Introduction
Natural products have proven to be a productive source of lead structures for the development of new antimicrobial agents.[1] Culture independent analyses of environmental samples suggest that traditional approaches used to identify microbial metabolites from laboratory grown microorganisms have likely missed the vast majority of bacterial natural products that exist in nature.[2,3] In most environments, microbes that have not yet been cultured are thought to outnumber their cultured counterparts by at least two to three orders of magnitude.[2–5] If the diversity of molecules discovered from cultured bacteria is any indication, as yet uncultured bacteria are likely to be a very rewarding source of previously unknown biologically active small molecules that could serve as novel anti-infective agents. New strategies using both culture-dependent and culture-independent methods are now being developed to access this untapped reservoir of chemical diversity. This review focuses primarily on recent culture-independent, or metagenomic, efforts to identify bioactive natural products and the biosynthetic gene clusters from which they are derived.
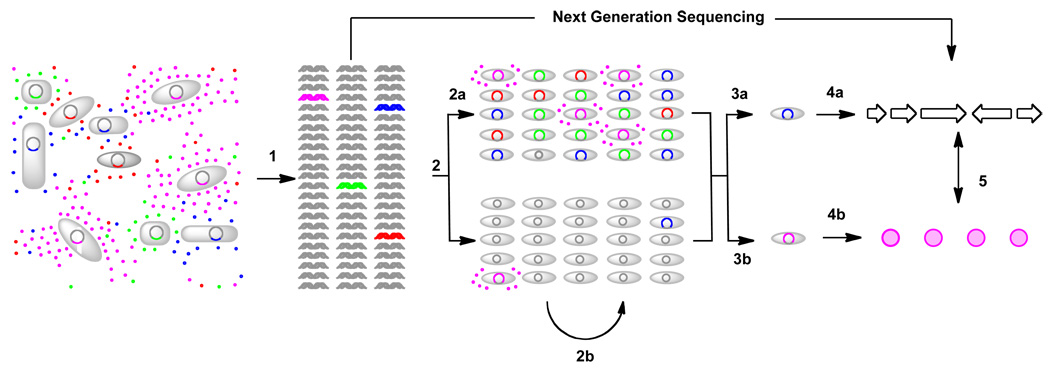
The foundation of all metagenomic approaches is the isolation and subsequent examination of DNA extracted directly from naturally occurring microbial populations (environmental DNA, eDNA), which avoids the difficulties associated with culturing environmental microbes (Figure 1).[6] Metagenomics is particularly appealing to natural product researchers because the genetic information needed to encode for the production of bacterial secondary metabolites is typically clustered on bacterial chromosomes. It is therefore possible to envision capturing complete small molecule biosynthetic gene clusters on individual or, at most, a small number of overlapping eDNA clones.[6] Both expression-dependent (functional) and expression-independent (homology) screening strategies have been used to identify eDNA clones that produce bioactive small molecules. In functional metagenomic studies, eDNA libraries are examined in simple high throughput assays designed to identify clones that have phenotypes traditionally associated with the production of small molecules, while in homology-based studies, libraries are probed to identify clones that contain conserved sequences traditionally associated with secondary metabolite biosynthesis. Hits identified in these initial high throughput assays are subsequently examined for the ability to confer the production of small molecules to model cultured heterologous hosts.
Figure 1.
Overview of metagenomic methods: Environmental DNA isolated directly from an environmental sample (1) is cloned into an easily cultured model bacterial host (2). Libraries (or eDNA) can then either be enriched for genes of interest (2a), transferred into another heterologous host (2b), or screened directly. The search for bioactive small molecules using a metagenomic approach has generally been conducted using either homology based methods (3a) or functional screening (3b). Novel sequences found in homology-based screens (4a) can be examined for the ability to encode the biosynthesis of novel small molecules in heterologous expression experiments (5). The characterization of hits from functional screens can lead directly to the identification of bioactive small molecules (4b) and their biosynthetic gene clusters (5).
