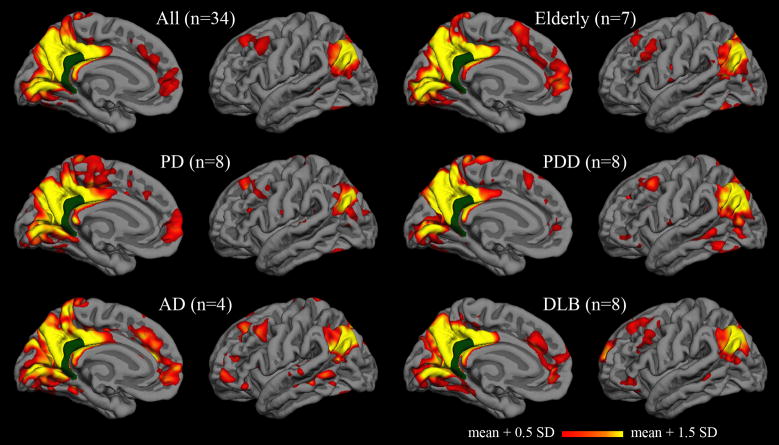
Functional correlation maps were calculated for five additional populations (Figure 5). The qualitative patterns in these group-average maps were similar for cognitively unimpaired elderly, Parkinson’s disease, Parkinson’s disease dementia, Alzheimer’s disease, and dementia with Lewy bodies. Characteristic features of the default network are observed in each group. Sample sizes from each group are insufficient for intergroup comparisons, but pooled analysis across groups demonstrates that some of the same regions remain most consistently correlated with the isthmus cingulate seed in native-space analysis (Table 3). As with the young subjects, the top two most consistently correlated regions in both hemispheres were inferior parietal and superior frontal.
Figure 5. Group correlation maps for elderly subjects and patients.
Vertex-wise average correlation maps after surface-based registration to the FreeSurfer “fsaverage” subject. The isthmus cingulate seed is shown in dark green. The upper left map is the average across all thirty-four elderly subjects and patients; the other five maps are averages within elderly and disease subgroups.
Table 3. Native-surface correlation analysis in elderly subjects and patients.
Regions most consistently correlated with the seed region in each hemisphere. A median rank of 2 for the superior frontal region in the left hemisphere indicates that this region is among the top 2 regions most correlated with the seed in at least 50% of subjects (interquartile ranges give analogous results for the 25th and 75th percentiles). Mean z and SE indicate the population mean z-transformed correlation coefficient and standard error, respectively. Sample size indicates the estimated sample size to detect a difference in mean z of 0.2 with 80% power and alpha value set to 0.05.
| Left Hemisphere | |||||
|---|---|---|---|---|---|
| region ranks | correlation coefficients | ||||
| Region name | median | quartiles | mean z | SE | sample size |
| Inferior Parietal | 2 | (1–4) | 0.84 | 0.05 | 36 |
| Superior Frontal | 4 | (2–5) | 0.77 | 0.05 | 30 |
| Caudal Middle Frontal | 6 | (4–14) | 0.67 | 0.05 | 38 |
| Hippocampus | 10 | (6–19) | 0.57 | 0.04 | 24 |
| Caudate | 10.5 | (7–17) | 0.59 | 0.04 | 27 |
| Right Hemisphere | |||||
| region ranks | correlation coefficients | ||||
| Region name | median | quartiles | mean z | SE | sample size |
| Inferior Parietal | 3 | (1–5) | 0.84 | 0.05 | 28 |
| Superior Frontal | 3 | (2–8) | 0.79 | 0.05 | 31 |
| Pericalcarine | 8.5 | (4–14) | 0.66 | 0.05 | 29 |
| Caudal Middle Frontal | 9 | (6–15) | 0.64 | 0.05 | 38 |
| Middle Temporal | 9 | (6–19) | 0.60 | 0.05 | 40 |
The effect of defining a seed in volume-atlas space rather than defining a seed on the native surface was quantified by calculating the correlation coefficient between the two average time series for each subject. For young subjects, the median correlation coefficient (and interquartile range) between the MNI isthmus cingulate and native-surface isthmus cingulate was 0.84 (0.81–0.86) for the left hemisphere and 0.87 (0.85–0.87) for the right hemisphere. The minimum coefficient among the young subjects was 0.63, and the maximum was 0.93. For healthy and impaired elderly subjects, the median correlation coefficient was 0.77 (0.65–0.85) for the left isthmus cingulate and 0.79 (0.66–0.85) for the right. The minimum coefficient among the elderly and patients was −0.09, and the maximum was 0.95.
Registration to an atlas volume also led to significant effects on correlation results throughout the rest of the cortex, even when the same (native-surface) seed time series was used for both methods. Correlation coefficients for MNI-transformed regions on the fsaverage surface differed from their native-surface homologues by a mean magnitude of 0.14 ± 0.06 (standard deviation) in young subjects. Paired t-tests for a non-zero magnitude difference between MNI and native regions gave p-values less than 0.005 for all regions in both hemispheres in the young subjects, and the median p-value was less than 10−4. In the elderly (including patients) group, the mean magnitude of the difference was 0.16 ± 0.04, which was significantly greater than in the young subjects (t-test, p < 0.05). Paired t-tests between MNI and native regions gave p-values less than 10−5 for all regions in both hemispheres in the elderly and patients group, and the median p-value was less than 10−7.
