Abstract
Recent studies suggest that innate immune responses by natural killer (NK) cells play a significant role in restricting human immunodeficiency virus type-1 (HIV-1) pathogenesis. Our aim was to characterize changes in NK cells associated with HIV-1 clade C disease progression. Here we used multiparametric flow cytometry (LSRII) to quantify phenotype and function of NK cells in a cross-sectional analysis of cryopreserved blood samples from a cohort of 41 chronically HIV-1-infected, treatment-naive adult South Africans. These individuals ranged in disease severity from early (CD4 count >500) to advanced HIV-1 disease (CD4 count <50). We found that the frequency of NK cells expressing KIR2DL1, an inhibitory receptor, and/or KIR2DS1, an activating receptor, tended to decrease with increasing HIV-1 viral load. We also discovered a significant increase (p < 0.05) in overall NK cell degranulation with disease progression. We found that acutely activated NK cells (CD69pos) were deficient in NKp46 expression ex vivo. In conclusion, we observed that with viremia and advanced HIV-1 disease, activated NK cells lack NKp46 expression, and KIR2DS1pos and/ or KIR2DL1pos NK cells are reduced in frequency. These findings suggest that modulation of receptor expression on NK cells may play a role in HIV-1 pathogenesis, and provide new insights on immunological changes in advanced HIV-1 disease.
Introduction
Increasing evidence suggests that innate immune responses, and in particular natural killer (NK) cells, can contribute to the control of human immunodeficiency virus type-1 (HIV-1) infection. Epidemiological studies have demonstrated that the expression of specific NK cell receptors, in conjunction with their HLA class I ligands, is associated with slower HIV-1 disease progression1,2 and functional studies have shown that NK cells from individuals that encode for these protective receptor/ligand combinations can restrict HIV-1 replication in vitro.3 In addition, decreased expression of several cell-surface receptors including Siglec-7 and CD56 marks dysfunctional NK cells in chronic HIV-1 infection.4–6 However, these recent studies were largely performed studying NK cells in HIV-1 clade B-infected caucasian populations, and very little is know about NK cell responses in sub-Saharan African populations, which are most affected by the HIV-1 epidemic. Recently, Eller and colleagues found increased degranulation and cytokine responses in NK cells from Ugandans infected with HIV-1 Clade A or Clade D viruses compared to HIV-uninfected individuals from the same region. In sub-Saharan Africa HIV-1 clade C is the predominant viral subtype7 and the most common ethnic groups there differ from other regions of world. Whereas limited studies have established genetic frequencies for some genes encoding surface receptors on NK cells from people of Zulu and Xhosa descent,8,9 common ethnic groups in South Africa, very little is known about NK cell profiles in these populations.
The function of NK cells is determined by the integration of signals from a number of activating and inhibitory receptors. These receptors allow NK cells to recognize virally infected cells that have downregulated the expression of HLA class I molecules and/or upregulated stress molecules on their surface that serve as ligands for activating NK cell receptors. Furthermore, NK cells play an important role in modulating the adaptive immune response to infection through their interaction with dendritic cells and the secretion of immunoregulatory cytokines. Recent studies have suggested that both the antiviral activity of NK cells and their immunoregulatory function can be impaired in HIV-1 infection.5,10
Here, we assessed the phenotype and function of NK cells in a cohort of chronically HIV-1 infected, treatment-naive adult South Africans, including individuals with high and very low CD4+ T cell counts. In individuals with advanced disease, we observed significant deregulation of NK cell subsets that included the downregulation of activating receptors. These findings suggest that modulation of receptor expression may play a role in HIV-1 pathogenesis, and provide new insights on immunological changes in advanced HIV-1 disease.
Materials and Methods
Study subjects
Forty-one antiretroviral (ARV) treatment-naive HIV-1-positive South African individuals were enrolled from the Sinikithemba HIV Clinic, McCord Hospital, in Durban, and the Edendale Hospital, in Pietermaritzburg; both located in KwaZulu-Natal province of South Africa. Exclusion criteria included concomitant tuberculosis infection. All HIV-1-seropositive patients tested positive for HIV-1 by the RAPID Determine HIV 1/2 test (Abbott Diagnostic Division, Hoofddorp, The Netherlands). Viral load was quantified by polymerase chain reaction (PCR) using the Roche COBAS Amplicore HIV-1 Monitor Test, version 1.5 (Roche, Basel, Switzerland). All individuals in this cohort were confirmed to be infected with HIV Clade C by near full-length viral sequencing performed previously in a separate study.11 CD4 T cell counts were determined from whole blood using the BD Trucount kit (BD Biosciences, San Jose, CA) on a FACSCalibur flow cytometer (BD Biosciences). KIR genotypes for the study participants were established from genomic DNA as previously described.12 The HLA (Supplemental Table 1; see www.liebertonline.com/aid) and the KIR genotype profiles (Table 1) of this cohort were representative of previous reports on populations from South Africa.8,9,13–16 The study protocol was approved by the Institutional Review Boards of the University of Kwa-Zulu-Natal, McCord's Hospital, Edendale Hospital, Kwa-Zulu-Natal Provincial Department of Health, Massachusetts General Hospital, and Harvard Medical School. All patients provided written informed consent prior to sample collection.
Table 1.