Functional metagenomic library screening
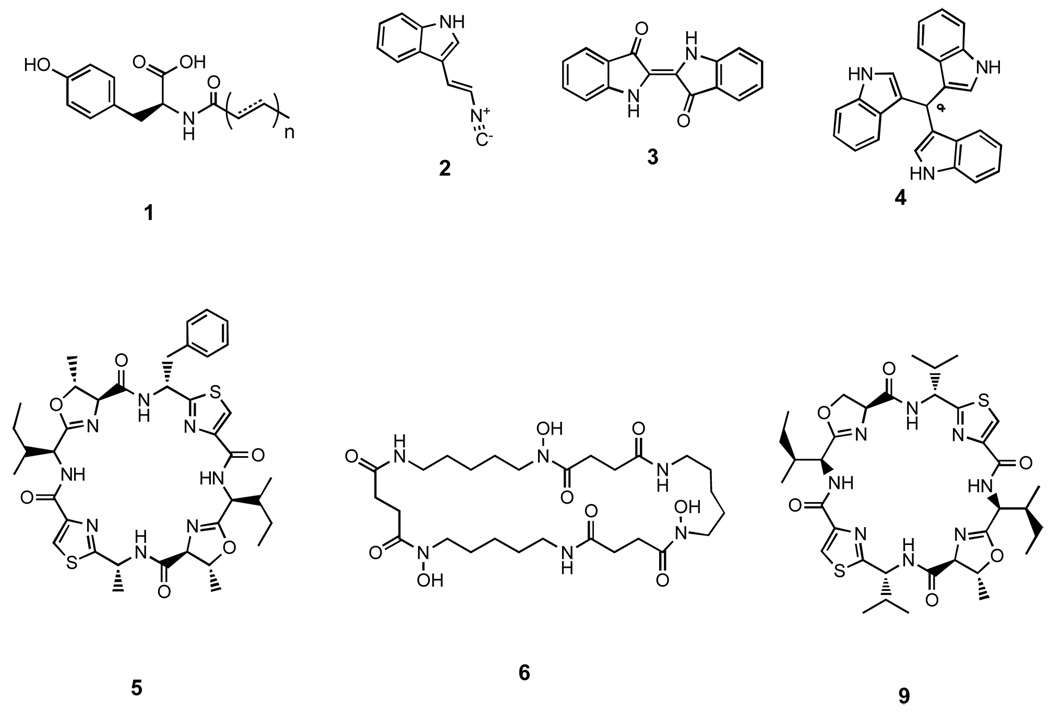
One of the simplest strategies used to detect eDNA clones that might produce small molecule antibiotics has been to screen libraries hosted in E. coli for clones that generate zones of growth inhibition against test microbes in top agar overlay assays. The isolation of clone specific metabolites produced by antibacterially active eDNA clones identified from bacterial top agar overlay assays has led to the characterization of a variety of new long-chain N-acylated amines (1), as well as a new isonitrile functionalized indole antibiotic (2) (Figure 2).[7,8] Small molecule antibiotics have also been found by examining pigmented eDNA clones, as well as through the direct examination of culture broth extracts from randomly selected clones.[9–14] Bioactive compounds identified from these types of studies include the antibacterially active pigments violacein, indigo (3) and the turbomycins (4), all recovered from soil libraries, as well as the known cyclic peptides patellamide D (5) and nocardamine (6), isolated from marine sponge and soil libraries, respectively.
Figure 2.
Representative natural products heterologously produced in model cultured bacteria from metagenomic derived genes and gene clusters. N-acyltyrosine (1), isocyanide functionalized indole (2), indigo (3), turbomycin A (4), cyanobactin patellamide D (5), nocardamine (6) and cyanobactin patellamide A (9).
Functional metagenomics has also been used to identify clones that produce proteins with potential anti-infective properties. Using an acylhomoserine lactone synthase promoter fused to a lacZ reporter, Schipper et al. identified three eDNA derived AHL lactonases that are capable of inhibiting biofilm formation by Pseudomonas aureginosa.[15] And, an examination of bacteriophage DNA isolated from bat guano and earthworm guts by Schmitz, et. al. led to the discovery of three new lysins capable of halting Bacillus anthracis proliferation.[16] In this work, the authors were able to functionally access phage lysins by inducing the expression of genes cloned from environmental samples using a vector associate araBAD promoter.