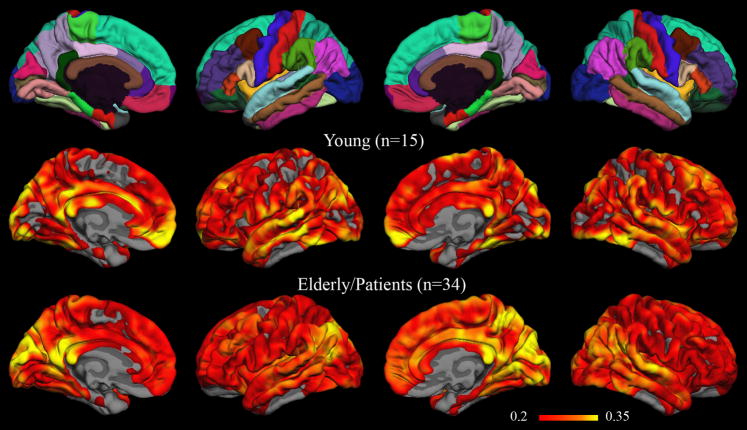
Vertex-wise group-level maps showing the root-mean-square difference between correlation coefficients obtained with MNI-transformed time series versus native-surface coefficients are displayed in Figure 6. The maps have a threshold RMS difference of 0.2, corresponding to the magnitude of the estimated population difference used in the power analysis above. Root-mean-square differences greater than 0.2 are widespread throughout both hemispheres in both young and elderly/diseased subjects. Among regions with the greatest RMS differences (greater than 0.35) are some associated with the default network, including the isthmus cingulate, inferior parietal, and medial prefrontal cortices.
Figure 6. Group RMS difference maps for volume-atlas versus native-surface correlation results.
The overlays in the bottom two rows show the root-mean-square difference between correlation results obtained after nonlinear transformation to the MNI152 volume-atlas and results obtained from analysis on each subject’s native surface. The minimum threshold for the RMS difference (0.2) matches the magnitude of estimated population difference used in power analyses (see Table 2 and Table 3). For reference, the fsaverage parcellation is included in the top row.
4 Discussion
Spontaneous BOLD correlation studies may afford opportunities to increase our understanding of how regions of the brain interact and to develop clinical tools for diagnosis or measurement of disease progression. Already, intriguing results have been reported in various diseases, including mild cognitive impairment and Alzheimer’s disease. Analysis in native space may improve accuracy, allow more rigorous investigation into resting-state correlation phenomena, and otherwise facilitate transition to clinical utility.
Reliance on warping to atlas space has the potential to critically influence results of functional correlation analyses. Despite the critical importance of accurate localization and known issues with warping, very few studies report on the accuracy of transformations, and transformed images are rarely included in published manuscripts. Many methods for warping to atlas volumes exist, and these usually have many user-selected parameters that affect the transformation but are not typically reported in methods descriptions. In the case of patients with atrophy or other structural abnormalities, these issues become more apparent and have been described previously (Bookstein, 2001; Ashburner and Friston, 2001; Senjem et al., 2005).
Figure 1 gives a striking example of the pitfalls of warping an atrophied brain to a normal template. Four methods from three standard software packages, using the recommended parameters, produced warped volumes that have obvious inconsistencies with the MNI template. Analysis of this subject with current methods might yield decreased correlations relative to controls, for example, in regions corresponding to the hippocampus or dorsal cortex. Following typical practices, statistical results would be overlaid on the atlas brain to show the effect. However, decreased correlations in this hypothetical example might be explained by the fact that relatively large portions of the MNI cortex correspond to cerebrospinal fluid in the patient’s warped brain, and therefore in the patient’s warped functional data.
Analysis on native surfaces avoids warping individual brains to atlas volumes and the accompanying issues. Figure 2 demonstrates how the model of the individual cortical surface can still readily capture the features of the severely atrophied brain from the previous example. Remaining within the individual anatomy rather than attempting to distort it through spatial normalization, reduces the risk of mistakenly analyzing correlations outside the gray matter, or outside the brain altogether. Images of other brains, especially those without atrophy, may transform more accurately to the template. Use of other warping algorithms, other warping parameters, or other templates may improve the registration of this, and other, imaged brain volumes. However, at a minimum, the accuracy of the transformations for brains with structural pathology must be assessed and reported alongside functional analyses that depend on that transformation. Moreover, it is possible even studies using young, healthy subjects with no pathology could benefit from analysis on individual surfaces.