Clinical Parameters and KIR Gene Repertoires of the South African Cohort (n = 41)a
| |
|
|
|
KIR gene |
||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| No. | PID | CD4 | VL | 2DL1 | 2DL2 | 2DL3 | 2DL4 | 2DL5 | 2DP1 | 2DS1 | 2DS2 | 2DS3 | 2DS4 | 2DS5 | 3DL1 | 3DS1 | 3DL2 | 3DL3 |
| 1 | SK011 | 325 | 123,000 | |||||||||||||||
| 2 | SK018 | 83 | 400,000 | |||||||||||||||
| 3 | SK021 | 163 | 387,000 | |||||||||||||||
| 4 | SK022 | 577 | 5,690 | |||||||||||||||
| 5 | SK036 | 32 | 313000 | |||||||||||||||
| 6 | SK060 | 909 | 941 | |||||||||||||||
| 7 | SK086 | 8 | 22400 | |||||||||||||||
| 8 | SK090 | 53 | 98,400 | |||||||||||||||
| 9 | SK102 | 196 | 298,000 | |||||||||||||||
| 10 | SK106 | 1020 | 59,200 | |||||||||||||||
| 11 | SK149 | 320 | 40800 | |||||||||||||||
| 12 | SK150 | 314 | 13900 | |||||||||||||||
| 13 | SK157 | 182 | 473,000 | |||||||||||||||
| 14 | SK171 | 117 | 750,000 | |||||||||||||||
| 15 | SK175 | 977 | 439 | |||||||||||||||
| 16 | SK183 | 189 | 750,000 | |||||||||||||||
| 17 | SK203 | 1466 | 50 | |||||||||||||||
| 18 | SK229 | 310 | 52300 | |||||||||||||||
| 19 | SK233 | 18 | 79700 | |||||||||||||||
| 20 | SK253 | 51 | 89,100 | |||||||||||||||
| 21 | SK256 | 861 | 15,000 | |||||||||||||||
| 22 | SK265 | 35 | 488000 | |||||||||||||||
| 23 | SK266 | 327 | 750000 | |||||||||||||||
| 24 | SK274 | 910 | 6,450 | |||||||||||||||
| 25 | SK280 | 738 | 663 | |||||||||||||||
| 26 | SK292 | 317 | 5070 | |||||||||||||||
| 27 | SK295 | 16 | 338000 | |||||||||||||||
| 28 | SK304 | 934 | 112,000 | |||||||||||||||
| 29 | SK307 | 805 | 441 | |||||||||||||||
| 30 | SK317 | 103 | 750,000 | |||||||||||||||
| 31 | SK326 | 328 | 273000 | |||||||||||||||
| 32 | SK327 | 174 | 157,000 | |||||||||||||||
| 33 | SK334 | 165 | 750,000 | |||||||||||||||
| 34 | SK356 | 309 | 120000 | |||||||||||||||
| 35 | SK358 | 316 | 59100 | |||||||||||||||
| 36 | SK392 | 169 | 750,000 | |||||||||||||||
| 37 | SK394 | 320 | 49300 | |||||||||||||||
| 38 | SK399 | 913 | 2,520 | |||||||||||||||
| 39 | SK410 | 310 | 234000 | |||||||||||||||
| 40 | SK428 | 306 | 79500 | |||||||||||||||
| 41 | SK444 | 325 | 123,000 | |||||||||||||||
PID, patient identification; CD4, CD4 cells/μl blood; VL, viral load; KIR gene present, filled square; KIR gene absent, open square.
Sample preparation
Blood samples were collected in EDTA tubes (BD Biosciences) and peripheral blood mononuclear cells (PBMCs) were isolated by density gradient centrifugation using Ficoll-Histopaque 1077 (Sigma-Aldrich, St. Louis, MO) within 6 h of blood collection. The PBMCs were cryopreserved in 90% fetal calf serum/10% dimethyl sulfoxide (DMSO) and stored in liquid nitrogen (–170°C) prior to analysis.
Multiparameter flow cytometric phenotypic analyses
Sample analysis was performed in Durban, South Africa, using a 16-color multiparametric LSRII flow cytometer (BD Biosciences). A minimum of 200,000 events was acquired in the lymphocyte gate based on forward and side scatter parameters. Dead cells were excluded with LIVE/DEAD Fixable Aqua Dead Cell Stain (Invitrogen, Frederick, MD). All antibodies were from BD Biosciences unless otherwise indicated. Allophycocyanin (APC)-conjugated anti-CD3 (clone UCH1), anti-CD14 (clone M5E2), and anti-CD19 (clone H1B19) antibodies were used to exclude non-NK cells (i.e., T cells, monocytes/ macrophages, and B cells, respectively) and phycoerythrin-Cy5 (PE-Cy5)-conjugated anti-CD56 (clone B159) and anti-CD16 (clone 3G8) antibodies were used to gate NK cells. In the panels used to identify specific NK cell subsets, Alexa 700-conjugated anti-CD56 (clone B159) and fluorescein isothiocyanate (FITC)-conjugated anti-CD16 (clone 3G8) antibody were used. In addition, the following antibodies were used: phycoerythrin (PE)-conjugated anti-KIR3DL1/S1 (clone Z27) (Beckman Coulter, Fullerton, CA), anti-KIR3DL1 (clone DX9), anti-NKp46 (clone 9E2), anti-CD158a (anti-KIR2DL1/KIR2DS1) (clone HP-3E4), and anti-CD158b (anti-KIR2DL2/KIR2DL3/KIR2DS2) (clone CH-L) antibodies; FITC-conjugated antibodies: anti-KIR3DL1 (clone DX9), anti-CD69 (clone FN50), and anti-CD158a (anti-KIR2DL1/KIR2DS1) (clone HP-3E4); and PECy5-conjugated anti-CD107a (clone H4A3).
Functional NK cell assays
We assessed the ability of NK cells to degranulate following stimulation in vitro as a surrogate for NK cell cytolysis as previously described.17 In brief, PBMCs were cocultured for 4 h at 37°C with 721.221 cells (ATCC), HLA class I-deficient B cells, at a 10:1 effector-to-target ratio in 200 μl of RPMI medium with 10% fetal bovine serum (FBS) (R10) in a 96-well round bottom plate. In addition, monensin (Golgi-Stop, BD Biosciences) was added to a final concentration of 6 μg/ml. NK cell degranulation was quantified by measuring the accumulation of CD107a expression by flow cytometry. As a positive control PBMCs alone were stimulated with phorbol-12-myristate-13-acetate (PMA) (1 μg/ml) and ionomycin (0.5 μg/ml) (Sigma-Aldrich) for 3 h.
Statistical analyses
Digital data of scatterplots from the flow cytometer were analyzed with FlowJo 8.8 (Tree Star, Ashland, OR) software. The Kruskal–Wallis test was used for the initial analysis of demographic data (e.g., viral loads) among the patient groups in the cohort. For analysis of the endpoints (i.e., cell frequencies), however, the data were analyzed collectively for the entire cohort, rather than between patient groups. Linear regressions were performed using Prism 5.0a software (GraphPad Software, La Jolla, CA). Cell frequencies between specific NK cell subsets (such as NKp46bright versus NKp46dull/neg) were compared by the Mann–Whitney U test.