Although nearly all small molecule focused functional metagenomic studies have been carried out in E. coli, it is clear that most of the biosynthetic diversity present in an environmental sample is unlikely to be functionally accessed using a single heterologous host. A computational analysis of promoters and ribosomal binding sites used by a taxonomically diverse group of sequenced bacteria found that at most, 40% of the enzymatic activities present within a typical metagenomic sample could be accessed using E. coli as a heterologous host.[17] The successful expression of entire biosynthetic gene clusters, which requires the coordinated production of multiple proteins, is likely to occur at an even lower frequency. Vector-host pairs that allow for the introduction and screening of metagenomic libraries in phylogenetically diverse bacteria have the potential to expand the number and type of compounds found from metagenomic studies. While cosmid and BAC vectors capable of replicating in a variety of Gram-positive and Gram-negative hosts have been described in the literature, until recently, none of these had been used in an extensive broad-host-range small molecule focused screen of metagenomic libraries.[18,19] Two RK2-derived broad-host-range vectors (pJWC1 and pRS44) were recently constructed with this specific purpose in mind.[20,21] Craig, et al. demonstrated the utility of pJWC1 by screening metagenomic libraries for eDNA clones that confer antibacterial activities to any of six different host Proteobacteria, including Agrobacterium tumefaciens, Burkholderia graminis, Caulobacter vibrioides, Escherichia coli, Pseudomonas putida, and Ralstonia metallidurans.[20,22] This study found that distinct collections of eDNA clones within the same metagenomic library are likely to confer detectable phenotypes to different hosts and that eDNA clones infrequently confer the same phenotype to two different hosts.
Homology-based metagenomic library screening
Cloning natural product gene clusters from uncultured symbionts
Many bioactive natural products that were originally isolated from extracts derived from mutlicellular organisms are now thought to be products of as yet uncultured microbial symbionts. Metagenomics provides a strategy for cloning the biosynthetic gene clusters of these metabolites, which may in turn provide a renewable source of compounds that have often been difficult to isolate in sufficient quantities to permit extensive biological testing.
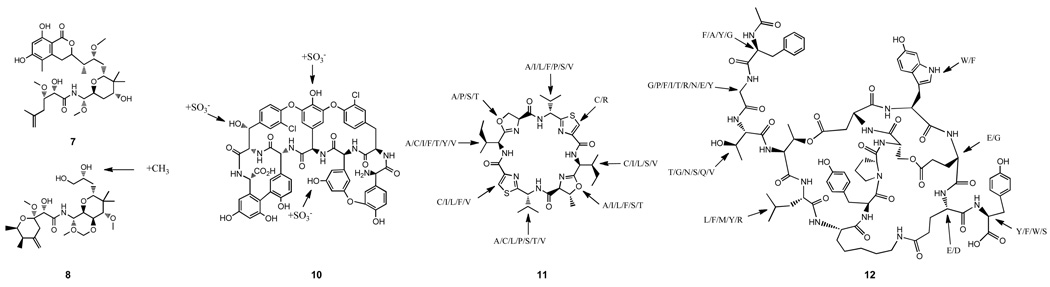
The biosynthetic gene cluster for pederin, an anticancer agent originally isolated from the beetle Paederus fuscipes, was recovered from a cosmid library constructed using beetle-derived metagenomic DNA and shown to originate from an uncultured symbiotic Pseudomonad. [23,24] As additional pederin-like structures had also been isolated from marine sponge extracts, it was hypothesized that these other molecules might originate from bacterial symbionts as well. In two separate studies, the Piel group reported the cloning of gene clusters encoding the biosynthesis of the pederin relatives onnamide and psymberin (7) from symbionts associated with field collected Demospongiae sponges (Figure 3).[25,26] While it has not yet been possible to heterologously express these gene clusters in the laboratory, Zimmermann et al., reported the use of a recombinant O-methyltransferase, PedO, from the pederin biosynthetic gene cluster to site-specifically methylate mycalamide A resulting in the production of a derivative (8) that exhibits enhanced antitumor activity.[27] In related work using libraries constructed from DNA extracted from uncultured cyanobacterial symbionts associated with marine Didemnidae sponges, two separate groups have reported the cloning and heterologous expression of biosynthetic gene clusters for a number of patellamides, cytotoxic cyclic peptides originally isolated from sponge extracts (5,9).[13,28]
Figure 3.
Natural product families that have been explored using metagenomic methods. Pederin (7), methylated mycalamide A (8), glycopeptides (10), patellamides (11) and microviridins (12). The cyanobactin and microviridin precursor peptide diversity found in metagenomic studies is displayed on patellamide A and microviridin B, respectively.