The effects of warping to a volume atlas on seed definition and on interregional correlations are quantified in this manuscript by direct comparison of native-surface results to results obtained after warping to the MNI152 volume atlas. In one such comparison, MNI transformation was shown to affect the isthmus cingulate seed time series, yielding MNI seed time series that were often poorly correlated with the seed defined on the subject’s native surface. 25% of the healthy and impaired elderly subjects had MNI-to-native isthmus cingulate correlation coefficients less than 0.66, and two subjects had coefficients less than 0.10. The weak correlation between the MNI and native time courses in this region suggests defining the seed on the native surface may impact resting correlation analyses independent of other methodological considerations. Moreover, a region-by-region comparison of volume-atlas and native-surface results revealed significant differences in every region tested, even when controlling for potential differences in the seed time series. This was true both in the young and elderly/patient groups. Finally, group maps of vertex-wise root-mean-square differences between the two methods also showed that MNI transformation led to sizeable effects throughout the cortex in both groups. Of concern is the observation that these group-level effects were as large as population differences reported in the literature, and regions highly correlated with the seed (in this case, default network regions) may be particularly vulnerable to modulation during transformation to an atlas volume. As the accuracy of registration to a volume atlas is logically dependent on the severity of structural pathology, studies of disease populations require special attention to these possible confounds.
Quantitative, native-surface parcellation analysis of spontaneous fMRI in young subjects highlights known default network regions when the isthmus cingulate is used as a seed (Table 2). Because the seed is defined on the native surface, individual vertex-wise maps can also be calculated to show both the similarities across subjects and the unique features of each subject’s data. Inter-individual variability in both anatomy and functional correlations is preserved in this method, and composite statistics still allow comparisons between groups and between sessions.
Association with other individual markers is one of the primary advantages of analysis in native space. Native-surface cortical analysis is especially convenient for comparison to cortical thickness, as many morphometry studies already use FreeSurfer to measure cortical thickness in the same parcellation regions used in the present study for functional analysis (for example, see Du et al., 2007; Desikan et al., 2010; Rimol et al., 2010; Liu et al., 2010). A potential confound in all functional MRI studies of populations with atrophy, including those using the present method, is that decreased tissue volume might lead to a decrease in BOLD signal-to-noise ratio. Therefore, while a decrease in functional correlations with atrophy is expected due to effects on neural communication, it is difficult to distinguish this neural effect from the signal-to-noise effect. While this limitation is not entirely addressed by analysis on native surfaces, the availability of regional cortical thickness and volume measurements allows local atrophy effects to be accounted for in each subject. This may be additionally advantageous in longitudinal studies of patients with neurodegenerative disorders where atrophy may give structural changes over the course of the experiment.
The power analysis for the native-surface parcellation analysis suggests that a moderate difference in correlation coefficient (with the isthmus cingulate seed) can be detected in any of the regions in Table 2 with practical sample sizes for neuroimaging studies and clinical trials (Jack et al., 2010). Sample sizes range from 16 to 42 subjects, depending on the region; this is consistent with another report of regional differences in variability of correlation strength (Chang and Glover, 2010). The actual sample size necessary to detect a change is highly dependent on the effect size between the populations (sample size is proportional to the square of the effect size). Both the intersubject variance and the effect size will be unique to each disease studied, so caution should be exercised when extrapolating the values presented in Table 2 to other populations. Further research is warranted to estimate population-specific sample sizes; these power analyses will provide additional quantitative information to guide inference and practical application of correlation measurements in those diseases. The value of the power analysis presented here is to demonstrate that the range of expected sample sizes suggests studies of diseases with moderate differences in BOLD correlations are feasible using the native-surface method.
In addition to the benefits of quantitative native-surface analysis of individual subject data, it is also valuable to display surface-based, group-level results. Integrating and interpreting large numbers of individual maps can be difficult, and space constraints will prevent many authors from including maps for each individual in publications. Comparison to other published group maps, particularly those of cortical thickness in aging and dementia (Salat et al., 2004; Dickerson et al., 2009), might also be useful. Registration of individual surface models to a surface average (e.g., the “fsaverage” subject in FreeSurfer) requires transformation to an atlas space, which raises some of the same concerns described for volume transformations, but evidence suggests that surface registration may be substantially more accurate (Fischl et al., 1999b). Importantly, since cortex is being registered to cortex, the risk of mapping an atlas coordinate to white matter or cerebrospinal fluid is greatly decreased.
Results from the patient and elderly subjects demonstrate that default network regions are highlighted in native-surface analysis in multiple populations and with relatively small sample sizes. The robustness of default network correlations to the effects of age, atrophy, and dementia (of at least two different presumed etiologies) is also evident. However, the group maps also serve as an illustration of the degree to which inter-individual differences can be hidden by group averages. The Alzheimer’s disease group map in Figure 5, for example, was generated from only four subjects. This map shows many of the characteristics of typical default network maps and is not remarkably different from the maps of the other groups. At least one of the four subjects, though, has a functional correlation map (Figure 2C, 2D) divergent from the typical default pattern. Investigation into the cognitive and pathological correlates of inter-individual differences in functional correlation patterns may prove enlightening, and native-surface analyses would be a powerful approach for such investigation.
