Abstract
Old World monkeys and, recently, African great apes have been shown, by serology and polymerase chain reaction (PCR), to harbor different γ2-herpesviruses closely related to Kaposi's sarcoma-associated Herpesvirus (KSHV). Although the presence of two distinct lineages of KSHV-like rhadinoviruses, RV1 and RV2, has been revealed in Old World primates (including African green monkeys, macaques, and, recently, mandrills), viruses belonging to the RV2 genogroup have not yet been identified from great apes. Indeed, the three yet known γ2-herpesviruses in chimpanzees (PanRHV1a/PtRV1, PanRHV1b) and gorillas (GorRHV1) belong to the RV1 group. To investigate the putative existence of a new RV2 Rhadinovirus in chimpanzees and gorillas we have used the degenerate consensus primer PCR strategy for the Herpesviral DNA polymerase gene on 40 wild-caught animals. This study led to the discovery, in common chimpanzees, of a novel γ2-herpesvirus belonging to the RV2 genogroup, termed Pan Rhadino-herpesvirus 2 (PanRHV2). Use of specific primers and internal oligonucleotide probes demonstrated the presence of this novel γ2-herpesvirus in three wild-caught animals. Comparison of a 1092-bp fragment of the DNA polymerase obtained from these three animals of the Pan troglodytes troglodytes subspecies, one from Gabon and the two others from Cameroon, revealed <1% of nucleotide divergence. The geographic colocalization as well as the phylogenetic “relationship” of the human and simian γ2-herpesviruses support the model according to which herpesviruses have diversified from a common ancestor in a manner mediating cospeciation of herpesviruses with their host species. By demonstrating the existence of two distinct Rhadinovirus lineages in common chimpanzees, our finding indicates the possible existence of a novel human γ2-herpesvirus belonging to the RV2 genogroup.
[The Herpesviral DNA polymerase sequence data determined herein have been deposited at the GenBank database under accession nos. AF290601, AF346488, AF346489, and AF346490.]
The members of the family Herpesviridae have been grouped into three subfamilies, designated Alphaherpesvirinae, Betaherpesvirinae, and Gammaherpesvirinae (Roizmann et al. 1992). Herpesviruses are widespread in vertebrate species, sharing several moderately to well conserved genes, as determined from amino acid identity comparisons (e.g., DNA polymerase and glycoprotein B). Among Gammaherpesvirinae, Epstein-Barr virus (EBV) and Kaposi's sarcoma-associated Herpesvirus (KSHV), also named Human herpesvirus 8 (HHV8), are the human prototypes of the Lymphocryptovirus genus and the Rhadinovirus genus, respectively. Both of these viruses play a critical role in human multistep carcinogenesis, especially in immunodeficiency patients, leading to Burkitt's lymphoma (Magrath and Judde 1996) and Kaposi's sarcoma (KS) (Chang et al. 1994; Schulz 1998), respectively. Rhadinoviruses, or γ2-herpesviruses, have also been found in several animal species including New World monkeys (Herpesvirus ateles and Herpesvirus saïmiri) (Albrecht and Fleckenstein 1990; Albrecht 2000) and Old World monkeys (macaques, African green monkeys, and recently mandrills) (Desrosiers et al. 1997; Rose et al. 1997; Auerbach et al. 2000; Greensill et al. 2000b; Lacoste et al. 2000c; Strand et al. 2000). Comparison and phylogenetic analyses of available sequences support the existence of two distinct genogroups among the Old World monkey rhadinoviruses, called RV1 and RV2 for Rhadinovirus genogroups 1 and 2 (Bosch et al. 1998; Greensill et al. 2000b; Lacoste et al. 2000c; Schultz et al. 2000). KSHV belongs to the RV1 genogroup, whereas no human virus has yet been discovered in the RV2 group.
Considering that KSHV and Kaposi's sarcoma are highly endemic in Central Africa (Schulz 1998; Gessain et al. 1999) and no γ2-herpesvirus sequence has been described in great apes (Sinkovics and Horvath 1999), the closest primate species to human in the animal kingdom, we decided to investigate the potential presence of KSHV-related viruses in chimpanzees and gorillas from Central Africa. Accordingly, we recently reported the detection and molecular characterization of the DNA polymerase gene fragment of three novel γ2-herpesviruses in these great apes (Lacoste et al. 2000b). These three new and different rhadinoviruses, two present in Pan troglodytes (PanRHV1a and PanRHV1b) and the latest in Gorilla gorilla (GorRHV1), were more closely related to KSHV (70%–85% identity at the nucleotide level) than any other previously described virus of this genus. These three novel γ2-herpesviruses belong to the RV1 genogroup as determined by phylogenetic analyses (Lacoste et al. 2000b). Moreover, an independent confirmation of the presence of PanRHV1a (named PtRV1) in a colony of captive common chimpanzees (Pan troglodytes troglodytes) was published recently (Greensill and Schulz 2000; Greensill et al. 2000a). The goals of the present study were therefore to search for other γ2-herpesviruses belonging especially to the RV2 genogroup in African great apes, chimpanzees, and gorillas, and to study the prevalence and the species specificity of such identified novel herpesviruses in wild-caught great apes from Central Africa.
RESULTS
To look for the presence of KSHV-related viruses in great apes, we first performed a serological analysis followed by a PCR-based study on the peripheral blood mononuclear cells (PBMCs) DNA. The plasma of 40 animals, 28 chimpanzees and 12 gorillas, mostly wild-caught and originating from the Western part of Central Africa, were tested by an immunofluorescence assay (IFA) that detects both latent and lytic KSHV antigens (Chatlynne et al. 1998). The results (Table 1) demonstrated a clear fluorescent reactivity to the KSHV antigens-producing cells (KS-1) in the plasma of 22/28 chimpanzees and 6/12 gorillas, with antibody titers ranging from 1/40 (initial dilution) to 1/640.
Table 1.