Most culture independent symbiont studies have focused on anticancer agents. In future studies, the same general approach will undoubtedly prove useful for investigating symbiont-derived antimicrobials. It was recently shown that a symbiotic Streptomyces species associated with leaf-cutting ants (Acromyrmex) produces the potent antifungal candicidin, which is active against the pathogenic fungus (Escovopsis), but non-active against the symbiotic fungus (Leucoagaricus) that the ants maintain as their main food source.[29] While the producing organism in this study could be cultured, collections of uncultured microbial symbionts that promise to be rich sources of future small molecule metagenomic studies have been found in environments ranging from the human gut to marine snails.[30,31]
Cloning gene cluster families from the environment
In contrast to culture-dependent studies that are often designed to explore the secondary metabolites produced by a single organism, culture-independent approaches provide the opportunity to investigate thousands of bacterial genomes simultaneously. This has led a number of groups to explore the possibility of using metagenomics to identify groups of related gene clusters that encode the biosynthesis of new structural variants of known secondary metabolites. When Banik, et al. screened a soil DNA cosmid library for clones containing genes associated with the biosynthesis of teicoplanin and vancomycin like glycopeptide antibiotics, they identified two new biosynthetic gene clusters.[32] One of these glycopeptide clusters contains unique genes encoding three sulfotransferases, a class of tailoring enzymes that has rarely been associated with glycopeptide biosynthesis. Using the teicoplanin aglycone as a substrate, seven new anionic glycopeptide congeners (10) were generated in vitro using the eDNA derived sulfotransferases. In an expansion of its earlier cyanobactin (patellamide) research [28], the Schmidt group recently reported the PCR amplification of 30 genes encoding novel patellamide-like precursor peptides from uncultured Prochloron spp. symbionts living in consortia with marine sponges (11).[33,34] In another PCR based study, in this case using DNA isolated from uncultured freshwater cyanobacteria of the genera Microcystis, Ziemert, et al. identified 15 new variants of the gene that encode for the precursor to the microviridin peptide (12).[35] Microviridins are ribosomally synthesized tricyclic depsipeptide proteases inhibitors produced by a number of cyanobacteria. The discovery of both the new microviridin and patellamide-like precursor peptides should aid in attempts to generate additional members of these two important cyclic peptide families.
Most homology-based screens have been carried out using degenerate PCR primers designed to recognize conserved sequences within secondary metabolite biosynthetic genes. As more metagenomic sequencing data appears in publicly available databases, it should also be possible to use purely bioinformatics based search strategies to identify new natural product biosynthetic enzymes and gene clusters. An in silico examination of data from the Global Ocean Metagenomic Survey for genes involved in the biosynthesis of lantibiotic type antibiotics uncovered more than 20 novel lantibiotic cyclases.[36] This class of peptide cyclase is used in the biosynthesis of potent cyclic peptide antibiotics (lantibiotics) from short linear ribosomally synthesized peptides. Environmental DNA derived lantibiotic cyclases could one day aid in the enzymatic synthesis of new lantibiotic variants. The examination of eDNA libraries and metagenomic sequencing data for relatives of known biosynthetic systems is likely to be a generally applicable strategy for identifying new structural variants of many bacterially derived antibiotics, potentially providing ready access to compounds with improved pharmacological properties and improved spectra of activity.
Library enrichment strategies
The development of generic gene enrichment strategies would undoubtedly simplify the screening of large eDNA libraries and likely increase the utility of metagenomics as a tool for the discovery of novel bioactive small molecules. A number of DNA hybridization strategies, including subtractive hybridization, PCR denaturing gradient gel electrophoresis (PCR-DGGE), "biopanning" and fluorescence in-situ hybridization coupled with cell sorting have been explored for enriching eDNA samples for genes of interests with varying degrees of success.[37–41] Zhang, et al. recently reported a phage display-based strategy for specifically enriching eDNA libraries for clones containing two important classes of natural product biosynthetic genes, polyketide synthases and non-ribosomal peptide synthetases (NRPS).[42] Their selection strategy takes advantage of the fact that PKS and NRPS proteins are posttranslationally modified with the addition of a phosphopantetheine prosthetic group. Phage that display either PKS or NRPS proteins on their surface could therefore be collected by first incubating the phage library with a recombinant phosphopantetheinyl transferase and a biotinylated phosphopantetheine analog, and then panning with streptavidin. At the moment, complete biosynthetic gene clusters are not accessible using this strategy. It does, however, provide a promising method for recovering individual NPRS and PKS megasynthases from complex eDNA samples.
