Abstract
CD4+ T-cell depletion is a characteristic of human immunodeficiency virus type 1 (HIV-1) infection. In this study, modulation of mRNA expression of 6800 genes was monitored simultaneously at eight time points in a CD4+ T-cell line (CEM-GFP) during HIV infection. The responses to infection included: (1) >30% decrease at 72 h after infection in overall host-cell production of monitored mRNA synthesis, with the replacement of host-cell mRNA by viral mRNA, (2) suppression of the expression of selected mitochondrial and DNA repair gene transcripts, (3) increased expression of the proapoptotic gene and its gene p53-induced product Bax, and (4) activation of caspases 2, 3, and 9. The intense HIV-1 transcription resulted in the repression of much cellular RNA expression and was associated with the induction of apoptosis of infected cells but not bystander cells. This choreographed host gene response indicated that the subversion of the cell transcriptional machinery for the purpose of HIV-1 replication is akin to genotoxic stress and represents a major factor leading to HIV-induced apoptosis.
The depletion of CD4+ T cells during human immunodeficiency virus type 1 (HIV-1) infection has been attributed to numerous mechanisms including apoptosis (Ameisen et al. 1991; Laurent-Crawford et al. 1991; Terai et al. 1991). Apoptosis is invariably accompanied by a disruption of inner mitochondrial transmembrane potential (Castedo et al. 1995). This contributes to the release of cytochrome c sequestered in the mitochondrion into the cytosol. In the cytoplasm, cytochrome c combines with APAF-1 and caspase 9 to constitute the apoptosome, which is responsible for the initiation of the caspase cascade triggering the intrinsic apoptosis pathway (Green et al. 1998; Roulston et al. 1999).
To analyze the effects of HIV-1 infection on T-cell transcription, we used a cell line to minimize heterogeneous host-cell responses to infection and to allow the precise enumeration of infected cells. CEM-GFP cells are a CD4+ lymphoblastoid T-cell line modified to express the green fluorescent protein upon productive HIV-1 infection (Gervaix et al. 1997). When infected by HIV-1, this cell line undergoes characteristic changes including cell-cycle retardation in G2/M, apoptosis, and high levels of virion production.
RESULTS
CEM-GFP cells were inoculated with HIV-1LAI at a multiplicity of infection of 0.5, an inoculum sufficient to ensure that every cell is contacted by virus particles. Aliquots of cells were obtained at 0.5, 2, 4, 8, 16, 24, 48, and 72 h after infection. A mock infection served as a control at each time point, essentially replacing the volume of viral input by an equivalent volume of culture medium from uninfected cells. Each sample was tested on two chips and the average taken. Normalization was done using global normalization and scaling.
At 72 h, HIV-1 transcripts represented more than 30% of all mRNAs present (identical amounts of labeled RNA for control or infected conditions were loaded onto the arrays). This was determined by assessing the level of GAPDH. The average difference value for GAPDH (spanning the whole length of the gene and representing the summation of 198 independent probes) at 30 min in the HIV-infected cells was 18 462 ±2027 compared to 12 246 ±1318 at 72 h. A similar reduction also was corroborated with β-actin probes. This reduction of mRNA expression is consistent with previously published results (Somasundaran et al. 1988). This intense viral replication was confirmed in the CEM-GFP cells and in primary-activated CD4+ T lymphocytes as monitored by real-time kinetic RT-PCR. The levels of transcription representative of all doubly and singly spliced (tat and vpr, respectively) as well as all unspliced (gag) messages were measured. Fold increases in the levels of expression of each of these three HIV-1 mRNAs were determined at 48 and 72 h for both infected CEM-GFP cells and primary CD4+ T cells and compared to that obtained at 24 h. The increased expression was similar in both cell types except that gag was expressed at higher levels in the CEM-GFP cells (Table 1). Tat was the gene with the greatest increase in expression, being up-regulated 315-fold at 72 h when compared to the level determined at 24 h.