Limitations of analysis on native surfaces include the computational cost of reconstructing and parcellating each subject’s surface, which can take approximately 24 hours processing time per node. Visual confirmation of automated pial and white matter boundaries in the T1-weighted volume is also recommended. In our opinion, however, checking the anatomical boundary definition is simpler than an equivalent check on a warping algorithm’s match of each gyrus in each slice in the volume. We also find it easier to correct small errors in the boundaries for the surface than to optimize nonlinear registration parameters for individual subjects. Analysis with this method requires a high-resolution T1-weighted volume for each subject, which may be viewed as a limitation to authors not routinely including this acquisition in their scanning protocols.
The parcellation atlas itself poses a limitation to the size and shape of the regions tested with the native-space parcellation analysis. Some applications may require more flexibility in region definitions, smaller regions, or specific subdivisions within some of the atlas regions. For others, there may be reason to question the implied assumption of relative functional homogeneity within the anatomically-defined regions. Exploratory studies may require functionally-defined regions or a vertex-wise analysis, which makes transformation to the atlas surface necessary for inter-subject comparisons. Investigators may also consider another atlas available in FreeSurfer for native surfaces that subdivides the regions shown in Figure 4 into gyral and sulcal cortex (Destrieux et al., 2010). Many other atlases exist, and custom atlases can be created within FreeSurfer or elsewhere; these additional atlases typically rely on registration to the average surface, though, and are not automatically produced in the reconstruction process within FreeSurfer. The primary advantages of the Desikan-Killiany atlas parcellation are its common use for cortical thickness studies and its definition based on landmarks that can be consistently identified in individual subjects’ anatomy.
An alternative to native-surface analysis is analysis within the native volumes of individual subjects. To avoid manual ROI selection, the seed region could be defined by a coordinate in the atlas volume and reverse-transformed to each subject by applying the inverse of the transformation matrix from the warping algorithm. Accuracy of seed placement in this case, however, is subject to the same limitations of the transformation matrix as analysis in atlas space. Seed location should be verified for each subject, most likely by visual inspection. A spherical region centered on a reverse-transformed atlas coordinate may increase the chances of including the desired cortex, but if the spheres are much larger than the thickness of the cortex, it is very likely voxels containing primarily white matter or cerebrospinal fluid will be included in the seed. Common diameters for spheres range from 8 mm to 12 mm or larger (Fox et al., 2005, 2006; Vincent et al., 2006; Hedden et al., 2009), compared to cortical thickness which is around 2 mm (Du et al., 2007; Liu et al., 2010).
Even if the reverse-transformed seed is sufficiently accurate, the native-surface functional correlation maps generated with it cannot be quantitatively compared across individuals unless additional regions are defined. One approach is to create more regions of interest from other reverse-transformed atlas volume coordinates, but each of these regions will be subject to the same limitations as the seed. Another approach is to use the reverse-transformed seed to perform seed-correlation analysis in the native brain. However, inter-subject comparisons of the native results in this voxel-wise approach still requires warping the functional data to an atlas volume, and it is unlikely that any advantage is gained by applying that transformation to final statistics rather than to the raw data. An analogous challenge faces independent component analysis (ICA) in native space: even if a suitable template is defined for each subject to identify the component of interest, warping the native-space components to atlas space is necessary for comparison across subjects or groups.
Future development of this method could focus on broadening the scope of its applications. Parcellation regions other than the isthmus cingulate might be used as seeds for other cortical networks that are associated with neurological or psychiatric diseases (Greicius, 2008; Seeley et al., 2009; Ebisch et al., 2010). Additional methods of analyzing functional correlations might also be adapted to native surfaces. Currently, methods such as ICA could be readily applied within the average surface space; however, ICA components are difficult to compare quantitatively across subjects without transforming to atlas space. One idea for native-space ICA is to use one or more of the anatomical parcellation regions highlighted in this study as a template for identifying the default network components. Independent components could then be compared across subjects within analogous parcellation regions. A similar approach could be taken to define templates for other resting-state network components. Regardless of analysis method, investigation of these resting-state functional patterns in disease can benefit from leveraging the underlying anatomy and pathology of individual subjects.
Supplementary Material
Research Highlights
Automated method using FreeSurfer for analyzing BOLD correlations on native surfaces
Surface parcellation allows comparisons of native-surface analyses across subjects
Method highlights default network regions in data from individual subjects
Results presented for young, elderly, Alzheimer’s, Parkinson-related dementias
Acknowledgments
We are grateful for the assistance of Donald J. Hagler, Jr. and Anders Dale for helpful suggestions for data analysis. We would also like to thank Elizabeth A. Murphy and Erik J. Kaestner for their help in recruiting and scanning the elderly subjects and patients, as well as the staff at the Center for Functional MRI and the Radiology Imaging Laboratory at UCSD for technical support during data acquisition. Funding for this work was generously provided by NIA 2P50AG005131, NINDS K02 NS067427, NIH 5 T32 GM007198-37.
