Abstract
We have used ceramic capillary tips generally used in the microelectronics industry for the production of DNA microarrays. The ceramic tips improve the morphology of microarray elements, allow higher element density, and increase printing tip life over the customary slotted stainless-steel pins. Ceramic tips are less expensive than steel pins and allow printing from 1536-well sample source plates. In this work, we describe experiments that establish printing performance of the ceramic tips and hybridization experiments that show that DNA hybridization is unaffected by the choice of tip material.
Over the past several years, the applications of DNA microarray technology have diversified from the initial gene expression studies in yeast (Schena et al. 1995; DeRisi et al. 1997) to include the characterization of human cancer tumor tissues (Perou et al. 2000), high-throughput SNP genotyping (Hacia et al. 1999), and the study of in vivo binding of transcription factor proteins (Ren et al. 2000). Two very different technical approaches to microarray production have enabled this rapid expansion, synthesis of short oligonucleotide probes directly on glass surfaces (in situ synthesis) and the deposition of probes (either PCR products or oligonucleotides) onto glass surfaces using either metal pins or modified ink jet systems (spotted arrays). A low-cost robotic system developed at Stanford University for producing spotted arrays has found application in many laboratories (Schena et al. 1995; Bowtell et al. 1999). This system uses stainless-steel printing tips, similar in design to a fountain pen, to aspirate and deposit DNA onto glass microscope slides. Through repeated cycles of sample aspiration, deposition onto a series of slides and cleaning of the printing tips, high-density microarrays are produced.
We have developed a microarray production method that uses hollow cylindrical ceramic capillary tips (ceramic tips) instead of stainless-steel tips. The ceramic tips are adapted from microelectronics applications, where they are typically used for bonding high-density interconnections (wires <.001‘ in diameter) during the assembly of integrated circuit packages. The ceramic tips are manufactured to very tight tolerances and are available commercially. Our data show that the ceramic tips improve the consistency of deposit morphology, resist deformation over long-term use, cost less, and offer the potential for significant improvement in deposit density.
RESULTS
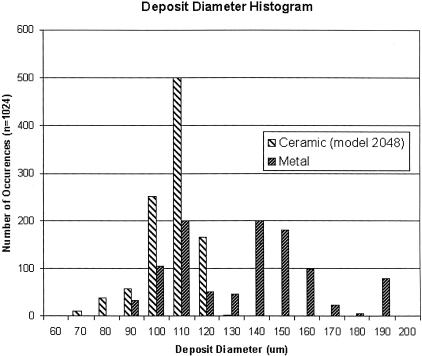
To evaluate the morphology of deposits spotted with ceramic tips, a solution of DNA amplified by PCR with nucleotides labeled with Cy3 fluorescent dye was printed onto standard microscope slides coated with poly-L-lysine (Schena et al. 1995). After spotting, the diameters of the deposits were measured by use of a scanning fluorescence imager. Figure 1 shows a histogram comparing the distribution of diameters of spots deposited by use of stainless-steel and ceramic tips (n = 1024 for each tip type). The ceramic tips yield more reproducible results, with a SD in spot diameter between tips of 10 microns (data not shown).
Figure 1.
Deposit diameter distributions for ceramic and stainless-steel printing pins. Microarrays were printed using either stainless-steel or ceramic tips. The histogram of the sizes of elements produced by each type of tip is shown.
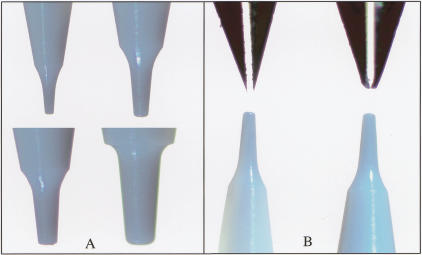
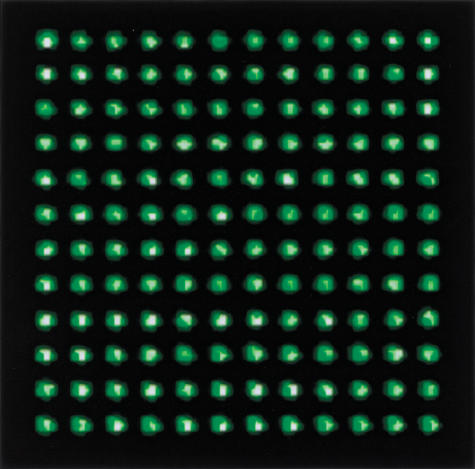
Similar experiments were conducted to assess the range of deposit diameters that can be applied by use of various sizes of ceramic tips. Figure 2A shows a selection of ceramic tip designs, with tip diameters ranging from 50 to 178 microns. Table 1 shows the spot diameters that were applied using various ceramic tip sizes, ranging from ∼60 to 130 microns in diameter. Spot size is proportional to the outer diameter of the ceramic tip, suggesting that further reductions in element size can be obtained by the production of smaller ceramic tips. To evaluate the potential for increased spot density using ceramic tips, the smallest available standard ceramic tip (with a tip dimension of 50 μm) was used to print labeled DNA at 100 micron spacing (Fig. 3), yielding a twofold higher density than we have been able to achieve with stainless-steel tips.