Epidemiological Data and Serological STLV-1/SIV and KSHV Results
| Name | Genus | KSHV serology titer | STLV-1/SIV serology | PCR and specific hybridization | Subspecies determination | Accession nos. | ||||
|---|---|---|---|---|---|---|---|---|---|---|
| PanRHV2 | PanRHV1a | PanRHV1b | GorRHV1 | |||||||
| Cameroon | PanCamCar | Pan troglodytes | 1/80 | −/− | − | − | + | − | vellerosus | |
| PanCamChe | Pan troglodytes | 1/320 | −/− | − | − | − | − | |||
| PanCamDja | Pan troglodytes | 1/40 | −/+ | + | + | − | − | troglodytes | AF346488 | |
| PanCamEko | Pan troglodytes | 1/80 | −/− | − | − | + | − | troglodytes | ||
| PanCamEpp | Pan troglodytes | − | −/− | − | − | − | − | |||
| PanCamEtr | Pan troglodytes | − | −/− | + | − | − | − | troglodytes | AF346489 | |
| PanCamEtu | Pan troglodytes | − | −/− | − | − | − | − | |||
| PanCamEwa | Pan troglodytes | 1/80 | −/− | − | + | − | − | vellerosus | ||
| PanCamJac | Pan troglodytes | 1/80 | −/+ | − | − | + | − | vellerosus | ||
| PanCamJab | Pan troglodytes | 1/320 | −/− | − | − | + | − | vellerosus | ||
| PanCamLou | Pan troglodytes | 1/160 | −/− | − | − | − | − | |||
| PanCamMac | Pan troglodytes | 1/160 | −/− | − | + | − | − | troglodytes | ||
| PanCamMay | Pan troglodytes | − | −/− | − | + | − | − | troglodytes | ||
| PanCamMok | Pan troglodytes | 1/160 | −/− | − | − | − | − | |||
| PanCamMuc | Pan troglodytes | 1/160 | −/− | − | + | − | − | vellerosus | ||
| PanCamPem | Pan troglodytes | 1/320 | −/− | − | − | − | − | |||
| PanCamPol | Pan troglodytes | 1/80 | −/− | − | − | − | − | |||
| PanCamSam | Pan troglodytes | 1/640 | −/− | − | − | + | − | troglodytes | ||
| PanCamSek | Pan troglodytes | 1/80 | −/− | − | + | − | − | troglodytes | ||
| PanCamSus | Pan troglodytes | 1/80 | −/− | − | − | − | − | |||
| PanCamTal | Pan troglodytes | − | −/− | − | − | − | − | |||
| PanCamWan | Pan troglodytes | 1/80 | −/− | − | + | − | − | vellerosus | ||
| Gabon | PanGabNte | Pan troglodytes | 1/160 | ind./− | − | − | − | − | ||
| PanGabNto | Pan troglodytes | 1/80 | ind./− | − | − | + | − | troglodytes | ||
| PanGabBel | Pan troglodytes | 1/320 | ind./− | + | − | − | − | troglodytes | AF290601, AF346490 | |
| GorGabOmo | Gorilla gorilla | 1/160 | ind./− | − | − | − | + | gorilla | ||
| GorGabCol | Gorilla gorilla | 1/40 | ind./− | − | − | − | − | gorilla | ||
| Cameroon | GorCamNya | Gorilla gorilla | − | −/− | − | − | − | − | gorilla | |
| GorCamEla | Gorilla gorilla | − | −/− | − | − | − | − | gorilla | ||
| GorCamEvi | Gorilla gorilla | − | −/− | − | − | − | − | gorilla | ||
| France | Gorph682 | Gorilla gorilla | 1/80 | −/− | − | − | − | − | gorilla | |
Epidemiological data include name, genus, and geographical origin. STLV-1 serology was determined by Immunofluorescence Assay (IFA) and Western blot, SIV serology by specific Enzyme-Linked Immunosorbent Assay (ELISA) and Western blot confirmation, and KSHV serology by IFA assay at a dilution of 1/40. Distribution of the different novel herpesviruses, in the 31 chimpanzees and gorillas for which DNA was available, was determined by specific oligonucleotide probe hybridization on the VYGA-GDTD1B nPCR products (and on Pp2is-Ppas nPCR products for PanRHV2). Shaded boxes indicate viruses for which herpesviral DNA polymerase GenBank accession numbers are available.
We then attempted to amplify a fragment of the very conserved Herpesvirus DNA polymerase gene from the PBMCs DNA of 31 out of the 40 great apes, by nested PCR with degenerate primers (Rose et al. 1997). Twenty DNA samples scored positive on the EtBr gel after the nested PCR. By using Southern blot analysis and specific probes for the recently described ape rhadinoviruses (PanRHV1a, PanRHV1b, and GorRHV1; Tables 1 and 2), we observed that 7 of the 31 studied animals were infected by a PanRHV1a, 6 by a PanRHV1b, and 1 by GorRHV1. Cloning and sequencing of two nested PCR products that did not hybridize with such specific viral probes revealed the presence of a novel Gammaherpesviral sequence. The 172-bp sequences (excluding primers) were identical to each other and exhibited 55%, 56%, 54%, and 59% nucleotide identity with the corresponding KSHV, PanRHV1a, PanRHV1b, and GorRHV1 fragments, respectively. Using the same nested PCR approach with, however, a specific reverse primer for the second PCR (Pan2as, designed from this novel sequence), we then amplified a second overlapping fragment of ∼400 bp (DFASA-Pan2as) from 3 of the 31 animals and obtained finally a 476-bp fragment of the DNA polymerase gene for two chimpanzees and a 400-bp fragment for one other (Table 2; Fig. 1). Another heminested PCR, using P2s (a new degenerate forward primer based on conserved amino acid motifs within the DNA polymerase of γ2-herpesviruses), P2eas, and P2ias (new virus-specific reverse primers designed from the DFASA-GDTD1B sequence; Table 2) yielded a further 870 bp of viral sequence. The resulting sequences were assembled to give a total of 1168 bp (excluding primers) of PanRHV2 for two chimpanzees and 1092 bp for one other.
Table 2.