Abbreviations
- BOLD
blood oxygenation level dependent
- fMRI
functional magnetic resonance imaging
- ICA
independent component analysis
- AD
Alzheimer’s disease
- PD
Parkinson’s disease
- PDD
Parkinson’s disease dementia
- DLB
dementia with Lewy bodies
- MNI
Montreal Neurological Institute
- RMS
root-mean-square
Footnotes
A number of brain regions exhibit greater activity in functional neuroimaging studies when subjects are permitted to rest than when they are instructed to engage in a specific cognitive task. These regions have been collectively deemed the “default network” (Raichle et al., 2001; Raichle and Snyder, 2007; Buckner et al., 2008)
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Contributor Information
Tyler M. Seibert, Email: tseibert@ucsd.edu.
James B. Brewer, Email: jbrewer@ucsd.edu.
References
- Allen G, Barnard H, McColl R, Hester AL, Fields JA, Weiner MF, Ringe WK, Lipton AM, Brooker M, McDonald E, Rubin CD, Cullum CM. Reduced hippocampal functional connectivity in Alzheimer disease. Arch Neurol. 2007;64:1482–1487. doi: 10.1001/archneur.64.10.1482. [DOI] [PubMed] [Google Scholar]
- Ashburner J, Friston KJ. Why Voxel-Based Morphometry Should Be Used. NeuroImage. 2001;14:1238–1243. doi: 10.1006/nimg.2001.0961. [DOI] [PubMed] [Google Scholar]
- Auer DP. Spontaneous low-frequency blood oxygenation level-dependent fluctuations and functional connectivity analysis of the ‘resting’ brain. Magn Reson Imaging. 2008;26:1055–1064. doi: 10.1016/j.mri.2008.05.008. [DOI] [PubMed] [Google Scholar]
- Bai F, Watson DR, Yu H, Shi Y, Yuan Y, Zhang Z. Abnormal resting-state functional connectivity of posterior cingulate cortex in amnestic type mild cognitive impairment. Brain Res. 2009;1302:167–174. doi: 10.1016/j.brainres.2009.09.028. [DOI] [PubMed] [Google Scholar]
- Biswal B, Yetkin FZ, Haughton VM, Hyde JS. Functional connectivity in the motor cortex of resting human brain using echo-planar MRI. Magn Reson Med. 1995;34:537–541. doi: 10.1002/mrm.1910340409. [DOI] [PubMed] [Google Scholar]
- Bookstein FL. “Voxel-Based Morphometry” Should Not Be Used with Imperfectly Registered Images. NeuroImage. 2001;14:1454–1462. doi: 10.1006/nimg.2001.0770. [DOI] [PubMed] [Google Scholar]
- Buckner RL, Andrews-Hanna JR, Schacter DL. The Brain’s Default Network: Anatomy, Function, and Relevance to Disease. Annals of the New York Academy of Sciences. 2008;1124:1–38. doi: 10.1196/annals.1440.011. [DOI] [PubMed] [Google Scholar]
- Buckner RL, Sepulcre J, Talukdar T, Krienen FM, Liu H, Hedden T, Andrews-Hanna JR, Sperling RA, Johnson KA. Cortical Hubs Revealed by Intrinsic Functional Connectivity: Mapping, Assessment of Stability, and Relation to Alzheimer’s Disease. J Neurosci. 2009;29:1860–1873. doi: 10.1523/JNEUROSCI.5062-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buckner RL, Vincent JL. Unrest at rest: Default activity and spontaneous network correlations. NeuroImage. 2007;37:1091–1096. doi: 10.1016/j.neuroimage.2007.01.010. [DOI] [PubMed] [Google Scholar]
- Chang C, Glover GH. Time-frequency dynamics of resting-state brain connectivity measured with fMRI. Neuroimage. 2010;50:81–98. doi: 10.1016/j.neuroimage.2009.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen J. Statistical power analysis for the behavioral sciences. Psychology Press; [Google Scholar]
- Cox RW, Jesmanowicz A. Real-time 3D image registration for functional MRI. Magn Reson Med. 1999;42:1014–1018. doi: 10.1002/(sici)1522-2594(199912)42:6<1014::aid-mrm4>3.0.co;2-f. [DOI] [PubMed] [Google Scholar]
- Dale AM, Fischl B, Sereno MI. Cortical surface-based analysis. I. Segmentation and surface reconstruction. Neuroimage. 1999;9:179–194. doi: 10.1006/nimg.1998.0395. [DOI] [PubMed] [Google Scholar]
- Dawson B, Trapp RG. Basic & clinical biostatistics. Lange Medical Books/McGraw-Hill; [Google Scholar]
- Desikan RS, Ségonne F, Fischl B, Quinn BT, Dickerson BC, Blacker D, Buckner RL, Dale AM, Maguire RP, Hyman BT, Albert MS, Killiany RJ. An automated labeling system for subdividing the human cerebral cortex on MRI scans into gyral based regions of interest. Neuroimage. 2006;31:968–980. doi: 10.1016/j.neuroimage.2006.01.021. [DOI] [PubMed] [Google Scholar]
