Summary
Francisella tularensis, the causative agent of tularemia, infects host macrophages, which triggers production of the proinflammatory cytokines interleukin 1 β (IL-1 β) and IL-18. We elucidate here how host macrophages recognize Francisella and elicit this pro-inflammatory response. Using mice deficient in the DNA-sensing inflammasome component AIM2, we demonstrate here that AIM2 is required for sensing Francisella. AIM2-deficient mice were extremely susceptible to Francisella infection with higher mortality and bacterial burden compared to wild-type mice. Caspase-1, activation, IL-1β secretion and cell death were absent in Aim2−/− macrophages in response to Francisella infection or presence of cytoplasmic DNA. This study identifies AIM2 as a crucial sensor of F. tularensis infection, and provides genetic proof for its critical role in the host innate immunity to intracellular pathogens.
The innate immune system is an evolutionary conserved first line of defense against invading organisms. It is now well established that the innate immune system uses molecular pattern recognition to detect infection and mount an immune response to eliminate the pathogen and infected cells. Specialized receptors collectively called pathogen- or pattern-recognition receptors (PRRs) are present on the cytoplasmic or endosomal membranes, or in the cell cytosol 1, 2. PRRs sense highly conserved molecular components of invading organisms known as pathogen-associated molecular patterns (PAMPs).
A recently identified PRR called AIM2 (absent in melanoma 2), was found to recognize cytoplasmic dsDNA through its HIN-200 domain3–6. AIM2 assembles a large inflammasome complex with cytosolic DNA, visible by confocal microscopy 3. The fully assembled AIM2 inflammasome then recruits and subsequently oligomerizes the caspase-1-activating adaptor protein ASC, which activates caspase-1, leading to production of the mature cytokines interleukin 18 (IL-18) and IL-1β and subsequent cell death 3, 7, 8.
Despite these recent advances in our understanding of how cytoplasmic DNA is recognized by the AIM2 inflammasome, little is known about the precise role of the AIM2 inflammasome in the innate immune defense against intracellular microbial and viral pathogens. Of particular interest is the facultative intracellular gram-negative bacteria Francisella tularensis, the causative agent of tularemia. Invasion of the cytosol by F. tularensis leads to type I interferon production as well as activation of caspase-1 and cell death 9, 10. F. tularensis activates the interferon response by a mechanism that is independent of signaling by plasma membrane or endosomal TLRs, or cytosolic RIG-I, MDA5, NOD1 or NOD2, but requires IRF3 signaling 10, 11. It has been proposed that the host cell may sense cytosolic F. tularensis DNA via unidentified cytosolic DNA sensors leading to induction of type I interferons as well as activation of caspase-1 and cell death pathways during F. tularensis infection 10, 11.
We describe here the generation of AIM2-deficient mice to investigate whether AIM2 plays an important role in the host proinflammatory innate immune response to F. tularensis specifically, and to cytosolic DNA in general. Our results show that AIM2-deficient macrophages are defective in caspase-1 activation, IL-1β secretion and cell death in response to cytosolic DNA, or infection with F. tularensis. Although the AIM2 deficiency does not affect the transcriptional type I interferon response to F. tularensis infection, AIM2-deficient mice are more susceptible to the lethal effect of F. tularensis than wild-type mice. Our results thus suggest that the type I interferon response is not sufficient on its own for protection against F. tularensis infection and that the AIM2 inflammasome activity is required for full innate immunity against this pathogen. Our work also indicates that among all known PRRs, AIM2 is uniquely required for the pro-inflammatory innate immune response to F. tularensis.
RESULTS
AIM2 is important for caspase-1 activation by DNA
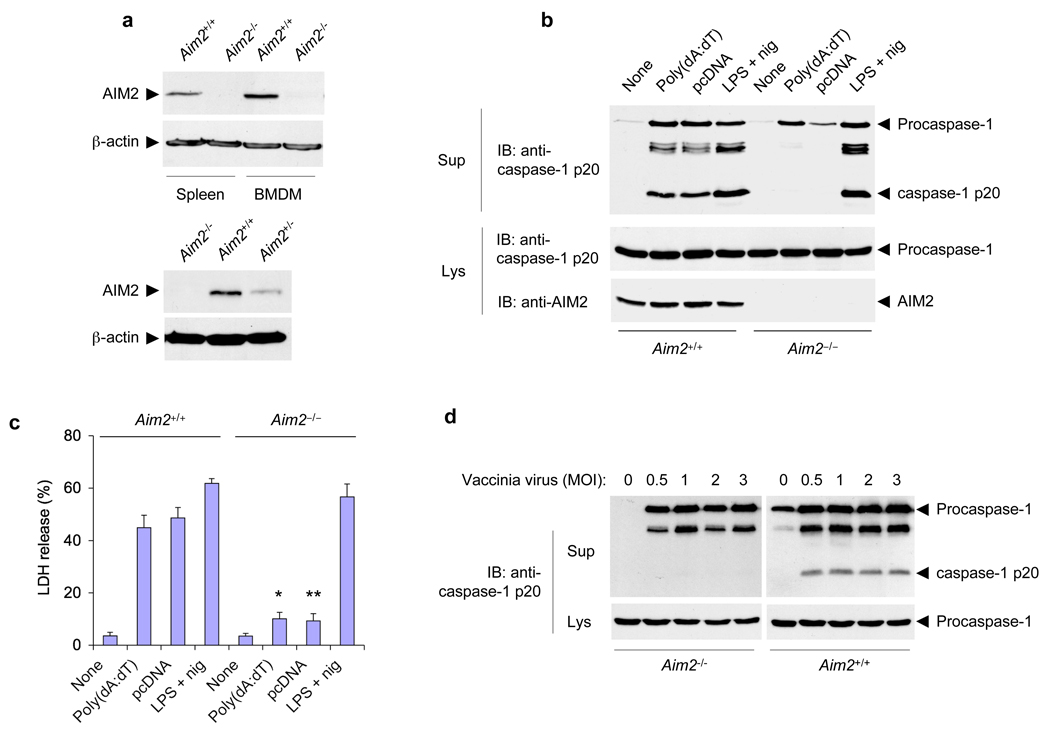
To investigate the precise role of AIM2 in the host innate immune defense against dangerous cytosolic DNA produced by intracellular viral and microbial pathogens, we generated AIM2-deficient mice using the gene trap technology (Supplementary Fig. 1a,b) 12, 13. Immunoblot analysis of spleen and bone marrow samples from an Aim2−/− mouse and its Aim2+/+ littermate revealed the presence of full length AIM2 protein in the Aim2+/+ mouse but not in the Aim2−/− littermate (Fig. 1a, upper panels), indicating that the insertion of the gene trap vector in the Aim2 gene resulted in disruption of Aim2. Heterozygous Aim2+/− mice expressed reduced amount of AIM2 in the spleen compared to Aim2+/+ mice (Fig. 1a, lower panels), indicating that the expression of AIM2 is gene-dose dependent. All Aim2−/− mice had no obvious phenotypic abnormalities and were morphologically indistinguishable from their heterozygous or WT littermates (Supplementary Fig. 1c), indicating that AIM2 deficiency does not have any apparent adverse effects on mouse development.
Figure 1.
Disruption of mouse Aim2 abolishes activation of the inflammasome by cytoplasmic DNA and vaccinia virus. (a) Immunoblot analysis of the expression of AIM2 in spleens and BMDMs from Aim2+/+ and Aim2−/− littermates (top) and in spleens from Aim2−/−, Aim2+/+ and Aim2+/− mice (bottom), assessed with an antibody specific for mouse AIM2. β-actin serves as a loading control. (b) Immunoblot (IB) analysis of mouse procaspase-1, caspase-1 (p20 subunit) and/or AIM2 in culture supernatants (Sup) and lysates (Lys) of Aim2+/+ and Aim2−/− BMDMs left untreated (None) or transfected with the synthetic DNA poly(dA:dT) or plasmid DNA (pcDNA), or treated for 5 h with LPS (500 ng/ml) followed by nigericin (2.5 µM) for 45 min (LPS + nig), assessed with monoclonal antibody to mouse caspase-1 p20 (anti-p20). (c) Release of LDH into culture supernatants of the BMDMs described in b, presented relative to the total cellular LDH content. *P < 0.05 and **P < 0.01, Aim2+/+ versus Aim2−/− (Student’s t-test). (d) Immunoblot analysis of mouse procaspase-1 and caspase-1 in culture supernatants and lysates of mouse Aim2−/− and Aim2+/+ BMDMs infected for 18 h with vaccinia virus (multiplicity of infection (MOI), above lanes). Data are representative of at least three experiments (mean and s.d. in c).
In vitro studies with bone marrow-derived macrophages (BMDM) from Aim2−/− and Aim2+/+ littermates revealed that liposome-delivered DNA activated caspase-1 and induced cell death in wild-type Aim2+/+, but not in Aim2−/− BMDM (Fig. 1b, c), indicating that AIM2 is indeed critical for sensing cytoplasmic DNA and activation of caspase-1. The AIM2 deficiency had no effect on caspase-1 activation by the Nlrp3 stimuli LPS plus ATP or LPS plus nigericin 14, or the Nlrp1 stimulus anthrax lethal factor 15 (Fig. 1b and Supplementary Fig. 2 a, b). Reconstitution of the Aim2−/− macrophages with a GFP-tagged AIM2 using retroviral transduction restored their responsiveness to transfected Cy3-labeled DNA as evidenced by oligomerization of the AIM2-GFP and the induction of pyroptotic cell death, which is dependent on caspase-1 activation (Supplementary Fig. 3a). There was no morphological evidence of pyroptotic cell death in the Aim2−/− macrophages expressing GFP alone (Supplementary Fig. 3b).