Figure 2.
Images of tip. (A) Ceramic tips with cross sectional diameters of 50, 83, 109, and 132 microns, all of which have been successfully used to fabricate microarrays. (B) Stainless-steel slotted tips and ceramic tips before (left) and after (right) printing ∼100,000 microarray elements. The stainless-steel tips are considerably deformed, but the ceramic tips are intact.
Table 1.
Tip Sizes and Spot Diameters
| Ceramic tip diameter (microns) | Mean spot diameter (microns) |
|---|---|
| 50 | 56 |
| 83 | 99 |
| 109 | 108 |
| 132 | 130 |
Figure 3.
Fluorescent image of labeled DNA deposited at 100 micron spacing.
The ceramic tips also exhibit superior resistance to deformation after repeated use. Figure 2B illustrates deformation of stainless-steel tips as a result of array printing (∼100,000 spots deposited), in contrast to ceramic tips, which show no visible wear. The stainless-steel tips tend to deform over time to an extent that is dependent upon the contact force experienced during microarray printing. This deformation leads to variation in spot diameter and morphology over time and from tip to tip. The ceramic tips do not deform, and therefore provide increased consistency in spot diameter and morphology. The tips used in this study have printed over 225,000 deposits each, and none of the 16 tips have broken due to the stresses and impact experienced during printing. The only tip breakage that has occurred has been due to operator error.
The ceramic tips are capable of printing >1000 deposits from a single aspiration (data not shown). The MicroQuill 2000 stainless-steel tips have the capability of printing >400 deposits per aspiration (http://www.majerprecision.com).
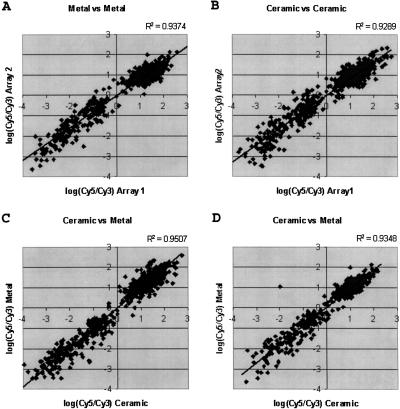
To demonstrate that probes deposited with ceramic tips hybridize similarly to those deposited with standard metal tips, hybridization experiments were conducted. A series of 1-kb PCR products, covering 2.8 Mb of the Adh region of Drosophila melanogaster, (Ashburner et al. 1999) were used to manufacture microarrays. Genomic DNA spiked with known amounts of DNA from BACs known to be included in this region was used as a probe. Probe DNA was amplified with random hexamers and Cy3-labeled nucleotides. Purified Drosophila genomic DNA was labeled with Cy5-labeled nucleotides and served as the reference sample. After hybridization, the microarrays were scanned in a fluorescent scanning imager. Deposits corresponding to the regions covered by the spiked BAC clones were higher in signal in the Cy3 channel (and thus a negative ratio), correctly indicating the increased abundance of DNA from these regions in the test sample. Figure 4, A and B show the correlation between two independent hybridizations using either metal or ceramic tips, whereas C and D show the correlation between equivalent spots on a microarray printed with both ceramic and metal pins. The strong correlation shows that the tips perform equivalently. We have obtained similar results using cDNA arrays to measure gene expression (data not shown).
Figure 4.
Comparison of log ratio of fluorescent hybridization signal between microarray features printed with metal and ceramic printing tips. Microarrays were manufactured (see text) using both steel and ceramic tips and hybridized with genomic DNA spiked with known BACs. The ratio for each element is plotted for two arrays printed with metal tips (A) Ceramic tips, (B), and one of each (C,D). The strong correlation in each of the plots shows that no significant difference exists between the two types of tips.
The time required to print microarrays is inversely proportional to the number of tips that can be used simultaneously in a given area. Traditionally we (and others) have manufactured microarrays using 16, 32, or 48 tips, with a tip density of ∼5 tips/cm2 (printed from 384-well microtiter plates). We have successfully printed microarrays using DNA samples (100 ng/μL sheared herring sperm DNA) arrayed in 1536-well source plates, potentially allowing a tip density of 20 tips/cm2. Hybridization experiments at this density have not yet been performed.