- Desikan RS, Sabuncu MR, Schmansky NJ, Reuter M, Cabral HJ, Hess CP, Weiner MW, Biffi A, Anderson CD, Rosand J, Salat DH, Kemper TL, Dale AM, Sperling RA, Fischl B the Alzheimer’s Disease Neuroimaging Initiative. Selective Disruption of the Cerebral Neocortex in Alzheimer’s Disease. PLoS ONE. 2010;5:e12853. doi: 10.1371/journal.pone.0012853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Destrieux C, Fischl B, Dale A, Halgren E. Automatic parcellation of human cortical gyri and sulci using standard anatomical nomenclature. Neuroimage. 2010;53:1–15. doi: 10.1016/j.neuroimage.2010.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickerson BC, Bakkour A, Salat DH, Feczko E, Pacheco J, Greve DN, Grodstein F, Wright CI, Blacker D, Rosas HD, Sperling RA, Atri A, Growdon JH, Hyman BT, Morris JC, Fischl B, Buckner RL. The cortical signature of Alzheimer’s disease: regionally specific cortical thinning relates to symptom severity in very mild to mild AD dementia and is detectable in asymptomatic amyloid-positive individuals. Cereb Cortex. 2009;19:497–510. doi: 10.1093/cercor/bhn113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du A, Schuff N, Kramer JH, Rosen HJ, Gorno-Tempini ML, Rankin K, Miller BL, Weiner MW. Different regional patterns of cortical thinning in Alzheimer’s disease and frontotemporal dementia. Brain. 2007;130:1159–1166. doi: 10.1093/brain/awm016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebisch SJH, Gallese V, Willems RM, Mantini D, Groen WB, Romani GL, Buitelaar JK, Bekkering H. Altered intrinsic functional connectivity of anterior and posterior insula regions in high-functioning participants with autism spectrum disorder. [Accessed December 4, 2010];Hum Brain Mapp. 2010 doi: 10.1002/hbm.21085. Available at: http://www.ncbi.nlm.nih.gov/pubmed/20645311. [DOI] [PMC free article] [PubMed]
- Fischl B, van der Kouwe A, Destrieux C, Halgren E, Segonne F, Salat DH, Busa E, Seidman LJ, Goldstein J, Kennedy D, Caviness V, Makris N, Rosen B, Dale AM. Automatically Parcellating the Human Cerebral Cortex. Cereb Cortex. 2004;14:11–22. doi: 10.1093/cercor/bhg087. [DOI] [PubMed] [Google Scholar]
- Fischl B, Salat DH, Busa E, Albert M, Dieterich M, Haselgrove C, van der Kouwe A, Killiany R, Kennedy D, Klaveness S, Montillo A, Makris N, Rosen B, Dale AM. Whole brain segmentation: automated labeling of neuroanatomical structures in the human brain. Neuron. 2002;33:341–355. doi: 10.1016/s0896-6273(02)00569-x. [DOI] [PubMed] [Google Scholar]
- Fischl B, Sereno MI, Dale AM. Cortical surface-based analysis. II: Inflation, flattening, and a surface-based coordinate system. Neuroimage. 1999a;9:195–207. doi: 10.1006/nimg.1998.0396. [DOI] [PubMed] [Google Scholar]
- Fischl B, Sereno MI, Tootell RB, Dale AM. High-resolution intersubject averaging and a coordinate system for the cortical surface. Hum Brain Mapp. 1999b;8:272–284. doi: 10.1002/(SICI)1097-0193(1999)8:4<272::AID-HBM10>3.0.CO;2-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fleisher AS, Sherzai A, Taylor C, Langbaum JBS, Chen K, Buxton RB. Resting-state BOLD networks versus task-associated functional MRI for distinguishing Alzheimer’s disease risk groups. Neuroimage. 2009;47:1678–1690. doi: 10.1016/j.neuroimage.2009.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox MD, Raichle ME. Spontaneous fluctuations in brain activity observed with functional magnetic resonance imaging. Nat Rev Neurosci. 2007;8:700–711. doi: 10.1038/nrn2201. [DOI] [PubMed] [Google Scholar]
- Fox MD, Snyder AZ, Vincent JL, Corbetta M, Van Essen DC, Raichle ME. The human brain is intrinsically organized into dynamic, anticorrelated functional networks. Proc Natl Acad Sci U S A. 2005;102:9673–8. doi: 10.1073/pnas.0504136102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox MD, Corbetta M, Snyder AZ, Vincent JL, Raichle ME. Spontaneous neuronal activity distinguishes human dorsal and ventral attention systems. Proceedings of the National Academy of Sciences. 2006;103:10046–10051. doi: 10.1073/pnas.0604187103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geser F, Wenning GK, Poewe W, McKeith I. How to diagnose dementia with Lewy bodies: state of the art. Mov Disord. 2005;20 (Suppl 12):S11–20. doi: 10.1002/mds.20535. [DOI] [PubMed] [Google Scholar]