Next we investigated the role of AIM2 in the innate immunity of macrophages against cytosolic DNA produced by infection with Vaccinia virus. In contrast to wild-type Aim2+/+ BMDMs, caspase-1 processing was completely absent in Aim2−/− BMDMs (Fig. 1d) 18 h post-infection with Vaccinia virus, providing definitive proof that the AIM2 inflammasome is indeed critical for detection of Vaccinia virus infection, as demonstrated previously by siRNA knockdown experiments 4.
AIM2 is important for caspase-1 activation by Francisella
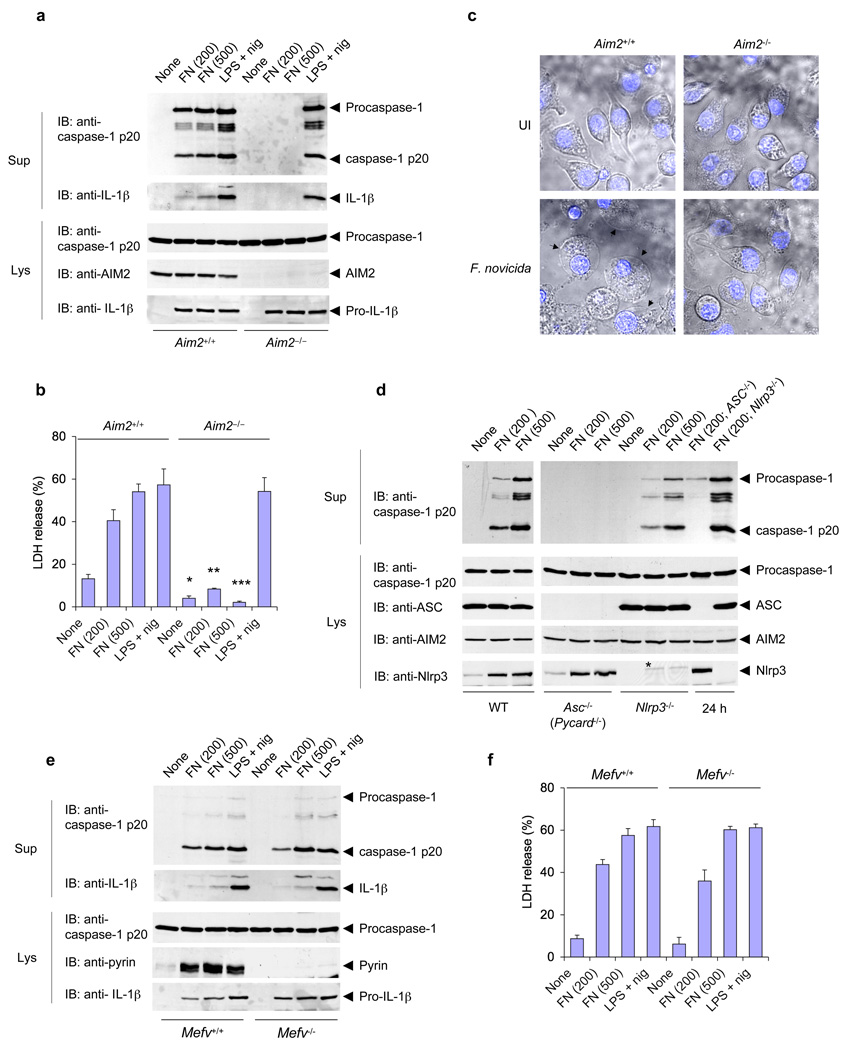
F. tularensis is a highly infectious bacterium that replicates in the cytoplasm of infected cells16, 17, leading to the activation of an IRF3-dependent and TLR-independent type I interferon response, as well as a Nlrp3-independent, but ASC-dependent inflammasome, that causes cell death 9, 10. These observations suggest that cytosolic DNA produced by Francisella during its escape from the phagosome might be the common ligand that activates both the type I interferon response, and possibly the AIM2 inflammasome pathways. To test this possibility, we analyzed inflammasome activation, and cell death by LDH release in macrophages from wild-type and Aim2−/− mice after infection with F. tularensis subspecies novicida (F. novicida). Indeed, processing of caspase-1, secretion of IL-1β, and LDH release were completely absent in Aim2−/− macrophages in response to F. novicida infection at 6 h post infection (Fig. 2a,b), even at high multiplicity of infection (Fig. 2a, 6th & 7th lane). Processing of caspase-1 and secretion of IL-1β were also absent in Aim2−/− macrophages at 24 h post infection, although there was LDH release at this time point (Supplementary Fig. 4a,b) likely due to caspase-1-independent cell death as previously reported in casp-1−/− macrophages 11. In contrast, normal processing of caspase-1, secretion of IL-1β, and LDH release were seen in wild-type macrophages (Fig. 2a,b, and Supplementary Fig. 4a,b). Processing of caspase-1 in response to Francisella infection or transfected DNA was also unimpaired in heterozygous Aim2+/− macrophages, although it was reduced compared to wild-type macrophages due to the reduced level of AIM2 in the Aim2+/− macrophages (supplementary Fig. 5a,b). Consistent with a critical role for AIM2 in the pyroptotic cell death pathway, morphological features of pyroptosis, including plasma membrane swelling and nuclear condensation, were very obvious in F. novicida-infected Aim2+/+, but not in Aim2−/− macrophages (Fig. 2c). Infection with Salmonella typhimurium which specifically activates the NLRC4 (Ipaf) inflammasome 18, resulted in normal processing of caspase-1 in both wild-type and Aim2−/− macrophages (Supplementary Fig. 4c), indicating that AIM2 is not involved in the pro-inflammatory response to Salmonella infection. Consistent with previous findings 10, the activation of caspase-1 by Francisella infection was dependent on ASC but not on Nlrp3 (Fig. 2d). Taken together, our results indicate that AIM2 is critical for the pro-inflammatory and cell death responses to Francisella infection, but dispensable for these processes in response to S. typhimurium infection.
Figure 2.
AIM2 is required for F. novicida–induced activation of the inflammasome. (a) Immunoblot analysis of mouse procaspase-1, caspase-1, IL-1β, AIM2 and/or pro-IL-1β in culture supernatants and lysates of mouse Aim2−/− and Aim2+/+ macrophages left untreated or infected for 6 h with F. novicida (FN; MOI in parentheses above lanes) or treated with LPS and nigericin as described in Figure 1b. (b) Release of LDH into culture supernatants of the macrophages in a. *P < 0.05, **P < 0.01 and ***P < 0.005, Aim2+/+ versus Aim2−/− (Student’s t-test). (c) Confocal live-cell microsopy of Aim2−/− and Aim2+/+ BMDMs left uninfected (UI) or infected for 6 h with F. novicida; nuclei were stained with Hoechst stain (blue). Images are merged differential interference contrast and Hoechst channels. Original magnification, x40. (d) Immunoblot analysis of mouse procaspase-1, caspase-1, ASC, AIM2 and/or Nlrp3 in culture supernatants and lysates of mouse wild-type, ASC-deficient (Pycard−/−; called ‘Asc−/−’ here) and Nlrp3−/− macrophages infected with F. novicida for 6 h or for 24 h (far right; MOI in parentheses above lanes). (e) Immunoblot analysis of mouse procaspase-1, caspase-1, IL-1β, pyrin and/or pro-IL-1β in culture supernatants and lysates of mouse pyrin-deficient (Mefv−/−) and pyrin-sufficient (Mefv+/+) macrophages infected for 6 h with F. novicida (MOI in parentheses above lanes) or treated with LPS and nigericin as described in a. (f) Release of LDH into culture supernatants of the macrophages in e. Data are representative of at least three experiments (mean and s.d. in b,f).
The familial Mediterranean fever protein pyrin, which can also form an inflammasome complex with ASC and caspase-1 19, 20, might be involved in caspase-1 activation by F. novicida in human macrophages 21. To test this possibility in mouse macrophages, we infected peritoneal macrophages derived from wild-type or pyrin-deficient (Mefv−/−) mice 22 with F. novicida or stimulated them with the Nlrp3 stimulus, LPS plus nigericin (Fig. 2e,f). Despite the notable upregulation of pyrin protein in the wild-type macrophages by these stimuli (2nd to 4th lanes, 4th panel from top), there was no notable difference in processing of caspase-1, secretion of IL-1β, or LDH release between the wild-type and Mefv−/− macrophages. These results indicate that pyrin does not play a role in the pro-inflammatory and cell death responses to Francisella infection in mice, or negatively regulate the activity of ASC or the inflammasome as previously proposed 22–24.