Results
The frequencies of NK cell subsets in peripheral blood change with HIV-1 disease progression in a cohort of HIV-1 chronically infected South African adults
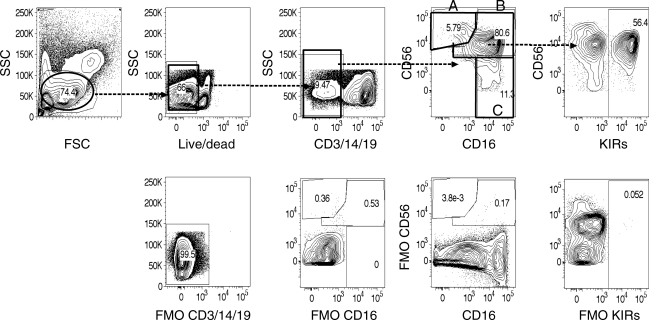
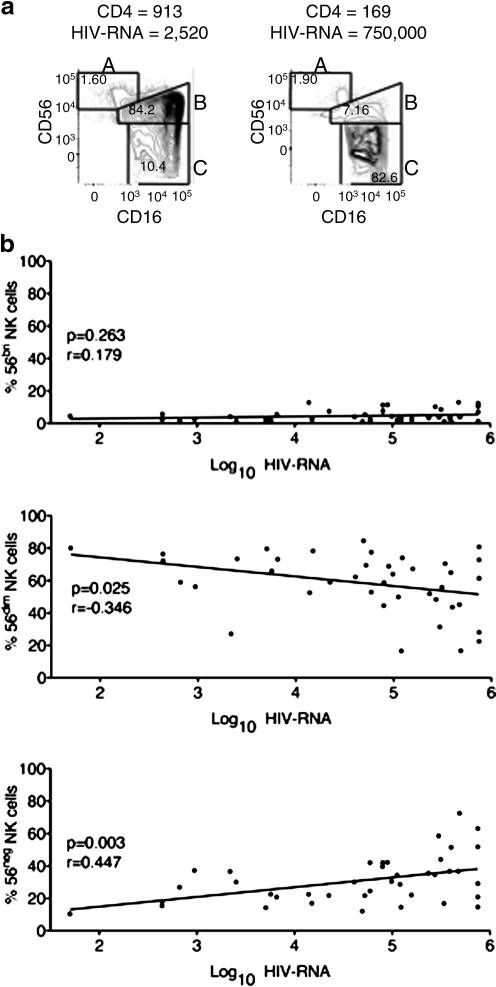
To test the hypothesis that the frequencies and phenotypes of NK cells differ in advanced HIV-1 disease among South Africans we conducted a cross-sectional analysis of a cohort of 41 HIV-1 chronically infected adults from KwaZulu-Natal, South Africa. These individuals, who were all ARV naive, ranged in disease progression from early disease (CD4 count >500) to advanced disease (CD4 count <50) (Table 1). The frequencies of KIR genes in this cohort were comparable to previously described KIR gene frequencies among South Africans.9 Using multiparametric flow cytometry (LSRII) we assessed the phenotypes and frequencies of NK cells in cryopreserved PBMC samples from these donors. We used fluorescence minus one (FMO) staining controls to establish the parameters for gating, excluded dead cells with a viability dye, and identified NK cell lymphocytes by a CD3negCD14negCD19neg and CD56pos and/or CD16pos phenotype (Fig. 1). We found that individuals with the most advanced HIV-1 disease had a significant expansion of CD56negCD16pos NK cells that correlated with viral load (p < 0.01) (Fig. 2). We also observed a similar trend in a negative correlation with CD4 count (data not shown). In addition to this increased frequency of CD56negCD16pos NK cells, we also noted a slight but significant (p < 0.05) decrease in the frequency of CD56dim NK cells (Fig. 2b). Our results confirmed previously described observations of alterations in NK cell subsets associated with advanced HIV-1 disease.18,19
FIG. 1.
Flow cytometric analysis of lymphocytes derived from peripheral blood mononuclear cells from HIV-1-infected individuals. In the gating strategy for NK cells, lymphocytes were first gated according to forward and side scatter and then for live cells based on the BD LIVE/DEAD Fixable Dead Cell Stain. Next, CD3, CD14, CD19-positive cells were excluded to remove T cells, monocytes, and B cells, respectively. Of the remaining cells, NK cells were identified by a CD56pos and/or CD16pos phenotype. To assess KIR expression we assessed KIR expression on all NK cells collectively, including CD56bright/CD16neg (Subset A), CD56dim/CD16neg (Subset B), and CD56neg/CD16pos (Subset C). Fluorescence minus one (FMO) staining controls were used to establish the gating parameters. The following antibodies were used for KIR detection: anti-KIR2DL1/S1 (clone HP-3E4), anti-KIR2DL2/S2/L3 (clone CH-L), anti-KIR3DL1 (clone NKB1), and anti-KIR3DL1/S1 (clone Z27).
FIG. 2.
The frequency of CD56negCD16pos NK cells increases with HIV-1 disease progression. (a) NK cell populations from the peripheral blood of two patients representing two extremes of HIV-1 disease severity. As in Fig. 1, NK cells were gated as lymphocytes by SSC and FSC, by live/dead stain-negative, CD3/CD14/CD19-negative, and then the remaining cells were gated to include CD56 and/or CD16-positive cells. Next the CD56 bright NK cells (Subset A) were gated based on high expression of CD56 without CD16. The CD56dim NK cells (Subset B) were gated based on coexpression of CD16 with low expression of CD56 above the FMO control. Likewise, CD56neg NK cells (Subset C) were gated based on expression of CD16 and gates were defined by the CD56 FMO control. The frequency of these subsets was based on the total NK cell population (CD56 and/or CD16-positive cells). (b) Correlation between log of viral load and the frequencies of 56bright, 56dim, and 56neg NK cells among the cohort of HIV-1-infected adults (n = 41).