Future prospects
The discovery of bioactive small molecules using metagenomic methods will undoubtedly benefit greatly from future advances in sequencing technology that allow for the comprehensive sequencing of complex microbiomes[43–46], as well as from increasing our understanding of the expression barriers encountered by foreign DNA in model laboratory grown bacterial hosts.[47,48] The TerraGenome project was established in 2008 in an effort to bring together sufficient sequencing power to sequence the first complete soil microbiome.[49] Although only a small number of compounds have been characterized to date using culture-independent methods, these initial studies indicate that as yet uncultured bacteria are likely to be a rich source of previously unstudied biologically active small molecules. Collaborative efforts involving individuals from many disparate fields including bacterial genetics, molecular biology, genomics, bioinformatics, robotics, synthetic biology and natural products chemistry (to name a few) will likely be necessary to effectively address this large scale and potentially very rewarding problem.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Newman DJ, Cragg GM. Natural products as sources of new drugs over the last 25 years. J Nat Prod. 2007;70:461–477. doi: 10.1021/np068054v. [DOI] [PubMed] [Google Scholar]
- 2.Hugenholtz P, Goebel BM, Pace NR. Impact of culture-independent studies on the emerging phylogenetic view of bacterial diversity. J Bacteriol. 1998;180:4765–4774. doi: 10.1128/jb.180.18.4765-4774.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rappe MS, Giovannoni SJ. The uncultured microbial majority. Annu Rev Microbiol. 2003;57:369–394. doi: 10.1146/annurev.micro.57.030502.090759. [DOI] [PubMed] [Google Scholar]
- 4.Torsvik V, Goksoyr J, Daae FL. High diversity in DNA of soil bacteria. Appl Environ Microbiol. 1990;56:782–787. doi: 10.1128/aem.56.3.782-787.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Torsvik V, Daae FL, Sandaa RA, Ovreas L. Novel techniques for analysing microbial diversity in natural and perturbed environments. J Biotechnol. 1998;64:53–62. doi: 10.1016/s0168-1656(98)00103-5. [DOI] [PubMed] [Google Scholar]
- 6.Handelsman J, Rondon MR, Brady SF, Clardy J, Goodman RM. Molecular biological access to the chemistry of unknown soil microbes: a new frontier for natural products. Chemistry & Biology. 1998;5:R245–R249. doi: 10.1016/s1074-5521(98)90108-9. [DOI] [PubMed] [Google Scholar]
- 7.Brady SF, Clardy J. Long-Chain N-Acyl Amino Acid Antibiotics Isolated from Heterologously Expressed Environmental DNA. Journal of the American Chemical Society. 2000;122:12903–12904. [Google Scholar]
- 8.Brady SF, Clardy J. Cloning and heterologous expression of isocyanide biosynthetic genes from environmental DNA. Angew Chem Int Ed Engl. 2005;44:7063–7065. doi: 10.1002/anie.200501941. [DOI] [PubMed] [Google Scholar]
- 9.Brady SF, Chao CJ, Handelsman J, Clardy J. Cloning and heterologous expression of a natural product biosynthetic gene cluster from eDNA. Org Lett. 2001;3:1981–1984. doi: 10.1021/ol015949k. [DOI] [PubMed] [Google Scholar]
- 10.Lim HK, Chung EJ, Kim JC, Choi GJ, Jang KS, Chung YR, Cho KY, Lee SW. Characterization of a forest soil metagenome clone that confers indirubin and indigo production on Escherichia coli. Appl Environ Microbiol. 2005;71:7768–7777. doi: 10.1128/AEM.71.12.7768-7777.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.MacNeil IA, Tiong CL, Minor C, August PR, Grossman TH, Loiacono KA, Lynch BA, Phillips T, Narula S, Sundaramoorthi R, et al. Expression and isolation of antimicrobial small molecules from soil DNA libraries. J Mol Microbiol Biotechnol. 2001;3:301–308. [PubMed] [Google Scholar]