Mechanism of Activation of AIM2 by Francisella
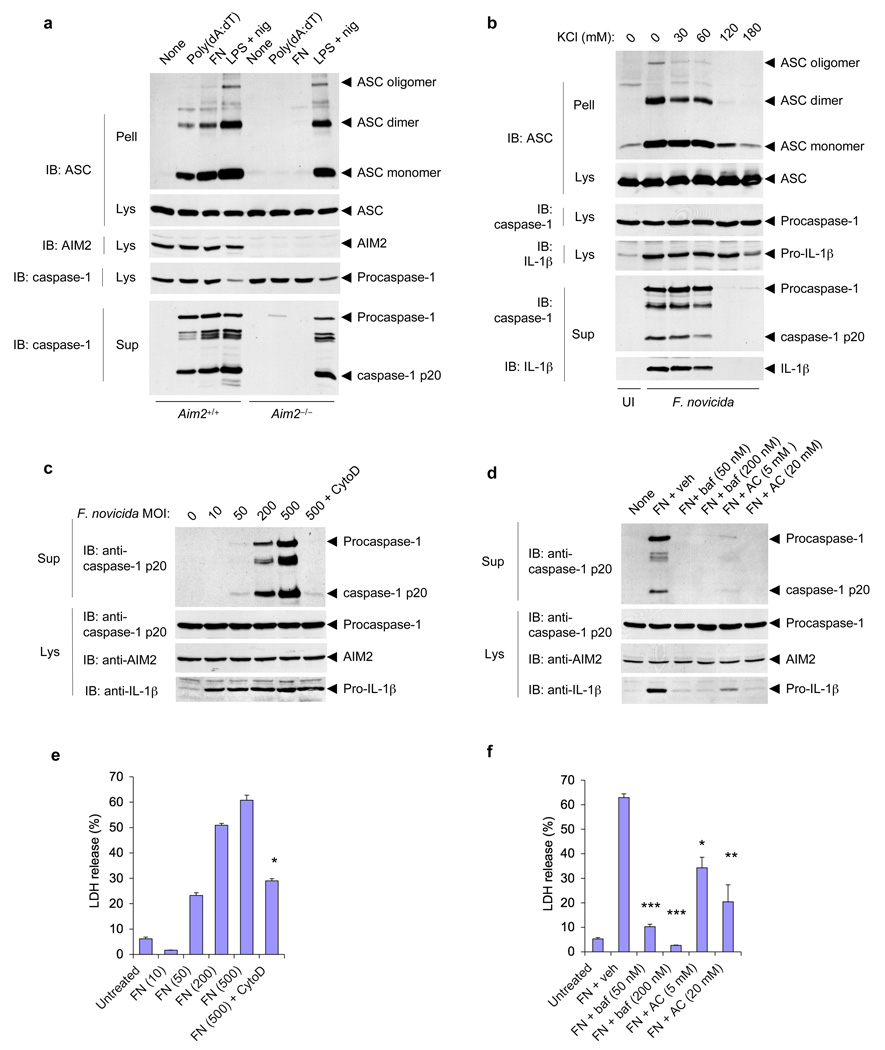
Engagement of AIM2 by cytoplasmic DNA leads to formation of the oligomeric ASC pyroptosome, which induces pyroptotic cell death by activating caspase-1 3, 8. To investigate whether Francisella infection induces AIM2-dependent formation of the ASC pyroptosome, we analyzed the presence of oligomeric ASC in cell pellets of Francisella-infected wild-type and Aim2−/− macrophages. As expected, Francisella-infection induced ASC pyroptosome formation in wild-type but not Aim2−/− macrophages (Fig. 3a, 3rd lane). Similarly, liposome-delivered cytoplasmic DNA induced ASC pyroptosome formation in wild-type but not Aim2−/− macrophages (Fig. 3a, 2nd lane). In contrast, LPS plus nigericin, which activates the Nlrp3 inflammasome, induced ASC pyroptosome formation in both wild-type and Aim2−/− macrophages (Fig. 3a, 4th and 8th lanes). Collectively, our data indicate that Francisella-infection induces pyroptotic cell death via AIM2-mediated ASC pyroptosome formation.
Figure 3.
Role of the ASC pyroptosome, potassium depletion, actin polymerization and lysosomal acidification in activation of the AIM2 inflammasome by F. novicida. (a) Immunoblot analysis of ASC pyroptosomes in Aim2+/+ and Aim2−/− BMDMs transfected with poly(dA:dT), infected for 6 h with F. novicida (MOI, 500), or treated with LPS and nigericin as described in Figure 1b; pellets of whole-cell lysates centrifuged at 3,800g followed by crosslinking with disuccinimidyl suberate (Pell), as well as cell lysates and culture supernatants, were hybridized with anti–mouse ASC, anti–mouse AIM2 and anti-caspase-1. (b) Immunoblot analysis of ASC pyroptosomes in Aim2+/+ BMDMs left uninfected or infected for 6 h with F. novicida (MOI, 500) in the presence of increasing concentrations of KCl in the culture medium, then fractionated and analyzed as in a. (c,d) Immunoblot analysis of mouse procaspase-1, caspase-1, AIM2 and/or pro-IL-1β in culture supernatants and lysates of Aim2+/+ BMDMs infected for 6 h with F. novicida (MOI, above lanes) in the presence (+ cytoD) or absence of cytochalasin D (c) or infected for 6 h with the F. novicida (MOI, 250) in the presence of vehicle (+ veh), bafilomycin (+ baf) or NH4Cl (+ AC; d). (e,f) Release of LDH into culture supernatants of the BMDMs in c (e) and d (f). (e) *P < 0.001, with versus without cytochalasin D (Student’s t-test); (f) *P < 0.05, **P < 0.01 and ***P < 0.001, F. novicida versus vehicle (Student’s t-test). Data are representative of two (a–d) or three (e,f) experiments (mean and s.d. in e,f).
To gain further insight into the signaling pathway by which Francisella infection activates the AIM2 inflammasome, we investigated the role of cytoplasmic potassium efflux, which is critical for formation of the ASC pyroptosome and recruitment of caspase-1 to ASC oligomers 8. Inhibition of potassium efflux by increasing potassium concentration in the culture medium completely blocked ASC pyroptosome formation, caspase-1 activation and IL-1β secretion by Francisella-infection (Fig. 3b), indicating that depletion of intracellular potassium is required for assembly of the AIM2 inflammasome and formation of the ASC pyroptosome. Activation of caspase-1 and cell death by Francisella-infection was markedly blocked by the actin oligomerization inhibitor cytochalasin D, and the endosomal acidification inhibitors bafilomycin A or NH4Cl (Fig. 3c–f), indicating that bacterial internalization and lysosomal acidification are required for recognition of Francisella-infection and activation of the AIM2 inflammasome. Inhibition of Francisella-induced caspase-1 activation by bafilomycin was not due to inhibition of bacterial uptake or replication as more intracellular bacteria were found in bafilomycin treated cells compared to the untreated control at 3 h post infection (supplementary Fig. 6). The increased bacterial count in bafilomycin treated cells is consistent with previous observations linking phagosomal acidification to killing and degradation of phagocytosed bacteria 25.
IRF3 is important for AIM2 activation by Francisella
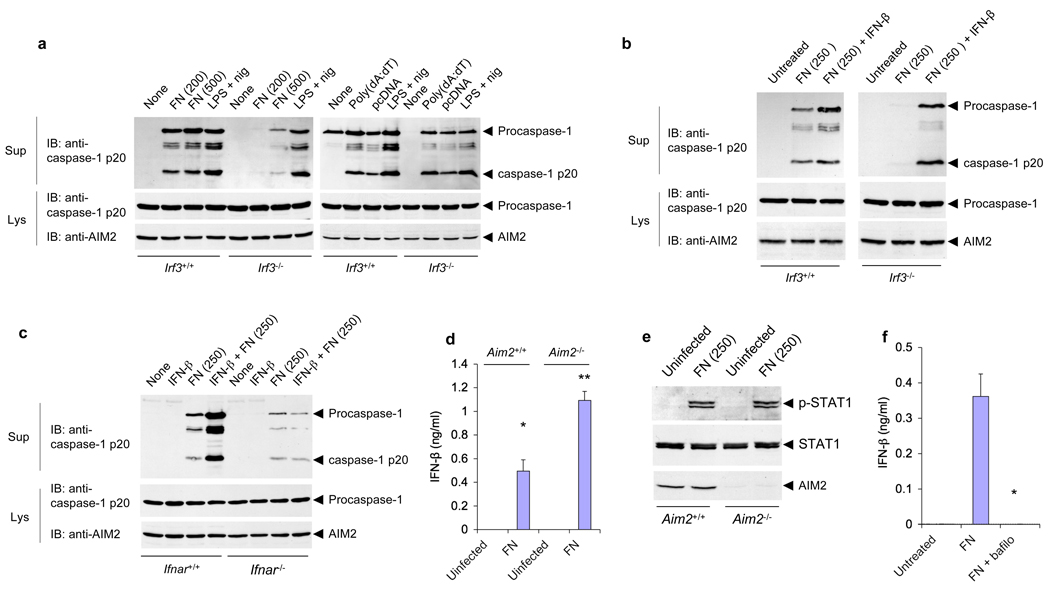
Activation of the inflammasome by Francisella infection requires an intact type I interferon response for efficient inflammasome activation 10. Consistent with these studies, Francisella infection of Irf3−/− macrophages, which are defective in the secretion of type I interferons in response to cytosolic DNA, resulted in less efficient activation of the AIM2 inflammasome compared with wild-type macrophages (Fig. 4a, left panels). In contrast, liposome-delivered DNA induced similar activation of the AIM2 inflammasome in both WT and Irf3−/− macrophages (Fig. 4a, right panels). Similar expression levels of AIM2 in wild-type and Irf3−/− macrophages (Fig. 4a, lower panel), were observed ruling out the possibility that Irf3−/− macrophages express less AIM2. To determine if type I interferon signaling through its IFNAR1 could restore AIM2 activation by Francisella infection in the Irf3−/− macrophages, we treated these macrophages with IFN-β at the time of infection and assayed caspase-1 activation at 6 h post-infection. Simultaneous treatment of Irf3−/− macrophages with both IFN-β and F. novicida restored Francisella–induced caspase-1 activation in these cells (Fig. 4b, right panels). Concomitant treatment with IFN-β at the time of infection slightly enhanced Francisella–induced caspase-1 activation in WT Irf3+/+ macrophages (Fig. 4b, left panels). Restoration of Francisella–induced caspase-1 activation in Irf3−/− macrophages required concurrent treatment with IFNβ and Francisella, as no restoration was observed if IFN-β treatment was done 2 h post-infection (supplementary Fig. 7a, left panels). No activation of caspase-1 was observed by IFN-β treatment alone (supplementary Fig. 7a, right panel). Additionally, concurrent treatment of Aim2−/− macrophages with IFN-β and Francisella did not restore Francisella–induced caspase-1 activation in these cells (supplementary Fig. 7b), indicating that IFN-β is not sufficient for Francisella-induced caspase-1 activation in the absence of AIM2.