Sequences of Oligonucleotide Primers and Probes Used for Herpesviral DNA Polymerase Consensus and Specific PCRs, Southern Blot Hybridizations and mtDNA PCR Amplification
| Oligonucleotide | Orientation | 5′–3′ sequence |
|---|---|---|
| DNA pol degenerate primers | ||
| DFASAa | + | GTG TTC GAC TTY GCN AGY YTN TAY CC |
| VYGAa | + | ACG TGC AAC GCG GTG TAY GGN KTN ACN GG |
| GDTD1Ba | − | CGG CAT GCG ACA AAC ACG GAG TCN GTR TCN CCR TA |
| P2s | + | GAG TTT CCK TCG GAR TAY GAC ATG |
| PanRHV2 specific primers | ||
| Pan2as | − | TCA CTT AAT GCG GTT GGA TCT AG |
| P2eas | − | CAT TGC GTG TGA CCT TGA TG |
| P2ias | − | TCT GTT TTC GCT TGC TCA AC |
| Pp2es | + | GCA TCT TTA TCA TAC GCT AAC GG |
| Pp2is | + | CGT TTG TTC TCA GTG GAG GAA AG |
| Pp2as | − | ATG GTG CGT CCC TGG AGC |
| Specific probes | ||
| PanRHV2-1 | + | CTG CAT TAC CTG TTG TCC TAA CGC C |
| PanRHV2-2 | + | ATA TGG TTT TAC TGG AGT AGC CAA CG |
| PanRHV1a | + | GTG GGT CTA CTG CGG AAG CCT ATA AAC GTC TC |
| PanRHV1b | + | TCT GTG CTT CTA CAT AGA CCA ATT GAG ACA CA |
| GorRHV1 | + | GCG AGT CTC CTG CAA AAG CCC ATA GAC GTT CCC A |
| mtDNA primers | ||
| MTD1S | + | CAC CAT TAG CAC CCA AAG CT |
| MTD1AS | − | CCT GAA GTA GGA ACC AGA TG |
Degenerate oligonucleotide primers described (Rose et al. 1997).
+, sense; −, antisense.
Positions of degeneracy are given. N = A, C, G and T; Y = C and T; R = A and G; K = G and T.
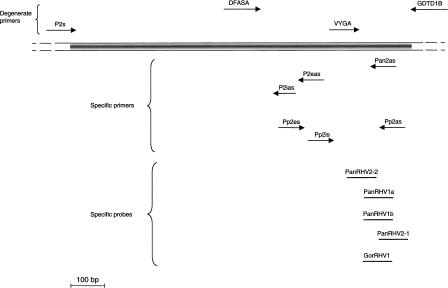
Figure 1.
Relative position and orientation of primers and probes used for consensus and virus-specific DNA polymerase PCR and for Southern blot hybridization. Primers above the Herpesviral DNA polymerase gene (ORF 9) sequence represent the initial herpesvirus degenerate primers used in an nPCR assay to identify novel DNA polymerase sequences as well as the P2s degenerate primer. Primers below the sequence represent specific primers used in a degenerate (DFASA or P2s)–nondegenerate nPCR assay used to amplify the upstream DNA polymerase sequences as well as specific primers used in an nPCR assay (Pp2es-Pp2as followed by Pp2is-Pp2as) to study the PanRHV2 prevalence. Relative positions of the specific oligonucleotide probes used for Southern blot hybridization on VYGA-GDTD1B nPCR products (PanRHV2-1, PanRHV1a, PanRHV1b, and GorRHV1) and also of PanRHV2-2 for hybridization on Pp2is-Pp2as nPCR products are shown. The sequences of the oligonucleotides are given in Table 2.
Database searches using the BLAST Web server demonstrated that these novel sequences were most similar to the DNA polymerases of the γ2-Herpesvirus subfamily. Comprehensive comparative analysis of these novel sequences with all the other available related Herpesviral sequences (Table 3; Figs. 2 and 3) revealed the existence of a novel and distinct chimpanzee KSHV-like viral strain that we propose to name PanRHV2 for Pan Rhadino-herpesvirus 2. This viral strain exhibited 63% identity at the nucleotide level and 73% identity at the amino acid level with KSHV in comparing a 454-bp DNA pol fragment (Table 3). Comparison of the 364 encoded amino acid sequences showed that the two Cameroonese PanRHV2 strains were 99.7% identical to each other, and the Gabonese viral strain compared to the two Cameroonese strains presents 99.2% and 99.5% amino acid identity, respectively.
Table 3.
Percent of Nucleotide and Amino Acid Identities between the Novel γ2-Herpesvirus PanRHV2 and the Other Primate Gammaherpesviruses
| Virus | PanRHV2a | |||
|---|---|---|---|---|
| % Nucleotide and amino acid sequence identity on | ||||
| 454bp | 1168bp | |||
| Nucleotide | Amino acid | Nucleotide | Amino acid | |
| KSHV | 63 | 73 | 66 | 74 |
| PanRHV1a | 62 | 72 | ||
| PtRV1b | 67 | 75 | ||
| PanRHV1b | 64 | 71 | ||
| GorRHV1 | 63 | 70 | ||
| RFHVMn | 62 | 70 | 66 | 73 |
| RFHVMm | 63 | 68 | 67 | 73 |
| MndRHV1 | 64 | 69 | ||
| ChRV1 | 60 | 68 | 66 | 73 |
| ChRV2 | 66 | 76 | ||
| MndRHV2 | 67 | 74 | ||
| MneRV2 | 67 | 76 | 71 | 79 |
| MGVMn | 67 | 76 | ||
| MGVMf | 67 | 75 | ||
| MGVMm | 67 | 74 | ||
| RRV | 67 | 74 | 71 | 78 |
| HVS | 60 | 68 | 64 | 71 |
| HVA3 | 60 | 66 | 64 | 70 |
| EBV | 57 | 59 | 61 | 60 |
PanRHV2 strain accession no. AF346490.
The comparison with the PtRV1 strain was performed only on the common 967-bp fragment.
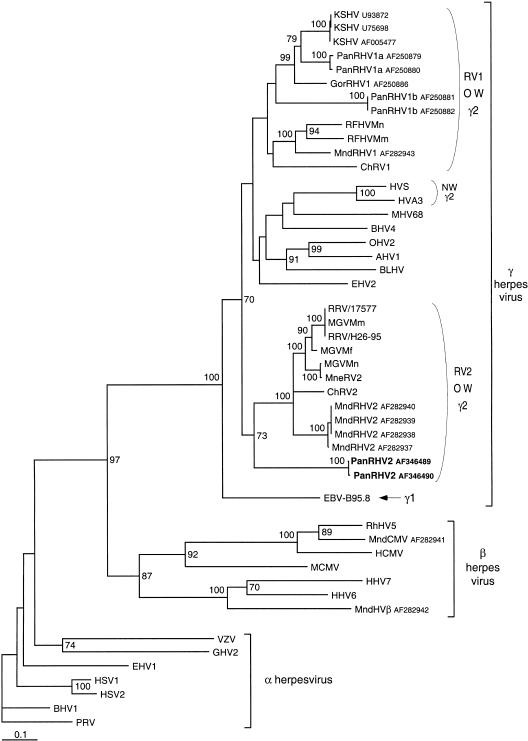
Figure 2.