- Glover GH, Li TQ, Ress D. Image-based method for retrospective correction of physiological motion effects in fMRI: RETROICOR. Magn Reson Med. 2000;44:162–167. doi: 10.1002/1522-2594(200007)44:1<162::aid-mrm23>3.0.co;2-e. [DOI] [PubMed] [Google Scholar]
- Greicius M. Resting-state functional connectivity in neuropsychiatric disorders. Curr Opin Neurol. 2008;21:424–430. doi: 10.1097/WCO.0b013e328306f2c5. [DOI] [PubMed] [Google Scholar]
- Greicius MD, Krasnow B, Reiss AL, Menon V. Functional connectivity in the resting brain: a network analysis of the default mode hypothesis. Proc Natl Acad Sci USA. 2003;100:253–258. doi: 10.1073/pnas.0135058100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greicius MD, Srivastava G, Reiss AL, Menon V. Default-mode network activity distinguishes Alzheimer’s disease from healthy aging: evidence from functional MRI. Proc Natl Acad Sci USA. 2004;101:4637–4642. doi: 10.1073/pnas.0308627101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hedden T, Van Dijk KRA, Becker JA, Mehta A, Sperling RA, Johnson KA, Buckner RL. Disruption of functional connectivity in clinically normal older adults harboring amyloid burden. J Neurosci. 2009;29:12686–12694. doi: 10.1523/JNEUROSCI.3189-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van den Heuvel MP, Hulshoff Pol HE. Exploring the brain network: a review on resting-state fMRI functional connectivity. Eur Neuropsychopharmacol. 2010;20:519–534. doi: 10.1016/j.euroneuro.2010.03.008. [DOI] [PubMed] [Google Scholar]
- Holland D, Kuperman JM, Dale AM. Efficient correction of inhomogeneous static magnetic field-induced distortion in Echo Planar Imaging. NeuroImage. 2010;50:175–183. doi: 10.1016/j.neuroimage.2009.11.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jack CR, Bernstein MA, Borowski BJ, Gunter JL, Fox NC, Thompson PM, Schuff N, Krueger G, Killiany RJ, Decarli CS, Dale AM, Carmichael OW, Tosun D, Weiner MW. Update on the magnetic resonance imaging core of the Alzheimer’s disease neuroimaging initiative. Alzheimers Dement. 2010;6:212–220. doi: 10.1016/j.jalz.2010.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koch W, Teipel S, Mueller S, Benninghoff J, Wagner M, Bokde ALW, Hampel H, Coates U, Reiser M, Meindl T. Diagnostic power of default mode network resting state fMRI in the detection of Alzheimer’s disease. [Accessed July 6, 2010];Neurobiol Aging. 2010 doi: 10.1016/j.neurobiolaging.2010.04.013. Available at: http://www.ncbi.nlm.nih.gov/pubmed/20541837. [DOI] [PubMed]
- Liu Y, Paajanen T, Zhang Y, Westman E, Wahlund L, Simmons A, Tunnard C, Sobow T, Mecocci P, Tsolaki M, Vellas B, Muehlboeck S, Evans A, Spenger C, Lovestone S, Soininen H. Analysis of regional MRI volumes and thicknesses as predictors of conversion from mild cognitive impairment to Alzheimer’s disease. Neurobiology of Aging. 2010;31:1375–1385. doi: 10.1016/j.neurobiolaging.2010.01.022. [DOI] [PubMed] [Google Scholar]
- McKeith I. Dementia with Lewy bodies and Parkinson’s disease with dementia: where two worlds collide. Practical Neurology. 2007;7:374–382. doi: 10.1136/jnnp.2007.134163. [DOI] [PubMed] [Google Scholar]
- McKhann G, Drachman D, Folstein M, Katzman R, Price D, Stadlan EM. Clinical diagnosis of Alzheimer’s disease: report of the NINCDS-ADRDA Work Group under the auspices of Department of Health and Human Services Task Force on Alzheimer’s Disease. Neurology. 1984;34:939–944. doi: 10.1212/wnl.34.7.939. [DOI] [PubMed] [Google Scholar]
- Pihlajamäki M, Jauhiainen AM, Soininen H. Structural and functional MRI in mild cognitive impairment. Curr Alzheimer Res. 2009;6:179–185. doi: 10.2174/156720509787602898. [DOI] [PubMed] [Google Scholar]
- Raichle ME, MacLeod AM, Snyder AZ, Powers WJ, Gusnard DA, Shulman GL. A default mode of brain function. Proc Natl Acad Sci USA. 2001;98:676–682. doi: 10.1073/pnas.98.2.676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raichle ME, Snyder AZ. A default mode of brain function: a brief history of an evolving idea. Neuroimage. 2007;37:1083–1090. doi: 10.1016/j.neuroimage.2007.02.041. discussion 1097–1099. [DOI] [PubMed] [Google Scholar]
- Rimol LM, Hartberg CB, Nesvåg R, Fennema-Notestine C, Hagler DJ, Jr, Pung CJ, Jennings RG, Haukvik UK, Lange E, Nakstad PH, Melle I, Andreassen OA, Dale AM, Agartz I. Cortical Thickness and Subcortical Volumes in Schizophrenia and Bipolar Disorder. Biological Psychiatry. 2010;68:41–50. doi: 10.1016/j.biopsych.2010.03.036. [DOI] [PubMed] [Google Scholar]