Changes in the frequencies of NK cells expressing selected KIRs during HIV-1 disease progression
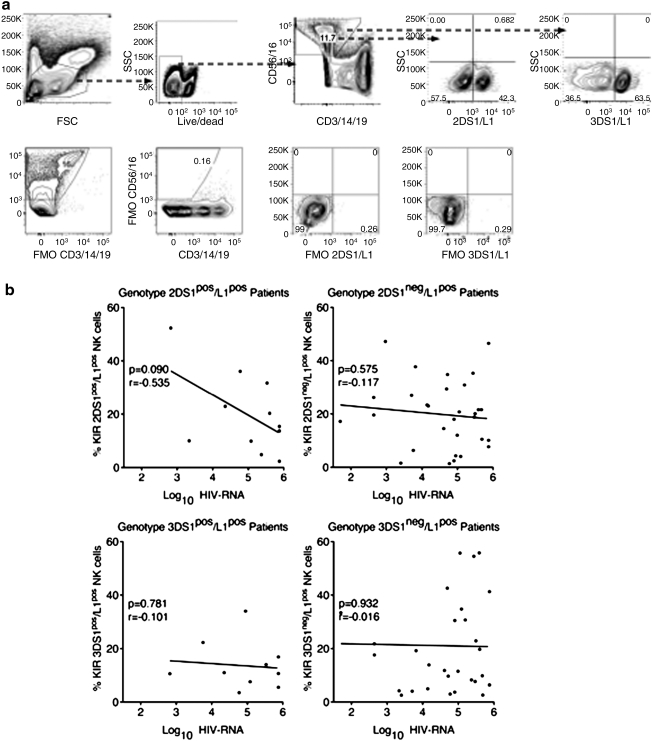
To further investigate these alterations in NK cells among individuals at various stages of HIV-1 disease we quantified the expression of some of the most commonly expressed KIRs. Specifically, we quantified the expression of activating KIRs, KIR2DS1, and KIR3DS1 and inhibitory KIRs, KIR2DL1, and KIR3DL1 by multiparametric flow cytometry on an LSRII. For our analysis, we compared KIR expression among donors of the same KIR genotype, which had been determined previously. Individuals who did not possess the respective KIR genes were excluded from the analysis. We found that in individuals who possessed both KIR2DL1 and KIR2DS1 genes the frequency of NK cells expressing one or both of these receptors tended to decrease with higher viral loads (Fig. 3). However, this trend did not reach statistical significance (p = 0.09, r = –0.535). Due to the limitation of antibody cross-reactivity with the HP-3E4 antibody that recognizes both KIR2DL1 and KIR2DS1 we were not able to determine whether this trend was due to a decreased frequency of KIR2DS1 or 2DL1 expressing cells or a decreased frequency of NK cells expressing both receptors. However, we did not observe a similar trend among the individuals who possessed only the KIR2DL1 gene (Fig. 3b). Also, we did not observe a similar trend in the frequency of KIR3DS1 and/or KIR3DL1-expressing NK cells as assessed by staining with a cross-reactive antibody (clone Z27) that recognizes both receptors. Using an antibody that reportedly recognizes only KIR2DL1, Eller and colleagues previously found a statistically significant lower frequency of KIR2DL1 expressing NK cells in chronically HIV-1-infected African adults compared to uninfected adults.20 Although they did not analyze their data by KIR genotypes, our results appear consistent.
FIG. 3.
Among individuals who possessed both KIR2DS1 and KIR2DL1 genes, the frequency of KIR2DS1pos and/or KIR2DL1pos NK cells tended to decrease with HIV-1 disease progression. (a) Gating strategy to quantify 2DS1pos/L1pos and 3DS1pos/L1pos NK cells using antibodies that are cross-reactive with both S1 and L1 isoforms. The gated populations were based on FMO staining controls as shown. (b) Correlations of the frequencies of KIRpos NK cells with HIV-1 viral load among donors with the indicated KIR genotype.
In the most advanced stages of HIV-1 disease, NK cells with an activated phenotype lacked NKp46 expression
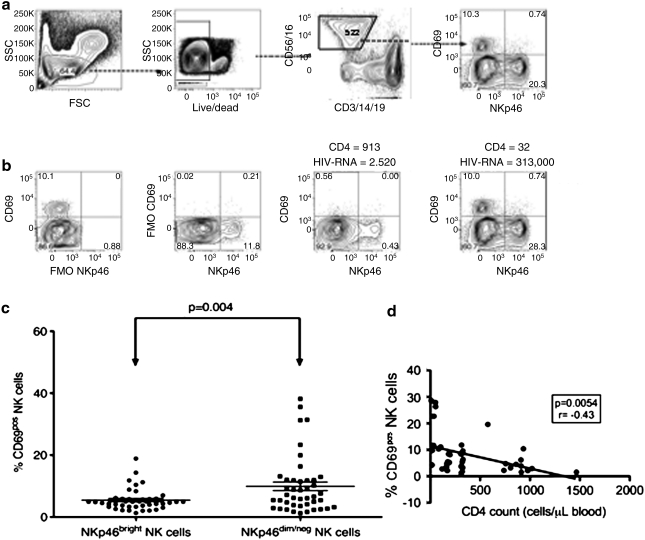
To further investigate the mechanisms underlying changes in NK cell phenotypes in advanced HIV-1 disease, we assessed the expression of CD69, a marker of acute activation, and NKp46, an activation receptor ubiquitously expressed on NK cells. We found that individuals with the lowest CD4 count and greatest viral load (i.e., most advanced HIV-1 disease) had the highest frequency of acutely activated (i.e., CD69 positive) NK cells (Fig. 4). Unexpectedly, we also discovered that these acutely activated NK cells lacked NKp46 expression. The level of expression of NKp46 on the CD69-positive NK cells did not differ from that of the FMO staining control (Fig. 4). Similarly, we found that the NKp46-positive NK cells were a separate and distinct population from the CD69-positive NK cells. Among the entire cohort, we found a significance difference (p < 0.01) in the frequency of CD69-positive cells among NK cells expressing NKp46 and those lacking in NKp46 expression, demonstrating that activated NK cells lacked NKp46 expression (Fig. 4c). Similarly, we found a significant negative correlation between the frequency of CD69pos NK cells and CD4 count (Fig. 4d). However, we found no significant correlation between CD4 count and the frequency of CD69pos NK cells among HIV-uninfected individuals of the same ethnic origins (Supplemental Fig. 1; see www.liebertonline.com/aid). These data suggest that the changes we observed in CD69 expression on NK cells are likely associated with HIV infection. Our results are consistent with a previous study of the activation status of NK cells in HIV-1-infected patients. Notably, Fogli and colleagues found previously that in HIV-1-infected patients, chronically activated (i.e., HLA-DR positive) NK cells lack NKp46 expression.21 However, our study is the first to describe such changes among acutely activated NK cells in chronically infected South African adults.
FIG. 4.