Phylogenetic tree resulting from analysis of selected 454-bp fragments (primers QAHNA and GDTD1B) (Rose et al. 1997) of the herpesvirus DNA polymerase gene, which is available for all viruses. The phylogeny was derived by the neighbor-joining method applied to pairwise sequence distances calculated using the Kimura two-parameter method (transition/transversion ratio set to 2). Horizontal branch lengths are drawn to scale with the bar indicating 0.1 nucleotide replacements per site. Numbers at each node indicate the percentage of bootstrap samples (out of 100) in which the cluster to the right is supported. Brackets on the right indicate previously defined subfamily and genus Herpesviral classification. Previously published sequences included and their accession numbers are as follows : HHV1/HSV1 (X04771), HHV2/HSV2 (M16321), HHV3/VZV (X04370), HHV4/Epstein-Barr Virus (V01555), HHV5/HCMV (M14709), HHV6A (X83413), HHV7 (U43400), KSHV/HHV8 (U75698, U93872, and AF005477), HVS (M31122), HVA3 (AF083424), PanRHV1a (AF250879 and AF250880), PanRHV1b (AF250881 and AF250882), GorRHV1 (AF250886), MndRHV1 (AF282943), MndRHV2 (AF282937–AF282940), MndHVβ (AF282942), MndCMV (AF282941), ChRV1( AJ251573), ChRV2 (AJ251574), RFHVMn (AF005478), RFHVMm (AF005479), RRV/17577 (AF083501), RRV/H26-95 (AF029302), MneRV2 (AF204167), Macaca γ virus strains Macaca mulatta (AF159033), Macaca fascicularis (AF159032), and Macaca nemestrina (AF159031) (named MGVMm, MGVMf, and MGVMn, respectively), PRV (L24487), BHV1 (Z78205), EHV1 (M86664), GHV2 (L40431), MCMV (U68299), RhHV5 (AF0033184), MHV68 (U97553), BHV4 (AF031811), EHV2 (U20824), BLHV (AF031808), AHV1 (AF005370), and OHV2 (AF031812).
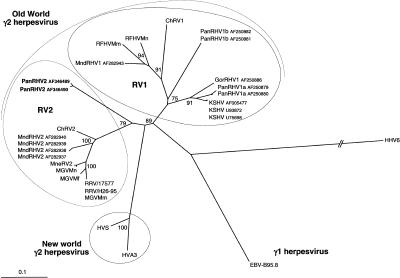
Figure 3.
Neighbor-joining protein distance tree for the 151 amino acid residues encoded by the 454-bp fragment (primers QAHNA and GDTD1B) (Rose et al. 1997) of DNA polymerase. Sequences were aligned by using ClustalW and analyzed by using the PROTDIST and NEIGHBOR programs in PHYLIP. One hundred replica samplings were subjected to bootstrap analysis (SEQBOOT). The branch lengths are proportional to the evolutionary distance (scale bar) between the taxa. Previously published sequences included and their accession numbers are as follows: HHV6A (X83413), HVS (M31122), HVA3 (AF083424), HHV4/Epstein-Barr Virus (V01555), KSHV/HHV8 (U75698, U93872, and AF005477), PanRHV1a (AF250879 and AF250880), PanRHV1b (AF250881 and AF250882), GorRHV1 (AF250886), MndRHV1 (AF282943), MndRHV2 (AF282937–AF282940), ChRV1 (AJ251573), ChRV2 (AJ251574), RFHVMn (AF005478), RFHVMm (AF005479), RRV/17577 (AF083501), RRV/H26-95 (AF029302), MneRV2 (AF204167), Macaca γ virus strains Macaca mulatta (AF159033), Macaca fascicularis (AF159032), and Macaca nemestrina (AF159031) (named MGVMm, MGVMf, and MGVMn, respectively).
Phylogenetic analyses using different methods (Neighbor Joining, DNA Maximum Parsimony) clearly placed this novel chimpanzee virus (PanRHV2) within the Rhadinovirus genus (Fig. 2). Significantly, even though only partial fragments of the DNA polymerases were studied, the phylogenetic analysis presented in Figure 2 is in close agreement with the known clustering of Herpesviruses into α, β, and γ subfamilies. Moreover, phylogenetic analyses performed on the 1168-bp DNA polymerase gene fragment provide an identical tree topology (Table 3; data not shown). PanRHV2 clusters with the macaque (RRV, MneRV2, MGVMn, MGVMm, and MGVMf), the African green monkey (ChRV2), and the recently reported mandrill (MndRHV2) viral strains in the RV2 genogroup.
A second analysis restricted only to all the available primate Rhadinovirus polymerase genes (Fig. 3) demonstrated the existence of three major distinct separate lineages, supported by high bootstrap values among γ2-herpesviruses. The first corresponds to the RV1 genogroup that comprises the human (KSHV), the chimpanzee (PanRHV1a and PanRHV1b), the gorilla (GorRHV1), the mandrill (MndRHV1) as well as the macaque (RFHVMm and RFHVMn), and the African green monkey (ChRV1) strains. This main lineage could be separated into three sublineages, each one being well supported (bootstrap values >75%). Among them, we can distinguish the lineage that contains only the Herpesviral strains from Old World monkeys (macaques, mandrills, and African green monkeys) from the two others constituted by Herpesviral strains of apes (gorilla, pan, and homo). The second main lineage, that is, the RV2 genogroup, comprises Old World monkeys strains (ChRV2, RRV, MneRV2, MGVMn, MGVMm, MGVMf, and MndRHV2) and the only new Herpesviral strain of apes, PanRHV2, present in common chimpanzees. At least, this RV2 lineage is more closely connected to the third lineage of New World monkey (Spider and Squirrel monkeys) rhadinoviruses than is the RV1 genogroup.
To study the prevalence of PanRHV2 infection, we hybridized the products of the heminested PCR (VYGA-GDTD1B) with a specific PanRHV2 oligonucleotide-labeled probe (PanRHV2-1). Three DNAs scored positive. We also developed a heminested PCR system with specific primers for the new PanRHV2 polymerase gene (Table 2). Using these primers in a nested PCR assay followed by hybridization of the PCR products with another internal specific oligonucleotide probe (PanRHV2-2), we detect the same three positive samples among the 31 DNAs.
Significantly, in our series only one case of multiple γ2-herpesviral infection was observed in a SIVcpz-infected chimpanzee among the 31 great apes we tested. This common chimpanzee, described as a natural host of HIV-1-related viruses (Corbet et al. 2000), is therefore also a natural host for distinct γ2-herpesviruses belonging to the two Rhadinovirus genogroups, PanRHV1a and PanRHV2.