Figure 4.
IRF3 signaling is required for activation of the AIM2 inflammasome by F. novicida but not by liposome-delivered DNA. (a,b) Immunoblot analysis of mouse procaspase-1, caspase-1 and/or AIM2 in culture supernatants and lysates of mouse Irf3−/− and Irf3+/+ macrophages infected for 6 h with F. novicida (MOI, in parentheses above lanes), treated with LPS and nigericin as described in Figure 1b, or transfected with poly(dA:dT) (a), or infected with F. novicida (MOI, 250) in the presence or absence of IFN-β (b). (c) Immunoblot analysis of mouse procaspase-1, caspase-1 and/or AIM2 in culture supernatants and lysates of Ifnar1−/− and Ifnar1+/+ macrophages left untreated or treated for 2 h with IFN-β alone or followed by infection for 6 h with F. novicida (MOI, in parentheses above lanes). (d) Enzyme-linked immunosorbent assay of IFN-β in culture supernatants of Aim2−/− and Aim2+/+ macrophages left uninfected or infected for 6 h with F. novicida (MOI, 250). *P < 0.05 and **P < 0.005 (Student’s t-test). (e) Immunoblot analysis of mouse STAT1 phosphorylated at Tyr701 (p-STAT1), total STAT1 and AIM2 in lysates of mouse Aim2−/− and Aim2+/+ macrophages left uninfected or infected with F. novicida (MOI, 250). (f) Enzyme-linked immunosorbent assay of IFN-β in culture supernatants of Aim2+/+ macrophages left untreated or infected for 6 h with F. novicida (MOI, 250) in the presence (+ bafilo) or absence of bafilomycin (50 nM). *P < 0.01 (Student’s t-test). Data are representative of two (a,c) or three (b,d–f) experiments (mean and s.d. in d,f).
To further investigate the role of type I interferons in the mechanism of activation of AIM2 by Francisella infection, we primed wild-type and Irf3−/− macrophages with IFN-β 2 h prior to infection with Francisella, and measured caspase-1 activation at different time points post-infection (supplementary Fig. 8). In the absence of IFN-β priming, caspase-1 processing in wild-type macrophages was barely detectable at 3 h post-infection. In contrast, maximum caspase-1 processing was observed at 3 h post infection in IFN-β-primed wild-type macrophages. Although un-primed Irf3−/− macrophages showed less efficient caspase-1 activation than wild-type macrophages at 3 and 5 h post-infection, they showed comparable caspase-1 activation to wild-type macrophages following IFN-β-priming. These results indicate that priming of macrophages with IFN-β accelerates and enhances the activation of the AIM2 inflammasome by Francisella infection not only in IRF3-deficient macrophages, but also in wild-type macrophages. These results also suggest that the delay in caspase-1 activation observed in un-primed WT macrophages might be due to the lack of type I interferons in the initial stages of Francisella infection.
Like IRF3-deficient macrophages, IFNAR1-deficient macrophages are also defective in activation of caspase-1 in response to Francisella infection (ref 10 and Fig. 4c). However, this defect cannot be corrected by IFN-β-priming indicating that signaling by IFNAR1 is required for efficient activation of the AIM2 inflammasome by Francisella infection.
The impaired AIM2 inflammasome activation in Ifnar1−/− macrophages was not caused by a lack of intracellular bacterial replication. On the contrary, the number of intracellular Francisella in Ifnar1−/− macrophages was notably higher than in wild-type macrophages at 5 h post-infection and reached to almost 50 fold higher at 24 h post infection (Supplementary Fig. 9). These results are consistent with a previous report10. Collectively these observations indicate that type I interferon signaling acts upstream of the AIM2 inflammasome, perhaps to enhance killing and lysis of Francisella in the phagosome to generate the cytosolic DNA that activates the AIM2 inflammasome (see model in supplementary Fig. 10, and discussion for details).
AIM2 is not critical for type I interferon production
To rule out the possibility that defective activation of the inflammasome in Aim2−/− macrophages is due to defective type I interferon production or signaling, we quantitated the amounts of secreted IFN-β in culture media of wild-type and Aim2−/− macrophages infected with F. novicida, and also determined the status of STAT-1 phosphorylation in lysates from these cells (Fig. 4d,e). Our results show that IFN-β production from Aim2−/− macrophages is actually higher than from wild-type macrophages in response to Francisella infection (Fig. 4d). However, there was no significant difference in STAT-1 phosphorylation between Francisella-infected wild-type and Aim2−/− macrophages (Fig. 4e). These results indicate that AIM2 is not critical for type I interferon production or signaling and that AIM2 deficiency does not negatively impact type I interferon response to F. novicida infection. In fact, AIM2 deficiency appears to enhance IFN-β production in response to F. novicida infection. This result is consistent with recent observations that siRNA-mediated knockdown of AIM2 potentiate IFN-β production in response to transfected cytosolic DNA 4. The production of IFN-β induced by Francisella-infection was completely blocked by bafilomycin A (Fig. 4f), indicating that lysosomal acidification is not only required for activation of the AIM2 inflammasome, but also necessary for Francisella-induced type I interferon production.
Francisella DNA induces AIM2 oligomerization
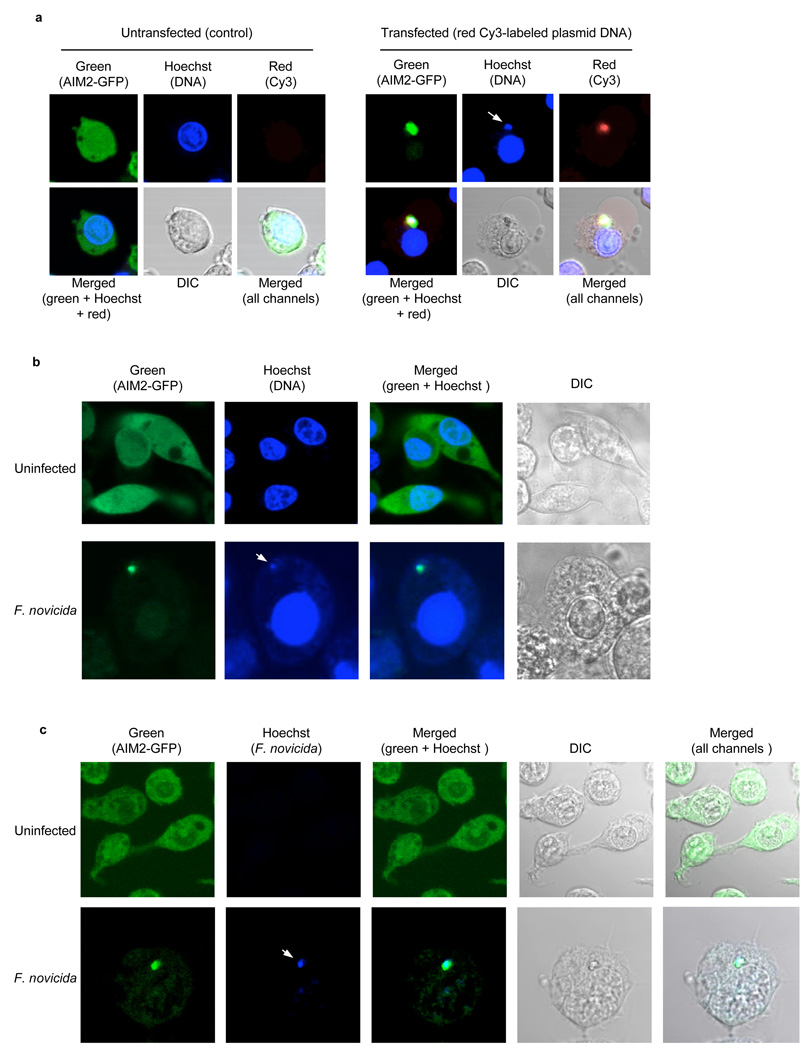
Cytoplasmic DNA activates AIM2 by directly binding to its HIN-200 domain leading to its oligomerization. This process can be visualized in live cells expressing a GFP-tagged AIM2 and a Cy3-labeled DNA (Fig. 5a and ref 3). Notably, the cytosolic DNA that is bound to AIM2 in the oligomeric AIM2-DNA complex can also be visualized with the DNA-specific Hoechst 33342 fluorescent stain (Fig. 5a). To formally demonstrate that AIM2 senses cytosolic DNA produced by Francisella infection, we infected the Nlrp3−/−-AIM2-GFP-N1 macrophages (a Nlrp3−/− cell line stably expressing a C-terminal GFP-tagged AIM2) 3 with F. novicida and subsequently stained the infected cells with Hoechst stain. The Nlrp3−/− AIM2-GFP-N1 macrophages were used in these experiments to rule out the involvement of Nlrp3 in the response to cytosolic DNA or Francisella infection. AIM2 was evenly distributed in the cytoplasm and nuclei of the uninfected cells (Fig. 5b, upper panels). Infection of these cells with F. novicida resulted in AIM2 oligomerization (Fig 5b, lower panels, and supplementary Fig. 11) as evidenced by the clustering of the AIM2-GFP. These AIM2-GFP clusters were also visible with the vital DNA-specific Hoechst stain indicating that AIM2 is clustered by binding to cytosolic DNA. The infected macrophages that contained the AIM2-DNA clusters showed clear features of pyroptotic cell death.