In advanced HIV-1 disease, acutely activated NK cells lacked NKp46 expression. (a) Gating strategy for the assessment of NKp46 and a marker of acute activation (CD69) on NK cells. (b) FMO controls and representative plots from two patients at different stages of HIV-1 disease. (c) In our cohort of chronically infected adults (n = 41), NKp46bright and NKp46dim/neg NK cells differed in their state of acute activation. The FMO control was used to delineate bright versus dim/negative expression where staining above the FMO was designated as bright and staining at or below the level of FMO was designated as dim/negative. (d) The frequency of NK cells expressing CD69 correlated inversely with CD4 count.
NK cells from individuals with the most advanced HIV-1 disease responded with the most degranulation from in vitro stimulation
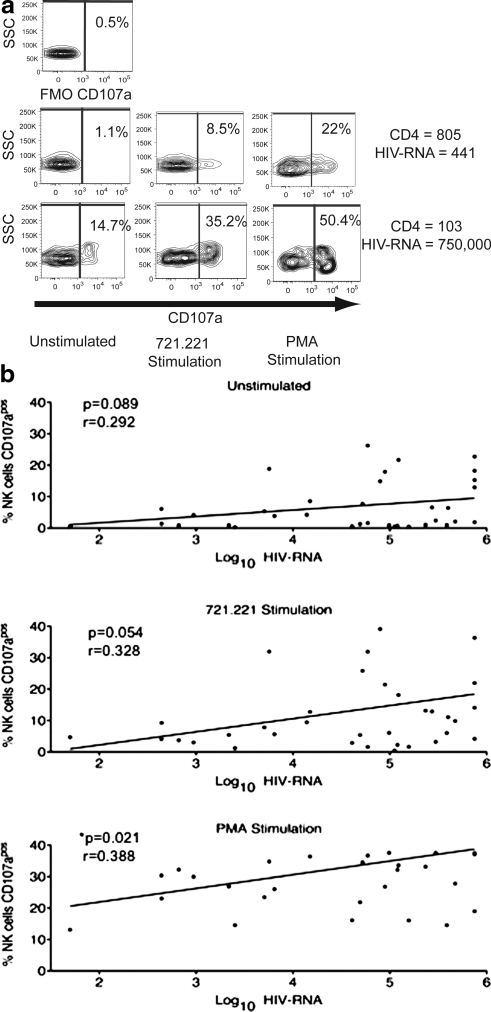
To assess the functional consequences of the phenotypic changes observed with advanced HIV-1 disease we evaluated ex vivo NK cell degranulation responses in coculture assays with 721.221 cells, a B-lymphoblastoid cell line that lacks HLA class I expression and has been previously shown to effectively elicit NK cell degranulation.22,23 We found that individuals with the highest viral load tended to have the greatest response to stimulation with 721.221 cells (Fig. 5), as measured by the expression of CD107a, a marker of NK cell degranulation. Whereas we found a significant positive correlation (p < 0.05) between viral load and NK cell degranulation following PMA-ionomycin stimulation, a positive control for NK cell activation, with 721.221-cell cocultures we observed a similar trend, but it did not reach statistical significance. Likewise, unstimulated NK cells from these donors also showed a trend of positive correlation between the patients' viral load and the extent of spontaneous degranulation. The higher background degranulation of unstimulated NK cells from donors with the highest viral loads most likely contributed to the same trend with 721.221-cell cocultures and PMA-ionomycin treatment. These findings are consistent with a previous report of greater NK cell activity with advanced HIV-1 disease among chronically HIV-1-infected adults.19
FIG. 5.
In response to in vitro costimulation, NK cell degranulation increases with HIV-1 disease progression. (a) Degranulation (CD107a expression) by NK cells in response to a 4-h coculture with a 721.221 HLA-deficient B cell line or PMA treatment in two representative patients at two different extremes of HIV-1 disease stage. (b) Correlations between log of viral load and percentages of CD107apos NK cells in unstimulated and 721.221- and PMA-stimulated samples.
However, our finding of greater NK cell degranulation with advanced HIV-1 disease is in contrast to a report by Mavilio and colleagues,18 who found that NK cells from viremic patients exhibited diminished lytic capacity compared to NK cells derived from aviremic patients. Previous studies suggest that degranulation may not necessarily correlate with lytic ability, particularly with the depletion of cytolytic granules in chronic HIV infection.19,24,25 Thus, differences in our methods of assessment might explain our contrasting conclusions. These differences in outcomes may also be attributed to differences in the patient characteristics in the respective studies. In the Mavilio et al.18 study viremic patients (nonvirus controllers) were compared to aviremic patients (virus controllers), whereas in our study we compared NK cell function among viremic patients at different stages of disease. Our more extensive examination of viremic patients revealed subtle differences among viremic patients that were not apparent previously.
Discussion
In HIV-1 pathogenesis, CD4 T cell count decline and high viral loads during chronic infection are hallmarks of HIV-1 clinical disease progression. In addition to these changes, researchers have also previously described alterations in the phenotypes and functions of NK cells that include an increase in the frequencies of CD56neg/CD16pos NK cells as well as a decline in the frequencies of CD56dim NK cells.18,26 In our cohort of chronically HIV-1-infected South African adults we confirmed this finding. We also extended this result by further assessing phenotypic changes associated with the most advanced HIV-1 disease by assessing individuals at various stages of disease ranging from CD4 counts >500 cells/μl blood to CD4 counts <50 cells/μl blood. In this cross-sectional study we found that the frequency of NK cells expressing either KIR2DL1 or KIR2DS1 tended to be lower among individuals with the most advanced stages of HIV-1 disease. The significance of this finding is limited by our small sample size (n = 41), inability to follow individual patients over time, and inability to distinguish between KIR2DL1 and KIR2DS1, due to cross-reactivity of the existing antibody against KIR2DL1 and KIR2DS1.