Table 1.
Expression of HIV Transcripts
| HIV transcript | Cell type | 24 h | 48 h | 72 h |
|---|---|---|---|---|
| tat | Primary | 1 | 12.9 | 315.6 |
| CEM-GFP | 1 | 9.4 | 211.0 | |
| vpr | Primary | 1 | 8.0 | 35.5 |
| CEM-GFP | 1 | 26.3 | 82.3 | |
| gag | Primary | 1 | 7.1 | 21.8 |
| CEM-GFP | 1 | 23.2 | 193.6 |
Real-time PCR quantification of tat, vpr, and gag. The level obtained at 24 h was arbitrarily given a value of 1.
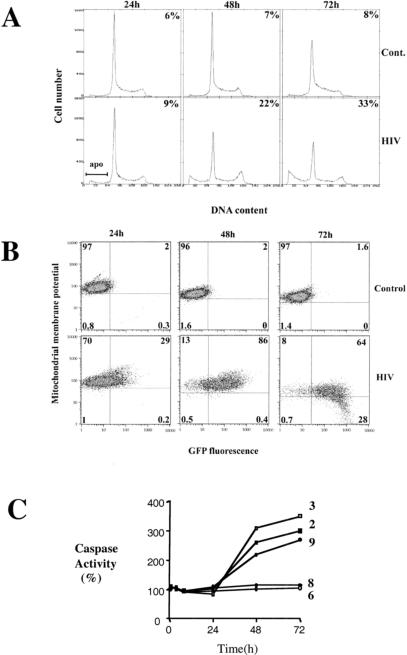
The percentages of CEM-GFP cells productively infected, the level of apoptosis, and cell-cycle profiles were assessed and collected at 24, 48, and 72 h after infection using flow cytometry. At 24 h, 29% of the cells were HIV-1-infected as reflected by green fluorescent protein (GFP) expression with ∼9% of the cells apoptotic as determined by the proportion of cells with sub-G1 DNA content. At 48 h, 86% of the cells were GFP positive and 22% apoptotic, while at 72 h, 92% were GFP positive and 33% apoptotic (Fig. 1A,B). Orange-fluorescent tetramethylrosamine was used to detect membrane potential changes in mitochondria, a marker of induction of apoptosis. Mitochondrial membrane potential was reduced over time, specifically in HIV-1-infected cells as defined by the intersection of GFP positivity and low orange-fluorescent tetramethylrosamine signal (CMTMRos, bottom right quadrant in Fig. 1B). Uninfected cells in the inoculated culture retained mitochondrial membrane potential, a clear indication that HIV-1 infection altered the membrane potential of mitochondria in infected but not bystander cells. At 72 h, 30% of GFP-positive cells had low mitochondrial transmembrane potential as measured by CMTMRos staining in contrast to <1% of GFP-negative cells (Fig. 1B). To confirm the apoptotic phenotype, caspase activity was assessed. Caspases 2, 3, and 9 but not 6 and 8 were found to be activated in HIV-1 inoculated cell population (Fig. 1C).
Figure 1.
Analysis of DNA content, cell cycle, percentage of infected cells, mitochondrial membrane potential, and caspase activity. (A) Cell cycle analysis and quantification of apoptosis. Control (mock-infected) and human immunodeficiency virus type 1 (HIV-1) -infected CEM-GFP cells were processed after 24 h, 48 h, and 72 h for DNA content using propidium iodide staining. An assessment of the percentage undergoing apoptosis (upper right corners) was performed by estimating the proportion of cells with subgenomic content of DNA (labeled apo in the figure). The percentages for 24 h, 48 h, and 72 h were 9%, 22%, and 33%, respectively. (B) Determination of the percentage of HIV-infected cells and evaluation of the mitochondrial membrane potential (Δψm). The percentage of GFP-positive cells (cumulative of the percentage present in top and bottom right quadrants) is an assessment of the number of cells productively infected. The percentages for 24 h, 48 h, and 72 h were 29%, 86%, and 92%, respectively. Cells were gated initially using forward and side scatter criteria to exclude dead cells. Orange-fluorescent tetramethylrosamine was used to assess the mitochondrial transmembrane potential. Gating on live cells only, 28% of cells that were GFP positive (92%) showed reduced staining at 72 h. (C) Cells were infected with HIV-1LAI at 0.5 MOI. At the indicated time points, 2 × 106 cells were washed with PBS and lysed in caspase buffer. Caspase activity of 10–20 μg of total protein was measured with specific substrates for caspase-2 (█), 3 (□), 6 ( ), 8 (⧫) and 9 (●) (100 μM) after 1 h incubation at 37°C.