Figure 5.
Cytoplasmic DNA secreted by Francisella induces AIM2 oligomerization. (a), Enlarged confocal live cell images of Nlrp3−/−-AIM2-EGFP-N1 BMDM following transfection with Cy-3™-labeled DNA (right panels) or nothing (control, left panels). The white arrow in the blue channel (right panels) indicates staining of the clustered cytoplasmic DNA with the blue Hoechst stain, which specifically stains DNA. (b), Enlarged confocal live cell images of Nlrp3−/−-AIM2-EGFP-N1 BMDM left uninfected (upper panels) or infected with F. novicida (lower panels) for 6 h and then stained with Hoechst stain before microscopy. The white arrow in the blue channel (lower panels) indicates staining of the AIM2-GFP cluster with the DNA-specific blue Hoechst stain. (c), Enlarged confocal cell images of Nlrp3−/−-AIM2-EGFP-N1 BMDM left uninfected (upper panels) or infected with Hoechst-labeled F. novicida (lower panels) for 6 h and then fixed on coverslips before confocal microscopy. The white arrow in the blue channel (lower panels) indicates the Hoechst-labeled Francisella cytoplasmic DNA. Additional representative confocal images of Nlrp3−/−-AIM2-EGFP-N1 BMDM infected with unstained or Hoechst-pre-stained F. novicida are shown in supplementary Figure 11. DIC, differential interference contrast. Original magnification, x40. Data are representative of at least three (a,b) or two (c) experiments.
To provide direct evidence that the cytosolic DNA responsible for clustering of AIM2-GFP in these cells is derived from Francisella, we labeled Francisella DNA with Hoechst stain by pre-staining F. novicida with Hoechst stain prior to using them to infect the Nlrp3−/− AIM2-GFP-N1 macrophages. As expected, infection of these cells with the Hoechst-labeled Francisella resulted in clustering of AIM2-GFP around the Hoechst-labeled DNA (Fig. 5c, lower panels, and supplementary Fig. 11b). Because only Francisella DNA is labeled with Hoechst stain, these results provide direct evidence that Francisella DNA is responsible for oligomerizing AIM2. Collectively, our results indicate that F. novicida infection of macrophages generates cytosolic DNA that specifically binds to and induces AIM2 oligomerization leading to the activation of caspase-1 and cell death (see model in Supplementary Fig. 10).
Innate immunity against Francisella infection requires AIM2
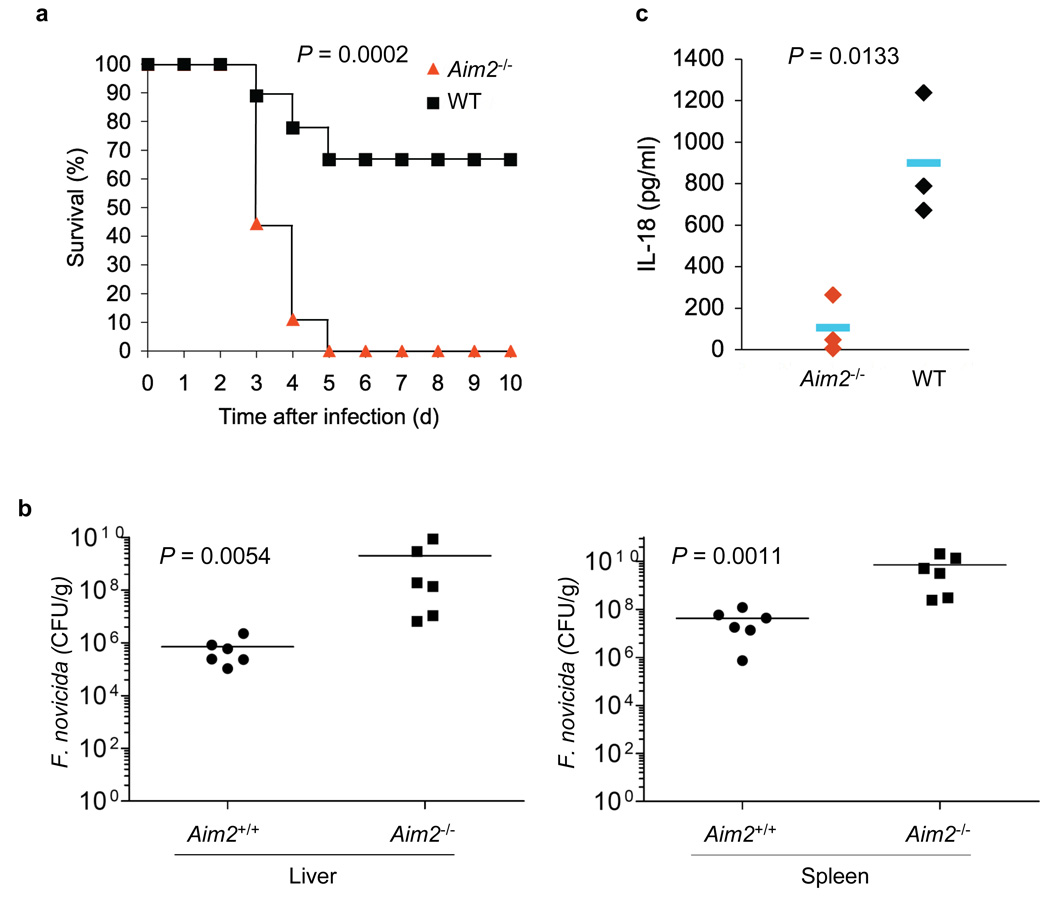
The above results indicate that AIM2 is important for the pro-inflammatory and cell death responses to Francisella-infection in vitro, through a mechanism that involves the recognition of cytoplasmic DNA produced by this intracellular pathogen. To assess the precise role of AIM2 in the host innate immune defense against Francisella-infection, wild-type and Aim2−/− mice were challenged subcutaneously with live F. novicida. Compared to wild-type mice, the subcutaneous infection of Aim2−/− mice with F. novicida resulted in dramatically higher rate of mortality (Fig. 6a). The mortality rate of Aim2−/− mice on day 5 was 100% compared with just 33% in wild-type mice. The remaining wild-type mice survived beyond 20 days post infection. Consistent with these results, the bacterial burdens in tissues of Aim2−/− mice were markedly higher than those in wild-type mice 48 h post infection (Fig. 6b). These results indicate that the AIM2 inflammasome plays a crucial role in the host defense against Francisella-infection likely by decreasing bacterial burden in tissues, thereby preventing systemic infection. Consistent with the critical role of AIM2 in the production of caspase-1-generated cytokines, the serum concentration of IL-18 was much higher in wild-type mice compared to Aim2−/− mice, 24 h post-infection (Fig. 6b) Taken together, these results indicate that the lack of AIM2 inflammasome activity increases the virulence of Francisella in mice due to defective caspase-1 activation, which is critical for the production of the caspase-1-generated cytokines such as IL-1β and IL-18, and induction of pyroptotic cell death of the infected macrophages.
Figure 6.
The AIM2 inflammasome is critical for innate immunity against Francisella infection. (a), Survival of Aim2+/+ and Aim2−/− mice injected subcutaneously with F. novicida (1.5 × 105 CFU) (Aim2+/+ n = 9, Aim2−/− n=9), and monitored over a period of 3 weeks. 66% of Aim2+/+ survived beyond 3 weeks post-infection. (b), Livers and spleens were harvested 48 h post-infection of mice subcutaneously with F. novicida (1.0 × 105 CFU) homogenized, and dilutions plated on Cystine Heart Agar plates for enumeration of CFU. Bacterial counts from the livers and spleens of the Aim2−/− were significantly higher compared with Aim2+/+ mice (P 0.0054, and 0.0011, respectively). (c) Enzyme-linked immunosorbent assay of IL-18 in serum from Aim2+/+ mice (n = 3) and Aim2−/− mice (n = 3) at 1 d after subcutaneous infection with F. novicida. In b,c, each symbol represents an individual mouse; small horizontal lines indicate the mean. P values, Kaplan-Meier log-rank test (a) or Student’s t-test (b,c). Data are representative of two experiments.
DISCUSSION
Recent biochemical evidence has implicated AIM2 in the recognition of, and innate immune response to cytosolic DNA 3–6. Recognition of cytosolic DNA by AIM2 activates the non-transcriptional inflammatory caspase-1 pathway in macrophages, leading to production of the potent inflammatory cytokines, IL-1β and IL-18, and cell death. We now provide genetic evidence that AIM2 is a critical PRR uniquely involved in the recognition and innate immune defense against infection with the potentially lethal intracellular pathogen Francisella tularensis.
Several proteins of the NLR family are involved in the activation of the inflammatory caspase-1 pathway in macrophages in response to pathogenic infection or products 26–28. For example, NLRC4 (also called Ipaf) is a PRR involved in detection of intracellular infection with Salmonella and activation of the caspase-1 pathway in response to this pathogen 18. Salmonella flagellin is the suggested molecular component responsible for activation of NLRC4 29, 30. Another PRR, Nlrp1, is involved in activation of caspase-1 in response to stimulation with anthrax lethal toxin 15. In contrast, the NLR protein Nlrp3 (also called Nlrp3/cryopyrin) is involved in caspase-1 activation following infection with a broad range of bacterial, viral, or fungal pathogens, and in response to different stress stimuli 28, 31. Nevertheless, the mechanism by which Nlrp3 recognizes such a broad range of pathogens and stimuli is still unclear.