Unlike previous studies that have found increased expression of inhibitory receptors such as inhibitory KIR18 and NKG2A,27 we found decreased expression of KIR2DL1 and/or KIR2DS1 in advanced HIV-1 disease in an African cohort. Our data were consistent with a study by Eller and colleagues, who found a decrease in the frequency of KIR2DL1pos NK cells among HIV-infected Ugandans.20 The change that we observed appeared to be unique to KIR2DS1/KIR2DL1, since we did not see a similar trend with KIR3DS1/KIR3DL1, which can also be expressed variably. Although the presence of KIR3DS1 has been associated with protection from HIV-1 disease progression among adults of European descent,1 this gene is much less prevalent among Africans compared to Europeans.8 Taking into consideration that KIR gene frequencies differ between African and European populations, our data imply that some KIR genes may play a unique role in HIV-1 pathogenesis among Africans. Likely explanations for a lower frequency of KIR2DL1pos and/ or KIR2DS1pos NK cells in more advanced HIV disease include a protective role for these cells in mitigating disease or a selective loss of these cells with disease progression. These findings warrant a more extensive analysis of KIR2DS1 and KIR2DL1 expression on NK cells among a larger cohort of native South Africans at various stages of HIV-1 disease progression. Based on our results we propose that NK cell receptor genes other than KIR3DS1 may play a more central role in HIV-1 pathogenesis among African populations.
Other recent studies indicate that ligands for KIRs other than KIR3DS1 play a significant role in HIV-1 pathogenesis. In a whole genome analysis Fellay and colleagues identified a single nucleotide polymorphism (SNP) associated with HLA-C expression, which explained 6.5% of the total variation in HIV-1 viral set point.28 The protective allele was associated with lower viral load and higher expression of the HLA-C gene. Interestingly, selected allotypes of HLA-C are well-established ligands for some KIR, KIR2DL1 and KIR2DS1, in particular.29 Our finding of a trend toward lower frequencies of KIR2DL1- and/or KIR2DS1-expressing NK cells in advanced HIV-1 disease further supports a role for HLA-C alleles in HIV-1 pathogenesis. Based on the protective effects of differential HLA-C expression, one potential mechanism of this protection may be through the modulation of NK cell responses. Because KIR2DL1 has a higher affinity for HLA-C than KIR2DS1,30 lower HLA-C expression would lead to preferential engagement of the inhibitory isoform and to the subsequent lack of expansion of NK cells expressing the activating receptor, KIR2DS1. An alternative mechanism would be selective immune activation and depletion of KIR2DL1- and/or KIR2DS1-expressing NK cells in peripheral blood with advancing HIV-1 disease. In support of this model, Stewart and colleagues have recently shown that some virally encoded peptides can modulate HLA-C recognition and alter the nature of NK cell responses triggered.31 More in-depth studies on HLA-C-dependent KIR-mediated recognition of HIV-1-infected cells are required to further test these models of protection from HIV-1 disease progression.
In this study, among individuals with the most advanced stages of disease we found a greater capacity in NK cell degranulation, a surrogate marker for NK cell cytotoxicity. This finding is consistent with previous reports of hightened NK cell activation profiles in HIV-1-infected subjects during viremia.19,20 However, this greater capacity to degranulate does not necessarily imply better immune control as studies by Mavillio and colleagues have shown that some NK cells expanded in HIV-1 disease are dysfunctional.26 Likewise, other studies have shown that NK cells express lower amounts of perforin cytolytic granules in chronic HIV-1 infection.24,25 We speculate that the greater capacity to degranulate of NK cells in advanced HIV-1 disease is a consequence of greater overall immune activation in advanced HIV-1 disease.
Our finding that unstimulated NK cells from individuals with the most advanced HIV-1 disease had the greatest background levels of degranulation further supports this assertion. Similarly, Eller and colleagues recently found that unstimulated NK cells isolated from HIV-infected adults from Uganda were more proinflammatory (e.g., MIP-1β) and expressed more degranulation than uninfected adults from the same region.20 Other recent studies have also shown that greater overall immune activation is associated with faster HIV-1 disease progression.32,33 Thus, these activated NK cells are likely contributing to an overall environment of greater immune activation, which further facilitates HIV-1 pathogenesis. This conclusion is consistent with our finding of a significant negative correlation between the frequency of NK cells expressing CD69 and CD4 T cell count. In advanced HIV-1 disease we speculate that viremia or opportunistic infections could drive NK cell immune activation; however, it remains undetermined whether this immune activation is characteristic of particular NK cell subpopulations.
In our further investigation of the phenotypes of activated NK cells in advanced HIV-1 disease we found that acutely activated NK cells (i.e., those NK cells that expressed CD69) generally lacked NKp46 expression. In a previous study, reduced surface expression of NKp46 was associated with impaired NK cell cytolytic function in HIV-1 viremic individuals.34 Furthermore, using HLA-DR expression as a marker of chronic activation, Fogli and colleagues also found that in HIV-1-infected patients chronically activated NK cells lacked NKp46 expression.21 In their study the diminished NKp46 expression did not impact overall NK cell activity. Fogli and colleagues found that activated NK cells (i.e., HLA-DRpos) had greater lytic activity than unactivated NK cells (i.e., HLA-DRneg) in redirected lysis assays in which anti-NKp46 antibodies were used to trigger responses. We also found greater overall degranulation activity among NK cells from patients with advanced HIV-1 disease. These results are in contrast to those of Mavilio and colleagues,26 who found that NK cells from viremic patients had less lytic capacity in redirected lysis assays in which anti-NKp46-specific antibodies were used to trigger responses.
We speculate that the differences in outcomes may be attributed to differences in the methods used in assessing NK activity or in the subsets of NK cells assessed. Degranulation is an indirect measure of NK cell lytic activity and therefore may not reflect lytic ability due to cytolytic granule depletion in chronic HIV-1 infection.24,25 In regard to the subsets of NK cells assessed, Mavillio and colleagues26 assessed responses by NKp46pos NK cells, whereas Fogli et al. and we assessed broader NK cell populations. Taken together, these results suggest that in advanced HIV-1 disease the overall activation state rather than the level of NKp46 expression may be a more accurate predictor of NK cell activity. Additional studies on the functional capacity of acutely activated (CD69pos) versus unactivated (CD69neg) NK cells from viremic individuals are needed to resolve this question.
Both in the study by Fogli and colleagues and in our present study, activated NK cells lacked NKp46 expression in advanced HIV-1 disease. We speculate that this lack of NKp46 expression may reflect receptor downregulation to modulate immune activation. The down regulation of receptor expression on activation is a common mechanism of immune regulation and has been well described, both for receptors on NK cells, such as CD16,35,36 as well as for the T cell receptor (TCR) on T cells.37 With the TCR this receptor downregulation occurs in a virus-specific manner.38 Whether similar mechanisms of immune regulation are occurring with NKp46 expression remains to be determined.