- 12.Gillespie DE, Brady SF, Bettermann AD, Cianciotto NP, Liles MR, Rondon MR, Clardy J, Goodman RM, Handelsman J. Isolation of antibiotics turbomycin a and B from a metagenomic library of soil microbial DNA. Appl Environ Microbiol. 2002;68:4301–4306. doi: 10.1128/AEM.68.9.4301-4306.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Long PF, Dunlap WC, Battershill CN, Jaspars M. Shotgun cloning and heterologous expression of the patellamide gene cluster as a strategy to achieving sustained metabolite production. Chembiochem. 2005;6:1760–1765. doi: 10.1002/cbic.200500210. [DOI] [PubMed] [Google Scholar]
- 14.Wang GY, Graziani E, Waters B, Pan W, Li X, McDermott J, Meurer G, Saxena G, Andersen RJ, Davies J. Novel natural products from soil DNA libraries in a streptomycete host. Org Lett. 2000;2:2401–2404. doi: 10.1021/ol005860z. [DOI] [PubMed] [Google Scholar]
- 15. Schipper C, Hornung C, Bijtenhoorn P, Quitschau M, Grond S, Streit WR. Metagenome-derived clones encoding two novel lactonase family proteins involved in biofilm inhibition in Pseudomonas aeruginosa. Appl Environ Microbiol. 2009;75:224–233. doi: 10.1128/AEM.01389-08. Acyl-homoserine lactonase homologs were identified during a functional metagenomic screen of a soil eDNA phagemid library. When heterologously expressed in Pseudomonas aureginosa, these enzymes prevented biofilm formation, a key stage in pathogenicity.
- 16.Schmitz JE, Daniel A, Collin M, Schuch R, Fischetti VA. Rapid DNA library construction for functional genomic and metagenomic screening. Appl Environ Microbiol. 2008;74:1649–1652. doi: 10.1128/AEM.01864-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gabor EM, Alkema WB, Janssen DB. Quantifying the accessibility of the metagenome by random expression cloning techniques. Environ Microbiol. 2004;6:879–886. doi: 10.1111/j.1462-2920.2004.00640.x. [DOI] [PubMed] [Google Scholar]
- 18.Courtois S, Cappellano CM, Ball M, Francou FX, Normand P, Helynck G, Martinez A, Kolvek SJ, Hopke J, Osburne MS, et al. Recombinant environmental libraries provide access to microbial diversity for drug discovery from natural products. Appl Environ Microbiol. 2003;69:49–55. doi: 10.1128/AEM.69.1.49-55.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Martinez A, Kolvek SJ, Yip CL, Hopke J, Brown KA, MacNeil IA, Osburne MS. Genetically modified bacterial strains and novel bacterial artificial chromosome shuttle vectors for constructing environmental libraries and detecting heterologous natural products in multiple expression hosts. Appl Environ Microbiol. 2004;70:2452–2463. doi: 10.1128/AEM.70.4.2452-2463.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Craig JW, Chang FY, Brady SF. Natural products from environmental DNA hosted in Ralstonia metallidurans. ACS Chem Biol. 2009;4:23–28. doi: 10.1021/cb8002754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Aakvik T, Degnes KF, Dahlsrud R, Schmidt F, Dam R, Yu L, Volker U, Ellingsen TE, Valla S. A plasmid RK2-based broad-host-range cloning vector useful for transfer of metagenomic libraries to a variety of bacterial species. FEMS Microbiol Lett. 2009;296:149–158. doi: 10.1111/j.1574-6968.2009.01639.x. [DOI] [PubMed] [Google Scholar]
- 22. Craig JW, Chang FY, Kim JH, Obiajulu SC, Brady SF. Expanding small-molecule functional metagenomics through parallel screening of broad-host-range cosmid environmental DNA libraries in diverse proteobacteria. Appl Environ Microbiol. 2010;76:1633–1641. doi: 10.1128/AEM.02169-09. The examination of metagenomic libraries constructed in a broad-host-range vector for phenotypic hits in a phylogenetically diverse collection of bacteria showed that individual metagenomic clones are likely to confer phenotypes to a phylogenetically narrow range of hosts.