None of the above mentioned NLRs or other known NLRs, however, are involved in sensing Francisella infection. Although macrophages from AIM2-deficient mice are not defective in sensing NLRC4, Nlrp1 and Nlrp3 stimuli, as evidenced by their normal caspase-1 activation response to Salmonella, anthrax lethal toxin, or LPS plus ATP or nigericin, or MSU, they are clearly unable to activate caspase-1 in response to Francisella infection. This suggests that Francisella may have developed mechanisms to evade detection of their non-nucleic acid PAMPs by the host cell inflammasome components such as NLRC4, Nlrp1 or Nlrp3 and probably by other as yet uncharactarized inflammasomes. Nevertheless, several studies suggest that Francisella infection activates TLR2 signaling via MyD88 leading to NF-κB-dependent transcriptional induction of pro-inflammatory cytokines 32–34. However, this response does not appear to be sufficient for activation of the Nlrp3 inflammasome because Aim2−/− macrophages are completely defective in caspase-1 activation in response to infection with Francisella.
Phagosomal disruption occurs during the escape of Francisella from the phagosome into the cytosol 35–37. This may be necessary for activation of caspase-1 and type I interferon production by Francisella because mutant strains that cannot disrupt the phagosome and escape into the cytosol are unable to induce type I interferons or activate caspase-1 10, 38. Despite previous observations of Nlrp3 inflammasome activation by phagosomal disruption 26, 39, 40, it is intriguing that phagosomal disruption during Francisella escape does not lead to Nlrp3 inflammasome activation as evidenced by the absence of caspase-1 activation in Francisella-infected AIM2-deficient macrophages. This defect in caspase-1 activation cannot be attributed to a defect in Nlrp3 signaling because these macrophages have normal amounts of caspase-1 activation in response to specific Nlrp3 stimuli such as LPS plus ATP or the pore forming toxin nigericin. Additionally, this defect is not likely due to a specific defect in Francisella-induced phagosomal disruption since infection of the AIM2-deficient macrophages with Francisella leads to even higher type I interferon production than that seen in wild-type macrophages, indicating normal phagosomal disruption and escape. Altogether, these observations suggest that phagosomal disruption generally does not induce Nlrp3 inflammasome activation and it is likely that a more specific stimulus or additional stimuli are required. Alternatively, Francisella might possess means to inhibit Nlrp3 inflammasome activation during its escape from the phagosome.
AIM2 inflammasome activation requires direct interaction of AIM2 with DNA 3. This interaction leads to oligomerization of AIM2 and the formation of a large AIM2-DNA complex visible by confocal microscopy 3. Our results show that infection of macrophages with Francisella also induces formation of a large AIM2-DNA complex, indicating that Francisella infection delivers DNA into the cytosol that is then recognized by AIM2. The exact mechanism by which Francisella delivers its DNA into the cytosol is currently unclear. Nevertheless, our results suggest that the DNA which activates AIM2, and likely type I interferon signaling, is produced by breakdown and digestion of killed Francisella by phagosomal enzymes. Supporting this conclusion, bafilomycin which can prevent killing and degradation of bacteria in the phagosome by inhibiting the activity of vacuolar-type ATPases that mediate lumen acidification 25, 41, completely inhibited AIM2 inflammasome activation and IFN-β production in wild-type macrophages by Francisella infection. Considering that bafilomycin does not prevent Francisella phagosomal permeabilization and escape into the cytosol 35, phagosomal escape by live Francisella and its replication in the cytosol is likely not the signal that activates AIM2 or type I interferon response. Rather killing and degradation of Francisella and the subsequent phagosomal disruption triggered by live Francisella escape is probably responsible for delivering Francisella DNA into the cytosol for recognition by AIM2 and type I interferon-signaling pathways. As a consequence we expect that more virulent strains of Francisella such as F. tularensis subsp. tularensis, which infects humans must have evolved mechanisms that protect them from phagosomal lysis, perhaps by altering the rate of acidification and maturation of their phagosomes 36. Therefore, these strains will be less efficiently detected by AIM2 due to decreased phagosomal release of killed bacterial DNA into the cytosol, and thereby would have a better chance of intracellular replication and an increased ability to cause a systemic infection.
IFN-β signaling through its IFNAR1 receptor is required for efficient caspase-1 activation by Francisella 10, 26. IFNAR1- or IRF3-deficient macrophages, which exhibit impaired IFN-β production, showed less efficient caspase-1 activation by Francisella compared to wild-type macrophages10, 26. Our data show that the reduced caspase-1 activation by Francisella in Irf3−/− or ifnar1−/− macrophages is not due to decreased AIM2 expression because these macrophages express comparable amounts of AIM2 as wild-type macrophages. Additionally, Irf3−/− and ifnar1−/− 42 macrophages exhibit normal caspase-1 activation in response to transfected DNA, ruling out the possibility that IFN-β signaling is required for transcriptional induction of a host factor critical for caspase-1 activation by the AIM2 inflammasome. Although both Francisella and transfected DNA both activate the AIM2 inflammasome IFN-β signaling might be required for efficient killing of Francisella in the phagosome and/or phagosomal permeablization and release of Francisella DNA into the cytosol. Indeed, ifnar1−/− macrophages harbor more bacteria 5–24h post-infection compared to wild-type macrophages, likely due to defective phagosomal bactericidal activity of the ifnar1−/− macrophages. Consequently, these macrophages might produce less cytosolic DNA, which could provide a rationale for the observed defective AIM2 inflammasome activation in these macrophages.
Based on current information on the life cycle of Francisella in macrophages 10, 26, 43, and our own data, we propose a two step mechanism by which Francisella infection of macrophages activates the AIM2 inflammasome. Shortly after Francisella enters the phagosome, the phagosome is rapidly acidified 17. Acidification causes lysis of some of the ingested bacteria and release of bacterial DNA into the lumen of the phagosome. During phagosomal escape of live Francisella, which occurs as early as 1 h post-infection 16, 17, the phagosome is ruptured releasing both live Francisella and undigested DNA of killed Francisella into the cytosol. This amount of cytosolic DNA might not be sufficient for AIM2 activation, but is likely sufficient for activation of an as yet unknown DNA sensor, which in turn activates IRF3 leading to production of type I interferons such as IFN-β. Bafilomycin treatment completely inhibited Francisella-induced IFN-β production, supporting this scenario. IFN-β binds to IFNAR1 resulting in the activation of this signaling pathway. This initial IFN-β signaling process acts like a positive feedback loop perhaps to increase phagosomal acidification and/or the bactericidal activity of the phagosomal enzymes thereby enhancing the release of more killed Francisella DNA into the cytosol. This is supported by the observations that prior priming of wild-type or Irf3−/− macrophages before infection with Francisella, or infection of these macrophages with Francisella together with IFN-β co-treatment accelerates and enhances AIM2 inflammasome activation, whereas inhibiting phagosomal acidification markedly reduces AIM2 inflammasome activation. In the second step of this process, the increased concentration of cytosolic DNA, as a result of IFN-β-induced phagosomal disruption, leads to full AIM2 activation by inducing its oligomerization. The oligomerized AIM2-DNA complex serves as a molecular platform to recruit ASC and facilitate its oligomerization into the large “ASC pyroptosome” 8. Consistently, Francisella infection or transfection with cytosolic DNA induces ASC pyroptosome formation in wild-type but not Aim2−/− macrophages, and can be completely blocked by ~120–180 mM extracellular KCl. It is likely that the ASC pyroptosome then activates procaspase-1 as described before 8, leading to pyroptotic cell death and production of the pro-inflammatory cytokines IL-1β and IL-18.
The phenotype of Francisella-challenged Aim2−/− provides further support that AIM2 is critical for sensing Francisella infection and activation of the ASC-caspase-1 pathway, not only in isolated macrophages, but also in a whole animal model. Indeed this phenotype is reminiscent of the phenotype seen in Francisella-challenged Casp-1−/− or Asc−/− (Pycard−/−) mice 9. In these mice strains as well as in Aim2−/− mice, Francisella infection is associated with increased lethality due to increased bacterial burden and systemic infection 9. The increased in bacterial burden in these mice is most likely due to decreased cell death of Francisella-infected macrophage, and decreased pro-inflammatory cytokine production. Francisella-induced macrophage cell death shares features of pyroptotic cell death induced by other pathogenic bacteria 9. However, Francisella-induced macrophage cell death is clearly dependent on AIM2, as well as on caspase-1 and ASC 9 providing further proof that AIM2, ASC and caspase-1 function in the same signaling pathway that recognizes Francisella infection.
In conclusion our data provides clear genetic and biochemical evidence that activation of the AIM2 inflammasome represents a crucial innate immune defense against Francisella infection. Future studies with the AIM2-deficient mice should clarify its role in the innate immune response to other intracellular microbial and viral pathogens as well as its involvement in nucleic acid-dependant autoimmune diseases such as systemic lupus erythematosus.
METHODS
Generation of AIM2 deficient mice
AIM2-deficient mice were generated by the gene trap method 13, from an ES cell clone obtained from the International Gene Trap Consortium 12. All mice were used in experiments following protocols approved by Institutional Animal Care and Use Committee, Thomas Jefferson University. See (Supplementary methods) for details.