In summary, in our cohort of chronically HIV-1-infected South Africans we confirmed several previously described changes in NK cell populations with HIV-1 disease progression. In addition, for the first time we describe decreased NKp46 expression on acutely activated NK cells among African patients with the most advanced HIV-1 disease. Based on this finding, a more in-depth analysis of the receptor expression profiles of activated NK cells among chronically HIV-1-infected patients with different clinical outcomes will likely yield further insights into innate immune modulation of HIV-1 pathogenesis among individuals in sub-Saharan Africa.
Supplementary Material
Acknowledgments
We thank Dudu Ndlovu and Pat Bartman for assistance in subject recruitment and sample collection. We thank Karen Bishop and the HPP Blood Processing Core Facility for assistance in sample processing. This work was supported by an NIH-FIC K01-TW007703-01A1 to W.H.C., HHMI Research Fellowship to A.W., Harvard CFAR grant to M.A., and also in part by NIH R01-A1067031 to M.A. M.A. is a Distinguished Clinical Scientist of the Doris Duke Charitable Foundation. This project also has been funded in part with federal funds from the National Cancer Institute, National Institutes of Health, under Contract No. HHSN261200800001E. The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products, or organizations imply endorsement by the U.S. Government. This research was supported in part by the Intramural Research Program of the NIH, National Cancer Institute, Center for Cancer Research.
Author Disclosure Statement
No competing financial interests exist.
References
- 1.Martin MP. Gao X. Lee JH, et al. Epistatic interaction between KIR3DS1 and HLA-B delays the progression to AIDS. Nat Genet. 2002;31(4):429–434. doi: 10.1038/ng934. [DOI] [PubMed] [Google Scholar]
- 2.Martin MP. Qi Y. Gao X, et al. Innate partnership of HLA-B and KIR3DL1 subtypes against HIV-1. Nat Genet. 2007;39(6):733–740. doi: 10.1038/ng2035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Alter G. Martin MP. Teigen N, et al. Differential natural killer cell-mediated inhibition of HIV-1 replication based on distinct KIR/HLA subtypes. J Exp Med. 2007;204(12):3027–3036. doi: 10.1084/jem.20070695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fu GF. Hao S. Zhao JL, et al. Changes in NK cell counts and receptor expressions and emergence of CD3(dim)/CD56 + cells in HIV-1 infected patients in China. Viral Immunol. 2009;22(2):105–116. doi: 10.1089/vim.2008.0081. [DOI] [PubMed] [Google Scholar]
- 5.Mavilio D. Lombardo G. Kinter A, et al. Characterization of the defective interaction between a subset of natural killer cells and dendritic cells in HIV-1 infection. J Exp Med. 2006;203(10):2339–2350. doi: 10.1084/jem.20060894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brunetta E. Fogli M. Varchetta S, et al. The decreased expression of Siglec-7 represents an early marker of dysfunctional natural killer-cell subsets associated with high levels of HIV-1 viremia. Blood. 2009;114(18):3822–3830. doi: 10.1182/blood-2009-06-226332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Puren AJ. The HIV-1 epidemic in South Africa. Oral Dis. 2002;8(Suppl 2):27–31. doi: 10.1034/j.1601-0825.2002.00007.x. [DOI] [PubMed] [Google Scholar]
- 8.Norman PJ. Carrington CV. Byng M, et al. Natural killer cell immunoglobulin-like receptor (KIR) locus profiles in African and South Asian populations. Genes Immun. 2002;3(2):86–95. doi: 10.1038/sj.gene.6363836. [DOI] [PubMed] [Google Scholar]
- 9.Middleton D. Meenagh A. Moscoso J. Arnaiz-Villena A. Killer immunoglobulin receptor gene and allele frequencies in Caucasoid, Oriental and Black populations from different continents. Tissue Antigens. 2008;71(2):105–113. doi: 10.1111/j.1399-0039.2007.00973.x. [DOI] [PubMed] [Google Scholar]
- 10.Fogli M. Mavilio D. Brunetta E, et al. Lysis of endogenously infected CD4 + T cell blasts by rIL-2 activated autologous natural killer cells from HIV-infected viremic individuals. PLoS Pathog. 2008;4(7):e1000101. doi: 10.1371/journal.ppat.1000101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rousseau CM. Birditt BA. McKay AR, et al. Large-scale amplification, cloning and sequencing of near full-length HIV-1 subtype C genomes. J Virol Methods. 2006;136(1–2):118–125. doi: 10.1016/j.jviromet.2006.04.009. [DOI] [PubMed] [Google Scholar]
- 12.Martin MP. Carrington M. KIR locus polymorphisms: Genotyping and disease association analysis. Methods Mol Biol. 2008;415:49–64. doi: 10.1007/978-1-59745-570-1_3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cao K. Moormann AM. Lyke KE, et al. Differentiation between African populations is evidenced by the diversity of alleles and haplotypes of HLA class I loci. Tissue Antigens. 2004;63(4):293–325. doi: 10.1111/j.0001-2815.2004.00192.x. [DOI] [PubMed] [Google Scholar]
- 14.Honeyborne I. Rathod A. Buchli R, et al. Motif inference reveals optimal CTL epitopes presented by HLA class I alleles highly prevalent in southern Africa. J Immunol. 2006;176(8):4699–4705. doi: 10.4049/jimmunol.176.8.4699. [DOI] [PubMed] [Google Scholar]
- 15.Williams F. Meenagh A. Darke C, et al. Analysis of the distribution of HLA-B alleles in populations from five continents. Hum Immunol. 2001;62(6):645–650. doi: 10.1016/s0198-8859(01)00247-6. [DOI] [PubMed] [Google Scholar]
- 16.Middleton D. Williams F. Meenagh A, et al. Analysis of the distribution of HLA-A alleles in populations from five continents. Hum Immunol. 2000;61(10):1048–1052. doi: 10.1016/s0198-8859(00)00178-6. [DOI] [PubMed] [Google Scholar]
- 17.Alter G. Malenfant JM. Altfeld M. CD107a as a functional marker for the identification of natural killer cell activity. J Immunol Methods. 2004;294(1–2):15–22. doi: 10.1016/j.jim.2004.08.008. [DOI] [PubMed] [Google Scholar]