To explore whether host-dependent evolution of chimpanzee rhadinoviruses exists, we determined the subspecies identity of the animals from which these novel viruses were derived. Four chimpanzee subspecies with nonoverlapping geographic ranges have been proposed on the basis of genetic differences in mitochondrial DNA sequences (Morin et al. 1994; Gonder et al. 1997; Gagneux et al. 1999). We amplified and sequenced a 498-bp fragment of mitochondrial DNA displacement loop (mtDNA D-loop) for the 15 chimpanzees infected by PanRHV1a, PanRHV1b, or PanRHV2. Comparison of these newly derived mtDNA sequences to representative sequences from the four different chimpanzee subspecies revealed that 9 out of the 15 infected chimpanzees belonged to the Pan troglodytes troglodytes subspecies, and the 6 others belonged to Pan troglodytes vellerosus (Table 1). Classification of the chimpanzees was unambiguous as their mtDNA sequences fell within well-defined subspecies clusters and was further corroborated by the known geographic origins of these animals (Cameroon and Gabon).
DISCUSSION
The novel data presented in this paper as well as recently published data (Greensill et al. 2000a; Lacoste et al. 2000b) indicate that chimpanzees and gorillas are natural hosts for at least four novel and distinct gammaherpesviruses belonging to the two known γ2-herpesvirus lineages. The new γ2-herpesvirus PanRHV2 described in this report corresponds to the first strain of the RV2 genogroup identified in great apes. In contrast, the three other DNA sequences previously detected in chimpanzees and gorillas (PanRHV1a/PtRV1, PanRHV1b, and GorRHV1) belonging to the RV1 genogroup are the closest known homologs to KSHV.
In instances in which particular animal species (macaques, Mandrills, and African green monkeys) have been thoroughly analyzed for the presence of γ2-herpesviruses, at least two distinct viruses, each one belonging to a particular genogroup RV1 or RV2, have been identified (Desrosiers et al. 1997; Rose et al. 1997; Auerbach et al. 2000; Greensill et al. 2000b; Lacoste et al. 2000c; Schultz et al. 2000). To date, therefore, five distinct macaque γ2-herpesviruses have been characterized, two in Macaca mulatta (RFHVMm and RRV/MGVMm), two in Macaca nemestrina (RFHVMn and MneRV2/MGVMn), one in Macaca fascicularis (MGVMf), and two in African green monkeys (ChRV1 and ChRV2) as well as in mandrills (MndRHV1 and MndRHV2). Among these viruses, RFHVMm, RFHVMn, ChRV1, and MndRHV1 belong to the RV1 genogroup, and RRV, MGVMm, MneRV2, MGVMn, MGVMf, ChRV2, and MndRHV2 belong to RV2. Data obtained here from great apes extend the existence of the two phylogenetically distinct groups of γ2-herpesviruses to the chimpanzees and demonstrate the existence of sublineages, within RV1 and RV2, of both Old World monkey and ape rhadinoviruses.
Regarding the comparison of the serological results for KSHV with the PCR detection of the new viruses, it is difficult to provide convincing epidemiological findings. This is because of the limited series of animal tested (40 by serology and 31 by nPCR), but also the fact that there is no serological assay specific for any of these new viruses. However, there is an overall good concordance, as seen in Table 1, between the serological results and the presence of PanRHV1a or PanRHV1b because 12 of 13 PCR positive samples were found in KSHV seropositive animals, but only one PCR positive DNA was detected among the 8 seronegative individuals.
We found that among the PanRHV1a-infected chimpanzees, there were 3 P. t. vellerosus and 4 P. t. troglodytes, and among the PanRHV1b-infected animals, there were 3 P. t. vellerosus and 3 P. t. troglodytes (Table 1). This indicates that for PanRHV1 viruses there is no specific association between a peculiar virus strain (1a or 1b) and a chimpanzee subspecies. Nevertheless, among the PanRHV2-infected chimpanzees, there were only 3 Pan t. troglodytes individuals. These data indicate a possible species specificity for this new virus, but such preliminary findings need to be confirmed on a larger series of animals.
It is worth noting that several of these animals, despite living in close contact in the same enclosure for several months or years, harbor different viruses. For example, PanCamDja and PanCamJac were housed in the same enclosure for 5 yr before being tested and they harbor different gammaherpesviruses. This suggests that viral infection took place before their arrival in the rescue center. Mother-to-offspring transmission is a possibility that has been suggested for KSHV in highly endemic areas of Central and East Africa (Plancoulaine et al. 2000).
Such data, taken as a whole, indicate that Central African great apes constitute an important reservoir of novel γ2-herpesviruses. Although there are no available data supporting this hypothesis, the close identity of these viruses with their human pathogenic counterpart KSHV, their presence in peripheral blood mononuclear cells, and the high genetic relationship between apes and humans indicate that they are potentially transmissible to humans.
Regarding the diseases associated with γ2-herpesviruses, RFHVMm and RFHVMn have been identified in retroperitoneal fibromatosis, a vascular fibroproliferative neoplasm with many morphological and histological similarities to Kaposi's sarcoma (Rose et al. 1997). RRV has also been isolated from simian immunodeficiency-virus-infected macaques with lymphoproliferative disorder reminiscent of multicentric Castleman's disease (Searles et al. 1999). Although the four γ2-herpesviruses found in chimpanzees and gorillas are closely related to their human pathogenic counterpart KSHV, no clinical pathology has yet been identified in association with infection. The question therefore remains as to whether there is any disease associated with these novel herpesviruses in their natural hosts and especially in the case of multiple infection by SIVcpz, PanRHV1a, and PanRHV2 as observed in one wild-caught animal of our series. Followup of both experimentally HIV-infected chimpanzees (Greensill et al. 2000a) and of naturally SIV-infected animals may therefore provide some important clues regarding the physiopathology and natural history of infection by these novel γ2-herpesviruses. Efforts are ongoing to establish a cell culture system for the propagation and extensive characterization of these viruses, which may allow the comparison with KSHV strains.