Antibodies and reagents
The polyclonal anti-AIM2 antibody was raised in rabbits (Invitrogen) against a mixture of recombinant mouse and human AIM2 proteins prepared in Alnemri laboratory. This antibody can detect endogenous mouse and human AIM2 proteins. The polyclonal anti-mouse pyrin was raised in rabbits (Invitrogen) against a truncated recombinant mouse pyrin protein (residues 1–349) prepared in our lab. The anti-IL-1β monoclonal antibody (3ZD) was obtained from the NCI preclinical repository, Biological resource branch. Other antibodies used against mouse Nlrp3 (polyclonal anti-Nlrp3 PYD; our lab), mouse ASC (polyclonal anti-mouse ASC; from Dr. Junji Sagara), and mouse caspase-1 p20 (monoclonal anti-mouse caspase-1 p20; from Dr. Junying Yuan) were described before 8, 20, 31. Antibody to STAT1 phosphorylated at Tyr701 (9171S) and antibody to mouse STAT1 (9172) were from Cell Signaling. ATP, nigericin, Poly (dA:dT) sodium salt, Bafilomycin A, and cytochalasin D were from Sigma-Aldrich. Ultrapure LPS was from Invivogen. Anthrax lethal factor and protective antigen were from List Biological Labs. Disuccinimidyl suberate (DSS) was from Thermo Fisher Scientific. CytoTox96 LDH-release kit was from Promega.
Macrophage cell culture and stimulation
Mouse bone marrow cells were isolated from mouse femurs with sterile Dulbecco modified Eagle medium (DMEM) and cultured in 6-well plates for 5 to 7 days in DMEM (GIBCO) supplemented with 10% L929 cell conditioned medium, 10% fetal bovine serum (FBS), and 1% penicillin-streptomycin. Infection of macrophages with Vaccinia virus (WR strain) were at the indicated MOI for 18 h. In some experiments, differentiated macrophages were treated with or without ultrapure LPS (500 ng/ml) in serum-free OPTI-MEM® I medium for 5 h followed by ATP (5 mM) or nigericin (2.5 µM) as indicated. Anthrax lethal toxin (protective antigen and lethal factor) treatment of macrophages were carried out at 5 µg/ml.
Infection of macrophages with F. novicida
Wildtype F. tularensis subspecies novicida (U112) was obtained from Dr. Denise M. Monack (Stanford University) and grown overnight with shaking in tryptic soy broth (Difco Laboratories) supplemented with 0.2% cysteine as described before 10. To determine the effect of AIM2 deficiency on F. novicida-induced caspase-1 activation, Aim2−/− and Aim2+/+ macrophages were seeded in 6-well plates at a density of 2 × 106 cells/well in OPTI-MEM® I serum free medium and allowed to attach for 2 hours. The cells were infected with different MOIs of F. novicida in 2 ml of OPTI-MEM® I medium for 6 hours as described before 10. The culture supernatants and cells were separated and then processed for immunoblot analysis as described below. In the experiments involving IFNβ priming, cells were pretreated with 250 U/mL of mouse IFN-β 2 h before infection. Cells were then infected with F. novicida. After 1 h, gentamycin (100 µg/mL) were added and incubated for 30 min. Cells were then washed with PBS and were further incubated in Opti-MEM containing a low concentration of gentamycin (2.5 µg/ml) for various periods of time until they were collected.
Immunoblot analysis
Proteins were precipitated from cell culture super¬natants by the addition of an equal volume of methanol and 0.25 volumes of chloroform as described 3, 40. Samples were separated by 12.5 % SDS-PAGE and were transferred onto nitrocellulose membranes. Blots were probed with rat monoclonal-anti-mouse caspase-1 p20 antibody or anti-IL-1β monoclonal antibody (3ZD). Total cell lysates were mixed with SDS sample buffer, fractionated on 12.5 % SDS-PAGE and then immunoblotted with the appropriate antibodies as described above. Each immunoblot in each figure is a representative of at least 3 independent experiments.
Assay of ASC pyroptosome formation in macrophages
These experiments were performed essentially as described recently 44, 45 and further described in (Supplementary methods).
Infection of mice with F. novicida
All mice were kept under specific pathogen-free conditions in filter-top cages at Thomas Jefferson University, and experimental studies were in accordance with the Institutional Animal Care and Use Guidelines. Mice were provided with sterile water and food ad libitum. Pairs of Aim2−/− and Aim2+/+ littermates 8–10 weeks of age were inoculated with the indicated dose of strain U112 subcutaneously in a 0.05-ml vol. Each pair of AIM2+/+ and AIM2−/− used in the experiment were siblings derived from the same parents. The mice were monitored for signs of distress/illness and lethality thrice daily for 3 weeks for the survival study. For determination of bacterial burden in mouse tissues, 6 pairs of Aim2−/− and Aim2+/+ littermates (siblings) 10–12 weeks of age were inoculated with the indicated dose of strain U112 subcutaneously in a 0.05-ml vol. Spleen and liver were harvested 48 h post-infection, homogenized, and dilutions were plated on Cystine Heart Agar plates and incubated at 37° C for 24 h, and CFU were enumerated and expressed as CFU/g tissue.
LDH release assay
Cell culture supernatants and cell pellets from treated macrophages were collected at the end of treatment and assayed for LDH activity with the CytoTox96 LDH-release kit (Promega), as described by the manufacturer’s protocols. Results are representative of at least 3 independent experiments.
Confocal microscopy
In the experiments using the Cy™3-labeled plasmid DNA, Nlrp3−/−-mAIM2-EGFP-N1 or Aim2−/−-mAIM2-EGFP-N1 macrophages were seeded on 35-mm glass-bottomed culture dishes (Mat Tek) and were allowed to attach for 24 h. Next day cells were transfected with Cy™3-labeled plasmid DNA (0.5 µg/dish) for 2–3 hours using Lipofectamine 2000 and then stained with Hoechst 33342 for 30 min. Cells were then observed using a Zeiss LSM 510 Meta confocal microscope at the Kimmel Cancer Center core facility. In the Francisella infection experiments, Nlrp3−/−-mAIM2-EGFP-N1 macrophages seeded on 35 mm cover glass bottom culture dishes were infected with un-stained or Hoechst-stained F. novicida (MOI 200:1) for 6 h. The un-stained F. novicida-infected cells were then stained with Hoechst 33342 stain, whereas the Hoechst-stained F. novicida-infected cells were not stained after infection. Cells were observed using a Zeiss LSM 510 Meta confocal microscope. The GFP (green) was excited with the 488 nm argon laser. The DNA Hoechst 33342 stain (blue) was excited with the 405 nm diode laser. The Cy™3 (red) was excited with the 543 nm He/Ne Laser. The Nlrp3−/−-mAIM2-EGFP-N1 and Aim2−/−-mAIM2-EGFP-N1 stable cell lines were generated by retroviral transduction, followed by sorting using Flow Cytometry at the Kimmel Cancer center core facility, as described before 3.
Supplementary Material
ACKNOWLEDGEMENTS
We thank D.L. Kastner (National Institutes of Health) for pyrin-deficient mice; J. Sagara (Shinshu University) for antibody to mouse ASC; J. Yuan (Harvard University) for antibody to mouse caspase-1; G. Nunez (University of Michigan) for S. typhimurium; D.M. Monack (Stanford University) for wild-type F. novicida; K. Fitzgerald (University of Massachusetts) for immortalized Ifnar1−/− and Irf3−/− mouse bone marrow macrophages; and M. Covarrubias for technical assistance with confocal microscopy. This work was supported by the National Institutes of Health (AG14357 and AR055398 to E.S.A.). L.Solorzano is supported by National Research Service Award T32-CA09678.
Footnotes
Note: Supplementary information is available on the Nature Immunology website.
AUTHOR CONTRIBUTIONS
T.F.-A. conceived this project and supervised the generation of the AIM2-deficient mice and together with J.-W.Y. performed most of the experiments and analyzed and interpreted the data. C. J. and L. H. performed Vaccinia virus experiments. L. E. provided Vaccinia virus and supervised Vaccinia studies and provided valuable advise. L. S. and S. K. performed the in vivo Francisella infection studies. C. L. and E. M. generated the AIM2-gene trapped chimeric mice. J. W., P. D. and M. M. provided technical assistance with genotyping and immunoblotting. E.S.A. directed the entire project and wrote the paper.