DISCUSSION
The two-fluor hybridization approach typically used in microarray experiments minimizes the effects of spot variations on experimental results; however, highly repeatable printing is preferable. Consistent, repeatable element size and morphology allow a more straightforward comparison of experimental results and improve the ease and accuracy of image analysis. The ceramic tips presented in this study provide more consistent deposit morphology and diameter, due in part to their resistance to deformation over time. The extent of deformation of stainless-steel tips is a function of printing parameters, including spring force, vertical printing speed, and coplanarity of the print head to the microarray substrate. As a result, each stainless-steel tip deforms differently, yielding variation in the contact area of the tip to the microarray substrate.
Ceramic tips are available in a wide range of designs and, as a result, offer significantly more flexibility than stainless-steel tips. The mean diameter of printed spots can be controlled by selecting an appropriately sized ceramic tip. This is an obvious advantage in terms of increasing the density of spots in microarray experiments by applying the smallest possible deposits. Initial experiments have shown the capability of printing deposits at 100-μm spacing, allowing at least a twofold increase in spot density. Our results suggest that array element spacing could be reduced to 75μm or less (yielding nearly a fourfold increase in density over arrays printed at 135-μm spacing). However, it may be advantageous to select tips that apply larger deposits in some applications, for example, in those in which the quantity of immobilized probe molecules is limiting.
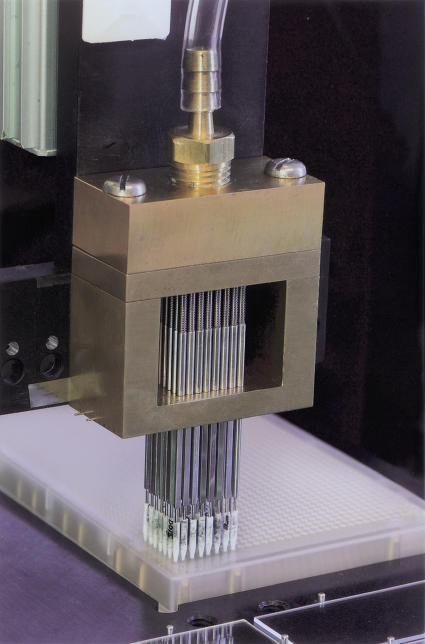
Implementation of ceramic tips requires custom tooling, consisting of a modified tip holder, printing pin assembly, and air reservoir. We have found that the ceramic tips are more sensitive to inaccuracies in leveling of the printing tip holder to the arrayer slide platter. As a result, the tip holder must be carefully aligned to prevent an increase in deposition failure. Another issue discovered early in the implementation was that ceramic tips are more difficult to dry after cleaning, and if they are not completely dry, the next DNA samples do not aspirate completely. We found that it was possible to dry the tips completely by adding a forced-air drying reservoir. Additionally, recent experiments have shown that cleaning the tips in warm water aids in preventing tip clogs. Figure 5 shows a picture of the tip assembly.
Figure 5.
The fully assembled print head. Ceramic tips (white) are mounted on hollow shafts held in place by a brass print head. The print head is covered by a pressurized manifold that forces air through the shaft and the ceramic tip for the purpose of tip drying.
Finally, the ability to print samples easily from 1536-well source plates will significantly reduce the amount of labor required for microarray production. Ceramic tips can aspirate liquids without being completely submerged. This, along with the tapered design of the tips, provides for easier alignment, as the tapered tip is much smaller in diameter than the source plate well. In the case of the stainless-steel tips, the tapered portion is significantly shorter than that of the ceramic tips. This limits the tolerance for misalignment of the pin to the source plate well, because the diameter of the pin at the top of the taper is much closer in size to the diameter of the source plate well.
METHODS
Ceramic Tip Availability and Tip Holder Assembly
Ceramic tips were purchased from K & S MicroSwiss. They are significantly lower in cost than commercial stainless-steel tips.
Stainless-Steel Pins
MicroQuill 2000 stainless-steel pins were used in this study (Majer Precision). They are manufactured from a 17–4 stainless steel rod, have an outer diameter of 1/16″, and a 40° sharp conical tip. A 38-mm wide slot is cut in the center of the tip using electrodischarge machining (http://www.majerprecision.com).
Fluorescent-Labeled PCR Products for Deposit Morphology and Diameter Tests
Labeled PCR products were produced by use of a standard protocol (http://genome-www.stanford.EDU/pbrown/) with template DNA from a BAC clone and a pair of unique primers. The labeled PCR product was then precipitated with a standard ethanol precipitation protocol and resuspended in 30 μL of deionized water. Prior to spotting onto microarray substrates, the products were diluted to 100 ng/μL in 3X SSC.