In the RV2 genogroup, only one viral sequence PanRHV2 has yet been identified among great apes. This sequence branches off alone in the RV2 group, independently of the Old World monkey viral strains, indicating that this sequence may represent the prototype strain of a great ape lineage within this group. These comparative data, obtained for all the nonhuman primate species, raise the possibility of the existence of another γ2-herpesvirus, belonging to the RV2 lineage, in humans, in which only KSHV, belonging to the RV1 genogroup, has been identified to date. The identification of novel RV2 γ2-herpesvirus sequences in other nonhuman primate species and the generation of new consensus degenerate primers targeted to the Herpesviral DNA polymerase may be helpful in the detection and identification of this putative human RV2 herpesvirus. Furthermore, the use of degenerate and consensus primers derived from all the primate γ-herpesvirus polymerase genes, including the four novel ones recently described, will allow us to understand the full extent of the natural infection by these viruses among great apes and the frequency of the eventual zoonotic transmission to humans. This virus hunt could be initially focused on captive or free living great apes and on persons at high risk through contact with such animals, including personnel of zoos and animal centers, as well as hunters and their relatives in Central Africa.
The dogma has always been that herpesviruses have diversified from a common ancestor, in a manner mediating cospeciation of herpesviruses with their host species through latent infection. Indeed, analyses of our phylogenetic results strongly support the notion of host-linked evolution of the γ2-herpesviruses, at least for the RV1 genogroup, because chimpanzees and gorillas, the nonhuman primate species closest to humans, are infected by the γ2-herpesvirus homologs closest to KSHV, the human γ2-herpesvirus prototype.
METHODS
Animals
Blood specimens from 40 great apes including 28 chimpanzees and 12 gorillas were studied. The larger series comprises 27 wild-born animals (23 chimpanzees and 4 gorillas), originating from different parts of Cameroon, where they were originally kept as pets after their mothers had been killed by hunters. They were then gathered in a wildlife rescue center in the South West province of Cameroon, in which some of them were kept in the same enclosure, often in close contact (Corbet et al. 2000). The second group (three chimpanzees and two gorillas) originated from the large animal center of the Centre International de Recherches Médicales, Franceville (CIRMF) in Gabon (Georges-Courbot et al. 1996). The other animals came from two different zoos in France, five gorillas and two chimpanzees from La Palmyre Zoo (kindly provided by T. Petit) and one gorilla from Saint Martin la Plaine Zoo (kindly provided by P. Thivillon). For all the animals from France, except one (Gorph682), only serum was available. All these great apes were seronegative for simian immunodeficiency viruses/human immunodeficiency viruses (SIV/HIV) except two animals from Cameroon (Cam 3 and Cam 4, named in our study PanCamDja and PanCamJac, respectively) from which a SIVcpz has recently been isolated and characterized (Corbet et al. 2000). Great apes from the CIRMF exhibited an HTLV Western blot seroindeterminate profile (Georges-Courbot et al. 1996), but all the 35 other animals were negative for HTLV-1/STLV-1 infection. Data in Table 1 concern only animals for which we had both serum and DNA.
KSHV Serological Analysis
All the plasma were tested, at a 1/40 dilution, for the KSHV-specific IgG, using an immunofluorescence assay (KSHV IFA, ABI). This assay, using the KS-1 cell line as the KSHV source of antigens, detects antibodies directed against both latent and lytic KSHV antigens, and is well adapted to conduct epidemiological works (Chatlynne et al. 1998; Plancoulaine et al. 2000). This assay does not detect any other human herpesviruses than KSHV.
DNA Extraction and Herpesvirus DNA Polymerase Gene Amplification
DNA was extracted from buffy coats with the QIAamp DNA Blood mini kit (QIAGEN) following the manufacturer's instructions.
Herpesvirus DNA polymerase gene sequences were amplified by consensus heminested PCR based on a previously described method (Rose et al. 1997). We slightly modified the reported cycling conditions as : 10 min at 94°C, 5 cycles of 30 sec at 94°C, 1 min at 60°C, 1 min at 72°C; followed by 30 cycles of 30 sec at 94°C, 30 sec at 46°C, 30 sec at 72°C. An extension of 10 min at 72°C was realized on the last cycle (Perkin Elmer GeneAmp PCR system 9600 thermal cycler). The initial round of PCR contained 500 ng of genomic DNA, 30 pmoles of degenerate primers, 2 mM MgCl2, 0.2 mM each dNTP, 5 μL of 10× PCR buffer, and 0.5 μL of Taq Gold DNA polymerase in a volume of 50 μL; 1 μL of this reaction was used in the heminested reaction. After two rounds of heminested PCR (GDTD1B and DFASA primers in the initial round followed by GDTD1B and VYGA), 172-bp fragments (excluding primer sequences) were obtained. Specific reverse primer (Pan2as) was designed from these sequences (Table 2; Fig. 1) and used in heminested PCRs with the primer DFASA to finally obtain a 476-bp fragment of viral DNA polymerase sequence (excluding primers).
Finally, we designed an additional primer (P2s) derived from a conserved amino acid motif within the DNA pol gene of herpesviruses to allow amplification of a longer DNA polymerase fragment (Table 2; Fig. 1). PCR cycling conditions were 10 min at 94°C, 30 cycles of 30 sec at 94°C, 30 sec at 52°C, 1 min at 72°C; followed by a final extension of 10 min at 72°C. After two rounds of heminested PCR (P2s and P2eas in the initial round then P2s and P2ias) a 870-bp fragment of viral sequence was obtained. Primers P2eas and P2ias were derived from the DFASA-GDTD1B sequences previously determined. For all experiments, stringent precautions against PCR contamination were taken. The amplification mixes were made in a special room physically separated from the laboratory, and at least two negative controls (mix, water, or cellular DNA prepared from a KSHV-negative sample) were included.
Specific PanRHV2 DNA Polymerase Gene Amplification
To determine the PanRHV2 prevalence, PanRHV2-specific PCR primers were designed from alignments of PanRHV2 sequences obtained by the degenerate DFASA-GDTD1B PCR procedure. PanRHV2-specific PCR outer primer pair Pp2es and Pp2as generates a 312-bp product, whereas the inner primer set Pp2is and Pp2as (Table 2; Fig. 1) amplifies a 274-bp product. Template volume and reagent concentrations were identical to the DNA pol consensus assay, and PCR conditions were 10 min at 94°C, 30 cycles of 30 sec at 94°C, 30 sec at 53°C, 30 sec at 72°C; and a final extension of 10 min at 72°C. Nested reactions used 2% of primary reaction product as template with the reaction component concentrations and PCR cycling conditions identical to the primary reaction.
Mitochondrial DNA (mtDNA) Amplification
Single-round PCR amplification and sequence analysis, without interim cloning, of chimpanzee mitochondrial (mt) DNA was performed on a 498-bp segment of the mitochondrial D-loop control region (corresponding to position 15998–16497 of the human mitochondrial sequence; Anderson et al. 1981) from PBMC DNA as previously described (Gao et al. 1999; Corbet et al. 2000). The sequences of the primers used for chimpanzee mtDNA amplification are given in Table 2.