- 23.Soldati M, Fioretti A, Ghione M. Cytotoxicity of pederin and some of its derivatives on cultured mammalian cells. Experientia. 1966;22:176–178. doi: 10.1007/BF01897720. [DOI] [PubMed] [Google Scholar]
- 24.Piel J. A polyketide synthase-peptide synthetase gene cluster from an uncultured bacterial symbiont of Paederus beetles. Proc Natl Acad Sci U S A. 2002;99:14002–14007. doi: 10.1073/pnas.222481399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Piel J, Hui D, Wen G, Butzke D, Platzer M, Fusetani N, Matsunaga S. Antitumor polyketide biosynthesis by an uncultivated bacterial symbiont of the marine sponge Theonella swinhoei. Proc Natl Acad Sci U S A. 2004;101:16222–16227. doi: 10.1073/pnas.0405976101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Fisch KM, Gurgui C, Heycke N, van der Sar SA, Anderson SA, Webb VL, Taudien S, Platzer M, Rubio BK, Robinson SJ, et al. Polyketide assembly lines of uncultivated sponge symbionts from structure-based gene targeting. Nat Chem Biol. 2009;5:494–501. doi: 10.1038/nchembio.176. Continued efforts to explore the biosynthetic potential of symbiont-rich marine eukaryotes led to the identification of the gene cluster encoding the biosynthesis of the antitumor agent psymberin.
- 27.Zimmermann K, Engeser M, Blunt JW, Munro MH, Piel J. Pederin-type pathways of uncultivated bacterial symbionts: analysis of o-methyltransferases and generation of a biosynthetic hybrid. J Am Chem Soc. 2009;131:2780–2781. doi: 10.1021/ja808889k. [DOI] [PubMed] [Google Scholar]
- 28.Schmidt EW, Nelson JT, Rasko DA, Sudek S, Eisen JA, Haygood MG, Ravel J. Patellamide A and C biosynthesis by a microcin-like pathway in Prochloron didemni, the cyanobacterial symbiont of Lissoclinum patella. Proc Natl Acad Sci U S A. 2005;102:7315–7320. doi: 10.1073/pnas.0501424102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Haeder S, Wirth R, Herz H, Spiteller D. Candicidin-producing Streptomyces support leaf-cutting ants to protect their fungus garden against the pathogenic fungus Escovopsis. Proc Natl Acad Sci U S A. 2009;106:4742–4746. doi: 10.1073/pnas.0812082106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Qin J, Li R, Raes J, Arumugam M, Burgdorf KS, Manichanh C, Nielsen T, Pons N, Levenez F, Yamada T, et al. A human gut microbial gene catalogue established by metagenomic sequencing. Nature. 2010;464:59–65. doi: 10.1038/nature08821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Peraud O, Biggs JS, Hughen RW, Light AR, Concepcion GP, Olivera BM, Schmidt EW. Microhabitats within venomous cone snails contain diverse actinobacteria. Appl Environ Microbiol. 2009;75:6820–6826. doi: 10.1128/AEM.01238-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Banik JJ, Brady SF. Cloning and characterization of new glycopeptide gene clusters found in an environmental DNA megalibrary. Proc Natl Acad Sci U S A. 2008;105:17273–17277. doi: 10.1073/pnas.0807564105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Donia MS, Hathaway BJ, Sudek S, Haygood MG, Rosovitz MJ, Ravel J, Schmidt EW. Natural combinatorial peptide libraries in cyanobacterial symbionts of marine ascidians. Nat Chem Biol. 2006;2:729–735. doi: 10.1038/nchembio829. [DOI] [PubMed] [Google Scholar]
- 34.Schmidt EW, Donia MS. Chapter 23. Cyanobactin ribosomally synthesized peptides--a case of deep metagenome mining. Methods Enzymol. 2009;458:575–596. doi: 10.1016/S0076-6879(09)04823-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Ziemert N, Ishida K, Weiz A, Hertweck C, Dittmann E. Exploiting the natural diversity of microviridin gene clusters for discovery of novel tricyclic depsipeptides. Appl Environ Microbiol. 2010;76:3568–3574. doi: 10.1128/AEM.02858-09. Gene sequences encoding 15 new predicted microviridin variants were PCR amplified both from cultured laboratory strains of Microcystis, as well as from uncultured, field collected isolates.