The data for Figure 1 was collected in two separate print runs. Each run was printed with 16 tips (stainless for first run, ceramic for second run), and each run had a total deposit count of 1024. The ceramic tips used had a tip diameter of 83 μm. Spot diameters were measured with a GenePix 4000A microarray scanner (Axon Instruments), which has a pixel resolution of 10 μm.
Microarray Hybridization Experiments
Microarrays representing the Adh region of the D. melanogaster genome were manufactured. The elements of the array correspond to nonoverlapping PCR fragments (made by standard protocols using BAC DNA as template) of ∼1 kb spanning the region. Fluorescent-labeled probes for hybridization were produced by Klenow labeling (Pollack et al. 1999) of either genomic DNA, or genomic DNA with three individual BACs added at different ratios. All samples were sheared by sonication to ∼300–800 bp. The BACs added were BACR18J08, BACR48E02, and BACR16P12.
Protocols for poly-L-lysine coating, probe preparation and arraying, array post-processing, DNA labeling, hybridization, washing, and scanning have been described elsewhere; detailed protocols are available at http://genome-www.stanford.EDU/pbrown/ and in Eisen et al. (1999).
Acknowledgments
This work was supported in part by a grant from the National Institutes of Health (Grant no. HG00750-09). P.T.S. is a National Science Foundation Biocomputing Postdoctoral Fellow. We thank Michael Holmes and Pavel Tomancek for performing experiments using arrays generated in this research, Catherine Nelson for editing assistance, Louis Curtis, Larry Peck, and Tony Freeman from the LBNL machine shop, and Gerry Rubin for encouragement and support.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL rgeorge@bdgp.lbl.gov; FAX (510) 486-6798
Article and publication are at http://www.genome.org/cgi/doi/10.1101/gr.190801.
REFERENCES
- Ashburner M, Misra S, Roote J, Lewis SE, Blazej R, Davis T, Doyle C, Galle R, George R, Harris N, et al. An exploration of the sequence of a 2.9-Mb region of the genome of Drosophila melanogaster: the Adh region. Genetics. 1999;153:179–219. doi: 10.1093/genetics/153.1.179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowtell DDL. Options available from start to finish for obtaining expression data by microarray. Nat Genet. 1999;21:25–32. doi: 10.1038/4455. [DOI] [PubMed] [Google Scholar]
- DeRisi JL, Iyer VR, Brown PO. Exploring the metabolic and genetic control of gene expression on a genomic scale. Science. 1997;278:680–686. doi: 10.1126/science.278.5338.680. [DOI] [PubMed] [Google Scholar]
- Eisen MB, Brown PO. DNA arrays for analysis of gene expression. Methods Enzymol. 1999;303:179–205. doi: 10.1016/s0076-6879(99)03014-1. [DOI] [PubMed] [Google Scholar]
- Hacia JG, Fan JB, Ryder O, Jin L, Edgemon K, Ghandour G, Mayer RA, Sun B, Hsie L, Robbins CM, et al. Determination of ancestral alleles for human single-nucleotide polymorphisms using high-density oligonucleotide arrays. Nat Genet. 1999;22:164–167. doi: 10.1038/9674. [DOI] [PubMed] [Google Scholar]
- Perou CM, Sorlie T, Eisen MB, van de Rijn M, Jeffrey SS, Rees CA, Pollack JR, Ross DT, Johnsen H, Akslen LA, et al. Molecular portraits of human breast tumours. Nature. 2000;406:747–752. doi: 10.1038/35021093. [DOI] [PubMed] [Google Scholar]
- Pollack JR, Perou CM, Alizadeh AA, Eisen MB, Pergamenschikov A, Williams CF, Jeffrey SS, Botstein D, Brown PO. Genome-wide analysis of DNA copy-number changes using cDNA microarrays. Nat Genet. 1999;23:41–46. doi: 10.1038/12640. [DOI] [PubMed] [Google Scholar]
- Ren B, Robert F, Wyrick JJ, Aparicio O, Jennings EG, Simon I, Zeitlinger J, Schreiber J, Hannett N, Kanin E, et al. Genome-wide location and function of DNA binding proteins. Science. 2000;290:2306–2309. doi: 10.1126/science.290.5500.2306. [DOI] [PubMed] [Google Scholar]
- Schena M, Shalon D, Davis RW, Brown PO. Quantitative monitoring of gene expression patterns with a complementary DNA microarray. Science. 1995;270:467–470. doi: 10.1126/science.270.5235.467. [DOI] [PubMed] [Google Scholar]