Southern Blot Analysis
Nested PCR products (VYGA-GDTD1B or Pp2is-Pp2as) were size-fractionated by 1.5% agarose gel electrophoresis. Following electrophoresis, gels were incubated for 30 min in 0.5 M NaOH–1.5 M NaCl and then for 30 min in 3 M sodium acetate (pH 5.0), after which they were transferred overnight by capillarity onto Biodyne A nylon membranes (Pall Corporation). DNA was cross-linked to the membranes by exposure to UV light in a UV Stratalinker (Stratagene Cloning Systems) and incubated for at least 6 h in hybridization buffer containing 6× Saline Sodium Phosphate EDTA (SSPE), 0.1% SDS, 5× Denhardt's solution, 50% deionized formamide, and 100 μg/mL of fragmented salmon sperm DNA at 42°C (prehybridization). Hybridizations were performed in the same buffer after the addition of the [γ32P]dATP end-labeled internal corresponding probes. The hydridized membranes were washed first for 1 h in 2× SSPE and 0.1% SDS and then for 15 min in 0.2× SSPE and 0.1% SDS at temperatures ranging from 45°C to 65°C, depending on the probe used. Washed membranes were exposed to phosphor screens and analyzed in a Phosphorimager (Molecular Dynamics, Amersham-France SA). The sequences of the probes are given in Table 2.
Cloning, DNA Sequencing, and Phylogenetic Analyses
The TA cloning procedure, DNA sequencing, as well as the phylogenetic tree constructions using the PHYLIP package have been described previously (Lacoste et al. 2000a, 2000c).
Regarding the names of the new primate herpesvirus described in this paper, we have tentatively and provisionally named it PanRHV2 for Pan Rhadino-herpesvirus 2. However, among the specialists in the field, there is discussion and some debate about a new proposal for primate Rhadinovirus nomenclature. When new names are approved by their consensus, we will naturally modify the names of these new herpesviruses.
Acknowledgments
V.L. is a fellow of the CANAM. This work was partly supported by grants from the Agence Nationale de Recherches sur le SIDA (ANRS), SIDACTION, Association de Recherches sur le Cancer (ARC), and Action Concertée from the network of the Institut Pasteur. We acknowledge Peter Jenkins and Liza Gadsby (the Pandrillus Directors) for their great help in obtaining some of the blood samples studied and their continuous interest in this work. We also thank Thierry Petit from la Palmyre Zoo and Pierre Thivillon from the Saint Martin la Plaine Zoo for providing some of the studied samples.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL agessain@pasteur.fr; FAX 33 0 140-61-34-65.
Article and publication are at http://www.genome.org/cgi/doi/10.1101/gr.158601.
REFERENCES
- Albrecht JC. Primary structure of the Herpesvirus ateles genome. J Virol. 2000;74:1033–1037. doi: 10.1128/jvi.74.2.1033-1037.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albrecht JC, Fleckenstein B. Structural organization of the conserved gene block of Herpesvirus saimiri coding for DNA polymerase, glycoprotein B, and major DNA binding protein. Virology. 1990;174:533–542. doi: 10.1016/0042-6822(90)90107-3. [DOI] [PubMed] [Google Scholar]
- Anderson S, Bankier AT, Barrell BG, de Bruijn MH, Coulson AR, Drouin J, Eperon IC, Nierlich DP, Roe BA, Sanger F, et al. Sequence and organization of the human mitochondrial genome. Nature. 1981;290:457–465. doi: 10.1038/290457a0. [DOI] [PubMed] [Google Scholar]
- Auerbach MR, Czajak SC, Johnson WE, Desrosiers RC, Alexander L. Species specificity of macaque Rhadinovirus glycoprotein B sequences. J Virol. 2000;74:584–590. doi: 10.1128/jvi.74.1.584-590.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bosch ML, Strand KB, Rose TM. Gammaherpesvirus sequence comparisons. J Virol. 1998;72:8458–8459. doi: 10.1128/jvi.72.10.8458-8459.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang Y, Cesarman E, Pessin MS, Lee F, Culpepper J, Knowles DM, Moore PS. Identification of herpesvirus-like DNA sequences in AIDS-associated Kaposi's sarcoma. Science. 1994;266:1865–1869. doi: 10.1126/science.7997879. [DOI] [PubMed] [Google Scholar]
- Chatlynne LG, Lapps W, Handy M, Huang YQ, Masood R, Hamilton AS, Said JW, Koeffler HP, Kaplan MH, Friedman-Kien A, et al. Detection and titration of human herpesvirus-8-specific antibodies in sera from blood donors, acquired immunodeficiency syndrome patients, and Kaposi's sarcoma patients using a whole virus enzyme-linked immunosorbent assay. Blood. 1998;92:53–58. [PubMed] [Google Scholar]
- Corbet S, Muller-Trutwin MC, Versmisse P, Delarue S, Ayouba A, Lewis J, Brunak S, Martin P, Brun-Vezinet F, Simon F, et al. env sequences of simian immunodeficiency viruses from chimpanzees in Cameroon are strongly related to those of human immunodeficiency virus group N from the same geographic area. J Virol. 2000;74:529–534. doi: 10.1128/jvi.74.1.529-534.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desrosiers RC, Sasseville VG, Czajak SC, Zhang X, Mansfield KG, Kaur A, Johnson RP, Lackner AA, Jung JU. A herpesvirus of rhesus monkeys related to the human Kaposi's sarcoma-associated herpesvirus. J Virol. 1997;71:9764–9769. doi: 10.1128/jvi.71.12.9764-9769.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gagneux P, Wills C, Gerloff U, Tautz D, Morin PA, Boesch C, Fruth B, Hohmann G, Ryder OA, Woodruff DS. Mitochondrial sequences show diverse evolutionary histories of African hominoids. Proc Natl Acad Sci USA. 1999;96:5077–5082. doi: 10.1073/pnas.96.9.5077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao F, Bailes E, Robertson DL, Chen Y, Rodenburg CM, Michael SF, Cummins LB, Arthur LO, Peeters M, Shaw GM, et al. Origin of HIV-1 in the chimpanzee Pan troglodytes troglodytes. Nature. 1999;397:436–441. doi: 10.1038/17130. [DOI] [PubMed] [Google Scholar]