- Rogers BP, Morgan VL, Newton AT, Gore JC. Assessing functional connectivity in the human brain by fMRI. Magn Reson Imaging. 2007;25:1347–1357. doi: 10.1016/j.mri.2007.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salat DH, Buckner RL, Snyder AZ, Greve DN, Desikan RSR, Busa E, Morris JC, Dale AM, Fischl B. Thinning of the cerebral cortex in aging. Cereb Cortex. 2004;14:721–730. doi: 10.1093/cercor/bhh032. [DOI] [PubMed] [Google Scholar]
- Seeley WW, Crawford RK, Zhou J, Miller BL, Greicius MD. Neurodegenerative diseases target large-scale human brain networks. Neuron. 2009;62:42–52. doi: 10.1016/j.neuron.2009.03.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Senjem ML, Gunter JL, Shiung MM, Petersen RC, Jack J. Comparison of different methodological implementations of voxel-based morphometry in neurodegenerative disease. NeuroImage. 2005;26:600–608. doi: 10.1016/j.neuroimage.2005.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheline YI, Raichle ME, Snyder AZ, Morris JC, Head D, Wang S, Mintun MA. Amyloid Plaques Disrupt Resting State Default Mode Network Connectivity in Cognitively Normal Elderly. Biological Psychiatry. 2010;67:584–587. doi: 10.1016/j.biopsych.2009.08.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorg C, Valentin R, Robert P, Alexander K, Afra WM. Impact of Alzheimer’s Disease on the Functional Connectivity of Spontaneous Brain Activity. [Accessed September 21, 2009];Curr Alzheimer Res. 2009 doi: 10.2174/156720509790147106. Available at: http://www.ncbi.nlm.nih.gov/pubmed/19747154. [DOI] [PubMed]
- Sorg C, Riedl V, Mühlau M, Calhoun VD, Eichele T, Läer L, Drzezga A, Förstl H, Kurz A, Zimmer C, Wohlschläger AM. Selective changes of resting-state networks in individuals at risk for Alzheimer’s disease. Proc Natl Acad Sci USA. 2007;104:18760–18765. doi: 10.1073/pnas.0708803104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Supekar K, Menon V, Rubin D, Musen M, Greicius MD. Network analysis of intrinsic functional brain connectivity in Alzheimer’s disease. PLoS Comput Biol. 2008;4:e1000100. doi: 10.1371/journal.pcbi.1000100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Dijk KRA, Hedden T, Venkataraman A, Evans KC, Lazar SW, Buckner RL. Intrinsic Functional Connectivity As a Tool For Human Connectomics: Theory, Properties, and Optimization. [Accessed January 10, 2010];J Neurophysiol. 2009 doi: 10.1152/jn.00783.2009. Available at: http://www.ncbi.nlm.nih.gov/pubmed/19889849. [DOI] [PMC free article] [PubMed]
- Vincent JL, Snyder AZ, Fox MD, Shannon BJ, Andrews JR, Raichle ME, Buckner RL. Coherent Spontaneous Activity Identifies a Hippocampal-Parietal Memory Network. J Neurophysiol. 2006;96:3517–3531. doi: 10.1152/jn.00048.2006. [DOI] [PubMed] [Google Scholar]
- Wang K, Liang M, Wang L, Tian L, Zhang X, Li K, Jiang T. Altered functional connectivity in early Alzheimer’s disease: a resting-state fMRI study. Hum Brain Mapp. 2007;28:967–978. doi: 10.1002/hbm.20324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Laviolette P, O’Keefe K, Putcha D, Bakkour A, Van Dijk KRA, Pihlajamäki M, Dickerson BC, Sperling RA. Intrinsic connectivity between the hippocampus and posteromedial cortex predicts memory performance in cognitively intact older individuals. [Accessed March 8, 2010];Neuroimage. 2010 doi: 10.1016/j.neuroimage.2010.02.046. Available at: http://www.ncbi.nlm.nih.gov/pubmed/20188183. [DOI] [PMC free article] [PubMed]
- Wang L, Zang Y, He Y, Liang M, Zhang X, Tian L, Wu T, Jiang T, Li K. Changes in hippocampal connectivity in the early stages of Alzheimer’s disease: evidence from resting state fMRI. Neuroimage. 2006;31:496–504. doi: 10.1016/j.neuroimage.2005.12.033. [DOI] [PubMed] [Google Scholar]
- Yan C, Liu D, He Y, Zou Q, Zhu C, Zuo X, Long X, Zang Y. Spontaneous brain activity in the default mode network is sensitive to different resting-state conditions with limited cognitive load. PLoS ONE. 2009;4:e5743. doi: 10.1371/journal.pone.0005743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Wang S, Liu B, Ma Z, Yang M, Zhang Z, Teng G. Resting brain connectivity: changes during the progress of Alzheimer disease. Radiology. 2010;256:598–606. doi: 10.1148/radiol.10091701. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.