- 36. Li B, Sher D, Kelly L, Shi Y, Huang K, Knerr PJ, Joewono I, Rusch D, Chisholm SW, van der Donk WA. Catalytic promiscuity in the biosynthesis of cyclic peptide secondary metabolites in planktonic marine cyanobacteria. Proc Natl Acad Sci U S A. 2010;107:10430–10435. doi: 10.1073/pnas.0913677107. An examination of publicly available metagenomic sequencing data led to the identification of new lantiobiotic cyclases.
- 37.Quinton CM, Stephanie GB, Don AC. Subtractive hybridization magnetic bead capture: A new technique for the recovery of full-length ORFs from the metagenome. Biotechnology Journal. 2007;2:36–40. doi: 10.1002/biot.200600156. [DOI] [PubMed] [Google Scholar]
- 38.Chew YV, Holmes AJ. Suppression subtractive hybridisation allows selective sampling of metagenomic subsets of interest. Journal of Microbiological Methods. 2009;78:136–143. doi: 10.1016/j.mimet.2009.05.003. [DOI] [PubMed] [Google Scholar]
- 39.Morimoto S, Fujii T. A new approach to retrieve full lengths of functional genes from soil by PCR-DGGE and metagenome walking. Appl Microbiol Biotechnol. 2009;83:389–396. doi: 10.1007/s00253-009-1992-x. [DOI] [PubMed] [Google Scholar]
- 40.Gray KA, Richardson TH, Robertson DE, Swanson PE, Subramanian MV, Allen I, Laskin JWBaGMG. Advances in Applied Microbiology. Volume 52. Academic Press; 2003. Soil-Based Gene Discovery: A New Technology to Accelerate and Broaden Biocatalytic Applications; pp. 1–27. [DOI] [PubMed] [Google Scholar]
- 41.Kalyuzhnaya MG, Zabinsky R, Bowerman S, Baker DR, Lidstrom ME, Chistoserdova L. Fluorescence In Situ Hybridization-Flow Cytometry-Cell Sorting-Based Method for Separation and Enrichment of Type I and Type II Methanotroph Populations. Applied and Environmental Microbiology. 2006;72:4293–4301. doi: 10.1128/AEM.00161-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Zhang K, He J, Yang M, Yen M, Yin J. Identifying natural product biosynthetic genes from a soil metagenome by using T7 phage selection. Chembiochem. 2009;10:2599–2606. doi: 10.1002/cbic.200900297. Phage display was used to enrich a metagenomic library for PKS and NRPS gene sequences.
- 43.Morozova O, Marra MA. Applications of next-generation sequencing technologies in functional genomics. Genomics. 2008;92:255–264. doi: 10.1016/j.ygeno.2008.07.001. [DOI] [PubMed] [Google Scholar]
- 44.Chan CK, Hsu AL, Halgamuge SK, Tang SL. Binning sequences using very sparse labels within a metagenome. BMC Bioinformatics. 2008;9:215. doi: 10.1186/1471-2105-9-215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yang B, Peng Y, Leung HC, Yiu SM, Chen JC, Chin FY. Unsupervised binning of environmental genomic fragments based on an error robust selection of I-mers. BMC Bioinformatics. 2010;11 Suppl 2:S5. doi: 10.1186/1471-2105-11-S2-S5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Glass EM, Wilkening J, Wilke A, Antonopoulos D, Meyer F. Using the metagenomics RAST server (MG-RAST) for analyzing shotgun metagenomes. Cold Spring Harb Protoc. 2010;2010 doi: 10.1101/pdb.prot5368. pdb prot5368. [DOI] [PubMed] [Google Scholar]
- 47.Komatsu M, Uchiyama T, Omura S, Cane DE, Ikeda H. Genome-minimized Streptomyces host for the heterologous expression of secondary metabolism. Proc Natl Acad Sci U S A. 2010;107:2646–2651. doi: 10.1073/pnas.0914833107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wang HH, Isaacs FJ, Carr PA, Sun ZZ, Xu G, Forest CR, Church GM. Programming cells by multiplex genome engineering and accelerated evolution. Nature. 2009;460:894–898. doi: 10.1038/nature08187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Vogel TM, Simonet P, Jansson JK, Hirsch PR, Tiedje JM, van Elsas JD, Bailey MJ, Nalin R, Philippot L. TerraGenome: a consortium for the sequencing of a soil metagenome. Nat Rev Micro. 2009;7:252–252. [Google Scholar]