- Georges-Courbot MC, Moisson P, Leroy E, Pingard AM, Nerrienet E, Dubreuil G, Wickings EJ, Debels F, Bedjabaga I, Poaty-Mavoungou V, et al. Occurrence and frequency of transmission of naturally occurring simian retroviral infections (SIV, STLV, and SRV) at the CIRMF Primate Center, Gabon. J Med Primatol. 1996;25:313–326. doi: 10.1111/j.1600-0684.1996.tb00023.x. [DOI] [PubMed] [Google Scholar]
- Gessain A, Mauclere P, van Beveren M, Plancoulaine S, Ayouba A, Essame-Oyono JL, Martin PM, de The G. Human herpesvirus 8 primary infection occurs during childhood in Cameroon, Central Africa. Int J Cancer. 1999;81:189–192. doi: 10.1002/(sici)1097-0215(19990412)81:2<189::aid-ijc4>3.0.co;2-e. [DOI] [PubMed] [Google Scholar]
- Gonder MK, Oates JF, Disotell TR, Forstner MR, Morales JC, Melnick DJ. A new West African chimpanzee subspecies? Nature. 1997;388:337. doi: 10.1038/41005. [DOI] [PubMed] [Google Scholar]
- Greensill J, Schulz TF. Rhadinoviruses (gamma2-herpesviruses) of Old World primates: Models for KSHV/HHV8-associated disease? AIDS. 2000;14:S11–19. [PubMed] [Google Scholar]
- Greensill J, Sheldon JA, Murthy KK, Bessonette JS, Beer BE, Schulz TF. A chimpanzee Rhadinovirus sequence related to Kaposi's sarcoma-associated herpesvirus/human herpesvirus 8: Increased detection after HIV-1 infection in the absence of disease. AIDS. 2000a;14:F129–135. doi: 10.1097/00002030-200012010-00001. [DOI] [PubMed] [Google Scholar]
- Greensill J, Sheldon JA, Renwick NM, Beer BE, Norley S, Goudsmit J, Schulz TF. Two distinct gamma-2 herpesviruses in African green monkeys: A second gamma-2 herpesvirus lineage among Old World primates? J Virol. 2000b;74:1572–1577. doi: 10.1128/jvi.74.3.1572-1577.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lacoste V, Judde JG, Briere J, Tulliez M, Garin B, Kassa-Kelembho E, Morvan J, Couppie P, Clyti E, Forteza Vila J, et al. Molecular epidemiology of human herpesvirus 8 in Africa: Both B and A5 K1 genotypes, as well as the M and P genotypes of K14.1/K15 loci, are frequent and widespread. Virology. 2000a;278:60–74. doi: 10.1006/viro.2000.0629. [DOI] [PubMed] [Google Scholar]
- Lacoste V, Mauclere P, Dubreuil G, Lewis J, Georges-Courbot MC, Gessain A. KSHV-like herpesviruses in chimps and gorillas. Nature. 2000b;407:151–152. doi: 10.1038/35025145. [DOI] [PubMed] [Google Scholar]
- Lacoste V, Mauclere P, Dubreuil G, Lewis J, Georges-Courbot MC, Rigoulet J, Petit T, Gessain A. Simian homologues of human gamma-2 and betaherpesviruses in mandrill and drill monkeys. J Virol. 2000c;74:11993–11999. doi: 10.1128/jvi.74.24.11993-11999.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magrath I, Judde J. Epstein-Barr virus and neoplasia. In: Bertino M, editor. Molecular biology of cancer. New York: Academic Press; 1996. [Google Scholar]
- Morin PA, Moore JJ, Chakraborty R, Jin L, Goodall J, Woodruff DS. Kin selection, social structure, gene flow, and the evolution of chimpanzees. Science. 1994;265:1193–1201. doi: 10.1126/science.7915048. [DOI] [PubMed] [Google Scholar]
- Plancoulaine S, Abel L, van Beveren M, Tregouet DA, Joubert M, Tortevoye P, de The G, Gessain A. Human herpesvirus 8 transmission from mother to child and between siblings in an endemic population. Lancet. 2000;356:1062–1065. doi: 10.1016/S0140-6736(00)02729-X. [DOI] [PubMed] [Google Scholar]
- Roizmann B, Desrosiers RC, Fleckenstein B, Lopez C, Minson AC, Studdert MJ. The family Herpesviridae: An update. The Herpesvirus Study Group of the International Committee on Taxonomy of Viruses. Arch Virol. 1992;123:425–449. doi: 10.1007/BF01317276. [DOI] [PubMed] [Google Scholar]
- Rose TM, Strand KB, Schultz ER, Schaefer G, Rankin GW, Jr, Thouless ME, Tsai CC, Bosch ML. Identification of two homologs of the Kaposi's sarcoma-associated herpesvirus (human herpesvirus 8) in retroperitoneal fibromatosis of different macaque species. J Virol. 1997;71:4138–4144. doi: 10.1128/jvi.71.5.4138-4144.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulz TF. Kaposi's sarcoma-associated herpesvirus (human herpesvirus-8) J Gen Virol. 1998;79:1573–1591. doi: 10.1099/0022-1317-79-7-1573. [DOI] [PubMed] [Google Scholar]
- Schultz ER, Rankin GW, Jr, Blanc MP, Raden BW, Tsai CC, Rose TM. Characterization of two divergent lineages of macaque rhadinoviruses related to Kaposi's sarcoma-associated herpesvirus. J Virol. 2000;74:4919–4928. doi: 10.1128/jvi.74.10.4919-4928.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Searles RP, Bergquam EP, Axthelm MK, Wong SW. Sequence and genomic analysis of a rhesus macaque Rhadinovirus with similarity to Kaposi's sarcoma-associated herpesvirus/human herpesvirus 8. J Virol. 1999;73:3040–3053. doi: 10.1128/jvi.73.4.3040-3053.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinkovics JG, Horvath JC. Kaposi's sarcoma: Breeding ground of herpesviridae: A tour de force over viral evolution. Int J Oncol. 1999;14:615–646. doi: 10.3892/ijo.14.4.615. [DOI] [PubMed] [Google Scholar]
- Strand K, Harper E, Thormahlen S, Thouless ME, Tsai C, Rose T, Bosch ML. Two distinct lineages of macaque gamma herpesviruses related to the Kaposi's sarcoma associated herpesvirus. J Clin Virol. 2000;16:253–269. doi: 10.1016/s1386-6532(99)00080-3. [DOI] [PubMed] [Google Scholar]