COMPETING INTERESTS STATEMENT
The authors declare no competing financial interests
REFERENCES
- 1.Akira S, Uematsu S, Takeuchi O. Pathogen recognition and innate immunity. Cell. 2006;124:783–801. doi: 10.1016/j.cell.2006.02.015. [DOI] [PubMed] [Google Scholar]
- 2.Kumar H, Kawai T, Akira S. Pathogen recognition in the innate immune response. Biochem J. 2009;420:1–16. doi: 10.1042/BJ20090272. [DOI] [PubMed] [Google Scholar]
- 3.Fernandes-Alnemri T, Yu JW, Datta P, Wu J, Alnemri ES. AIM2 activates the inflammasome and cell death in response to cytoplasmic DNA. Nature. 2009;458:509–513. doi: 10.1038/nature07710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hornung V, et al. AIM2 recognizes cytosolic dsDNA and forms a caspase-1-activating inflammasome with ASC. Nature. 2009;458:514–518. doi: 10.1038/nature07725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Roberts TL, et al. HIN-200 proteins regulate caspase activation in response to foreign cytoplasmic DNA. Science. 2009;323:1057–1060. doi: 10.1126/science.1169841. [DOI] [PubMed] [Google Scholar]
- 6.Burckstummer T, et al. An orthogonal proteomic-genomic screen identifies AIM2 as a cytoplasmic DNA sensor for the inflammasome. Nat Immunol. 2009;10:266–272. doi: 10.1038/ni.1702. [DOI] [PubMed] [Google Scholar]
- 7.Srinivasula SM, et al. The PYRIN-CARD protein ASC is an activating adaptor for caspase-1. J Biol Chem. 2002;277:21119–21122. doi: 10.1074/jbc.C200179200. Epub 22002 Apr 21119. [DOI] [PubMed] [Google Scholar]
- 8.Fernandes-Alnemri T, et al. The pyroptosome: a supramolecular assembly of ASC dimers mediating inflammatory cell death via caspase-1 activation. Cell Death Differ. 2007;14:1590–1604. doi: 10.1038/sj.cdd.4402194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mariathasan S, Weiss DS, Dixit VM, Monack DM. Innate immunity against Francisella tularensis is dependent on the ASC/caspase-1 axis. J Exp Med. 2005;202:1043–1049. doi: 10.1084/jem.20050977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Henry T, Brotcke A, Weiss DS, Thompson LJ, Monack DM. Type I interferon signaling is required for activation of the inflammasome during Francisella infection. J Exp Med. 2007;204:987–994. doi: 10.1084/jem.20062665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Henry T, Monack DM. Activation of the inflammasome upon Francisella tularensis infection: interplay of innate immune pathways and virulence factors. Cell Microbiol. 2007;9:2543–2551. doi: 10.1111/j.1462-5822.2007.01022.x. [DOI] [PubMed] [Google Scholar]
- 12.Skarnes WC, et al. A public gene trap resource for mouse functional genomics. Nat Genet. 2004;36:543–544. doi: 10.1038/ng0604-543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Araki M, Araki K, Yamamura K. International Gene Trap Project: towards gene-driven saturation mutagenesis in mice. Curr Pharm Biotechnol. 2009;10:221–229. doi: 10.2174/138920109787315006. [DOI] [PubMed] [Google Scholar]
- 14.Mariathasan S, et al. Cryopyrin activates the inflammasome in response to toxins and ATP. Nature. 2006;440:228–232. doi: 10.1038/nature04515. [DOI] [PubMed] [Google Scholar]
- 15.Boyden ED, Dietrich WF. Nalp1b controls mouse macrophage susceptibility to anthrax lethal toxin. Nat Genet. 2006;38:240–244. doi: 10.1038/ng1724. [DOI] [PubMed] [Google Scholar]
- 16.Checroun C, Wehrly TD, Fischer ER, Hayes SF, Celli J. Autophagy-mediated reentry of Francisella tularensis into the endocytic compartment after cytoplasmic replication. Proc Natl Acad Sci U S A. 2006;103:14578–14583. doi: 10.1073/pnas.0601838103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Santic M, Asare R, Skrobonja I, Jones S, Abu Kwaik Y. Acquisition of the vacuolar ATPase proton pump and phagosome acidification are essential for escape of Francisella tularensis into the macrophage cytosol. Infect Immun. 2008;76:2671–2677. doi: 10.1128/IAI.00185-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mariathasan S, et al. Differential activation of the inflammasome by caspase-1 adaptors ASC and Ipaf. Nature. 2004;430:213–218. doi: 10.1038/nature02664. Epub 2004 Jun 2009. [DOI] [PubMed] [Google Scholar]
- 19.Yu JW, et al. Cryopyrin and pyrin activate caspase-1, but not NF-kappaB, via ASC oligomerization. Cell Death Differ. 2006;13:236–249. doi: 10.1038/sj.cdd.4401734. [DOI] [PubMed] [Google Scholar]
- 20.Yu JW, et al. Pyrin activates the ASC pyroptosome in response to engagement by autoinflammatory PSTPIP1 mutants. Mol. Cell. 2007;28:214–227. doi: 10.1016/j.molcel.2007.08.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gavrilin MA, et al. Pyrin critical to macrophage IL-1beta response to Francisella challenge. J Immunol. 2009;182:7982–7989. doi: 10.4049/jimmunol.0803073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chae JJ, et al. Targeted disruption of pyrin, the FMF protein, causes heightened sensitivity to endotoxin and a defect in macrophage apoptosis. Mol Cell. 2003;11:591–604. doi: 10.1016/s1097-2765(03)00056-x. [DOI] [PubMed] [Google Scholar]
- 23.Chae JJ, et al. The B30.2 domain of pyrin, the familial Mediterranean fever protein, interacts directly with caspase-1 to modulate IL-1beta production. Proc Natl Acad Sci U S A. 2006;103:9982–9987. doi: 10.1073/pnas.0602081103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Papin S, et al. The SPRY domain of Pyrin, mutated in familial Mediterranean fever patients, interacts with inflammasome components and inhibits proIL-1beta processing. Cell Death Differ. 2007;14:1457–1466. doi: 10.1038/sj.cdd.4402142. [DOI] [PubMed] [Google Scholar]
- 25.Frankenberg T, Kirschnek S, Hacker H, Hacker G. Phagocytosis-induced apoptosis of macrophages is linked to uptake, killing and degradation of bacteria. Eur J Immunol. 2008;38:204–215. doi: 10.1002/eji.200737379. [DOI] [PubMed] [Google Scholar]
- 26.Brodsky IE, Monack D. NLR-mediated control of inflammasome assembly in the host response against bacterial pathogens. Semin Immunol. 2009;21:199–207. doi: 10.1016/j.smim.2009.05.007. [DOI] [PubMed] [Google Scholar]
- 27.Ting JP, et al. The NLR gene family: a standard nomenclature. Immunity. 2008;28:285–287. doi: 10.1016/j.immuni.2008.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Martinon F, Mayor A, Tschopp J. The inflammasomes: guardians of the body. Annu Rev Immunol. 2009;27:229–265. doi: 10.1146/annurev.immunol.021908.132715. [DOI] [PubMed] [Google Scholar]
- 29.Miao EA, et al. Cytoplasmic flagellin activates caspase-1 and secretion of interleukin 1beta via Ipaf. Nat Immunol. 2006;7:569–575. doi: 10.1038/ni1344. [DOI] [PubMed] [Google Scholar]
- 30.Franchi L, et al. Cytosolic flagellin requires Ipaf for activation of caspase-1 and interleukin 1beta in salmonella-infected macrophages. Nat Immunol. 2006;7:576–582. doi: 10.1038/ni1346. [DOI] [PubMed] [Google Scholar]
- 31.Bauernfeind FG, et al. Cutting edge: NF-kappaB activating pattern recognition and cytokine receptors license NLRP3 inflammasome activation by regulating NLRP3 expression. J Immunol. 2009;183:787–791. doi: 10.4049/jimmunol.0901363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Abplanalp AL, Morris IR, Parida BK, Teale JM, Berton MT. TLR-dependent control of Francisella tularensis infection and host inflammatory responses. PLoS One. 2009;4:e7920. doi: 10.1371/journal.pone.0007920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Malik M, et al. Toll-like receptor 2 is required for control of pulmonary infection with Francisella tularensis. Infect Immun. 2006;74:3657–3662. doi: 10.1128/IAI.02030-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Katz J, Zhang P, Martin M, Vogel SN, Michalek SM. Toll-like receptor 2 is required for inflammatory responses to Francisella tularensis LVS. Infect Immun. 2006;74:2809–2816. doi: 10.1128/IAI.74.5.2809-2816.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Clemens DL, Lee BY, Horwitz MA. Francisella tularensis phagosomal escape does not require acidification of the phagosome. Infect Immun. 2009;77:1757–1773. doi: 10.1128/IAI.01485-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Clemens DL, Lee BY, Horwitz MA. Virulent and avirulent strains of Francisella tularensis prevent acidification and maturation of their phagosomes and escape into the cytoplasm in human macrophages. Infect Immun. 2004;72:3204–3217. doi: 10.1128/IAI.72.6.3204-3217.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chong A, et al. The early phagosomal stage of Francisella tularensis determines optimal phagosomal escape and Francisella pathogenicity island protein expression. Infect Immun. 2008;76:5488–5499. doi: 10.1128/IAI.00682-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Santic M, Molmeret M, Klose KE, Jones S, Kwaik YA. The Francisella tularensis pathogenicity island protein IglC and its regulator MglA are essential for modulating phagosome biogenesis and subsequent bacterial escape into the cytoplasm. Cell Microbiol. 2005;7:969–979. doi: 10.1111/j.1462-5822.2005.00526.x. [DOI] [PubMed] [Google Scholar]
- 39.Halle A, et al. The NALP3 inflammasome is involved in the innate immune response to amyloid-beta. Nat Immunol. 2008;9:857–865. doi: 10.1038/ni.1636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hornung V, et al. Silica crystals and aluminum salts activate the NALP3 inflammasome through phagosomal destabilization. Nat Immunol. 2008;9:847–856. doi: 10.1038/ni.1631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bidani A, et al. Bactericidal activity of alveolar macrophages is suppressed by V-ATPase inhibition. Lung. 2000;178:91–104. doi: 10.1007/s004080000012. [DOI] [PubMed] [Google Scholar]
- 42.Muruve DA, et al. The inflammasome recognizes cytosolic microbial and host DNA and triggers an innate immune response. Nature. 2008;452:103–107. doi: 10.1038/nature06664. [DOI] [PubMed] [Google Scholar]
- 43.Ray K, Marteyn B, Sansonetti PJ, Tang CM. Life on the inside: the intracellular lifestyle of cytosolic bacteria. Nat Rev Microbiol. 2009;7:333–340. doi: 10.1038/nrmicro2112. [DOI] [PubMed] [Google Scholar]
- 44.Juliana C, et al. The anti-inflammatory compounds parthenolide and Bay 11–7082 are direct inhibitors of the inflammasome. J Biol Chem. doi: 10.1074/jbc.M109.082305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Fernandes-Alnemri T, Alnemri ES. Chapter Thirteen Assembly, Purification, and Assay of the Activity of the ASC Pyroptosome. Methods Enzymol. 2008;442:251–270. doi: 10.1016/S0076-6879(08)01413-4. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.