- 18.Mavilio D. Benjamin J. Daucher M, et al. Natural killer cells in HIV-1 infection: Dichotomous effects of viremia on inhibitory and activating receptors and their functional correlates. Proc Natl Acad Sci USA. 2003;100(25):15011–15016. doi: 10.1073/pnas.2336091100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Alter G. Malenfant JM. Delabre RM, et al. Increased natural killer cell activity in viremic HIV-1 infection. J Immunol. 2004;173(8):5305–5311. doi: 10.4049/jimmunol.173.8.5305. [DOI] [PubMed] [Google Scholar]
- 20.Eller MA. Eller LA. Ouma BJ, et al. Elevated natural killer cell activity despite altered functional and phenotypic profile in Ugandans with HIV-1 clade A or clade D infection. J Acquir Immune Defic Syndr. 2009;51(4):380–389. doi: 10.1097/QAI.0b013e3181aa256e. [DOI] [PubMed] [Google Scholar]
- 21.Fogli M. Costa P. Murdaca G, et al. Significant NK cell activation associated with decreased cytolytic function in peripheral blood of HIV-1-infected patients. Eur J Immunol. 2004;34(8):2313–2321. doi: 10.1002/eji.200425251. [DOI] [PubMed] [Google Scholar]
- 22.Litwin V. Gumperz J. Parham P. Phillips JH. Lanier LL. Specificity of HLA class I antigen recognition by human NK clones: Evidence for clonal heterogeneity, protection by self and non-self alleles, and influence of the target cell type. J Exp Med. 1993;178(4):1321–1336. doi: 10.1084/jem.178.4.1321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Draghi M. Yawata N. Gleimer M. Yawata M. Valiante NM. Parham P. Single-cell analysis of the human NK cell response to missing self and its inhibition by HLA class I. Blood. 2005;105(5):2028–2035. doi: 10.1182/blood-2004-08-3174. [DOI] [PubMed] [Google Scholar]
- 24.Portales P. Reynes J. Pinet V, et al. Interferon-alpha restores HIV-induced alteration of natural killer cell perforin expression in vivo. AIDS. 2003;17(4):495–504. doi: 10.1097/00002030-200303070-00004. [DOI] [PubMed] [Google Scholar]
- 25.Alter G. Suscovich TJ. Kleyman M, et al. Low perforin and elevated SHIP-1 expression is associated with functional anergy of natural killer cells in chronic HIV-1 infection. AIDS. 2006;20(11):1549–1551. doi: 10.1097/01.aids.0000237371.31315.48. [DOI] [PubMed] [Google Scholar]
- 26.Mavilio D. Lombardo G. Benjamin J, et al. Characterization of CD56-/CD16 + natural killer (NK) cells: A highly dysfunctional NK subset expanded in HIV-infected viremic individuals. Proc Natl Acad Sci USA. 2005;102(8):2886–2891. doi: 10.1073/pnas.0409872102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhang R. Xu J. Hong K, et al. Increased NKG2A found in cytotoxic natural killer subset in HIV-1 patients with advanced clinical status. AIDS. 2007;21(Suppl 8):S9–S17. doi: 10.1097/01.aids.0000304691.32014.19. [DOI] [PubMed] [Google Scholar]
- 28.Fellay J. Shianna KV. Ge D, et al. A whole-genome association study of major determinants for host control of HIV-1. Science. 2007;317(5840):944–947. doi: 10.1126/science.1143767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Biassoni R. Falco M. Cambiaggi A, et al. Amino acid substitutions can influence the natural killer (NK)-mediated recognition of HLA-C molecules. Role of serine-77 and lysine-80 in the target cell protection from lysis mediated by “group 2” or “group 1” NK clones. J Exp Med. 1995;182(2):605–609. doi: 10.1084/jem.182.2.605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Biassoni R. Cantoni C. Falco M, et al. The human leukocyte antigen (HLA)-C-specific “activatory” or “inhibitory” natural killer cell receptors display highly homologous extracellular domains but differ in their transmembrane and intracytoplasmic portions. J Exp Med. 1996;183(2):645–650. doi: 10.1084/jem.183.2.645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Stewart CA. Laugier-Anfossi F. Vely F, et al. Recognition of peptide-MHC class I complexes by activating killer immunoglobulin-like receptors. Proc Natl Acad Sci USA. 2005;102(37):13224–13229. doi: 10.1073/pnas.0503594102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Catalfamo M. Di Mascio M. Hu Z, et al. HIV infection-associated immune activation occurs by two distinct pathways that differentially affect CD4 and CD8 T cells. Proc Natl Acad Sci USA. 2008;105(50):19851–19856. doi: 10.1073/pnas.0810032105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Perreau M. Pantaleo G. Kremer EJ. Activation of a dendritic cell-T cell axis by Ad5 immune complexes creates an improved environment for replication of HIV in T cells. J Exp Med. 2008;205(12):2717–2725. doi: 10.1084/jem.20081786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.De Maria A. Fogli M. Costa P, et al. The impaired NK cell cytolytic function in viremic HIV-1 infection is associated with a reduced surface expression of natural cytotoxicity receptors (NKp46, NKp30 and NKp44) Eur J Immunol. 2003;33(9):2410–2418. doi: 10.1002/eji.200324141. [DOI] [PubMed] [Google Scholar]
- 35.Harrison D. Phillips JH. Lanier LL. Involvement of a metalloprotease in spontaneous and phorbol ester-induced release of natural killer cell-associated Fc gamma RIII (CD16-II) J Immunol. 1991;147(10):3459–3465. [PubMed] [Google Scholar]
- 36.Grzywacz B. Kataria N. Verneris MR. CD56(dim)CD16(+) NK cells downregulate CD16 following target cell induced activation of matrix metalloproteinases. Leukemia. 2007;21(2):356–359. doi: 10.1038/sj.leu.2404499. author reply 359. [DOI] [PubMed] [Google Scholar]
- 37.von Essen M. Bonefeld CM. Siersma V, et al. Constitutive and ligand-induced TCR degradation. J Immunol. 2004;173(1):384–393. doi: 10.4049/jimmunol.173.1.384. [DOI] [PubMed] [Google Scholar]
- 38.Bonefeld CM. Haks M. Nielsen B, et al. TCR down-regulation controls virus-specific CD8 + T cell responses. J Immunol. 2008;181(11):7786–7799. doi: 10.4049/jimmunol.181.11.7786. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.