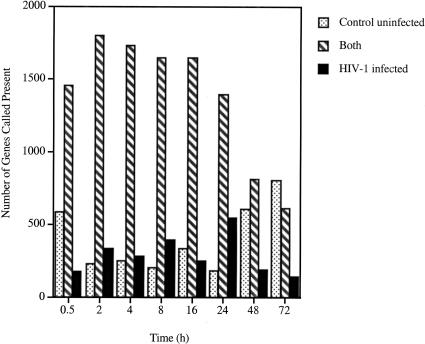
Total RNA isolated at each time point for both control and infected CEM-GFP was reverse transcribed, and labeled RNA (cRNA) was generated. cRNA (15μg) was hybridized to HuGeneFL 6800 array. Genes expressed in infected or uninfected cell populations and those common to both were determined for each of the eight time points using GeneChip analysis suite 3.1 and 2HAPI (High-density Array Pattern Interpreter) software packages (Fig. 2). At 30 min after inoculation, 181 genes were expressed only in the HIV-inoculated cells, representing genes that were induced by HIV-1 binding or entry. In contrast, 588 genes were expressed only in the control population, representing genes that are suppressed by exposure to HIV-1. Most of the modulation in gene expression affected genes expressed at lower copy number (the data can be viewed and queried at our 2HAPI web site located at http://2hapi.ucsd.edu). A general downregulation of gene expression occurred over time, especially in the HIV-1 infected population. At 72 h, transcripts of 760 genes were called present in the HIV-1-infected cells (summation of genes expressed solely in HIV-infected cells and genes common to both control and HIV-infected cells) and 805 genes were detected in the control cells representing a downmodulation compared to the earlier time points. Two independent processes are operating; in the infected population, the cells are killed by HIV infection and in the control, the culture reaches confluence. This also was confirmed by the hierarchical cluster analysis expressing ratios of expression of HIV over controls. An impressive shutdown of numerous genes was observed.
Figure 2.
Genes expressed in control uninfected and HIV-1-infected cells. Stringent criteria were used for calling a gene present (i.e., a gene that can be reliably detected and quantitated with accuracy). A general down-regulation, especially with the HIV-infected population, can be noted. Genes that are expressed in control only increase from 184 at 24 h to 805 at 72 h, while genes specific for HIV-infected cells number 549 at 24 h and 148 at 72 h. Genes expressed in both populations diminish over time as more genes are shut down in the HIV-infected cell population.
Genes expressed early in HIV-1 infection (0.5, 2, 4, and 8 h) included some that were characteristic of cellular defense mechanisms. These included members of the interferon alpha family (accession no. J00212) at 2 h, 4 h, and 8 h, and MxB (accession no. M30818), an antiviral interferon-alpha/beta-inducible protein belonging to the family of large GTPases, at 30‘ and 2 h (Melen et al. 1996). Genes were grouped in clusters according to patterns of coordinated expression over time (Tamayo et al. 1999). A group of 12 representative clusters represented the best fit with the most distinctive patterns of expression (see supplemental information Fig. A). Cluster 9 shows an impressive increase in later time points for 333 genes. Conversely, cluster 3 represents genes that were down-regulated with time (421 genes).
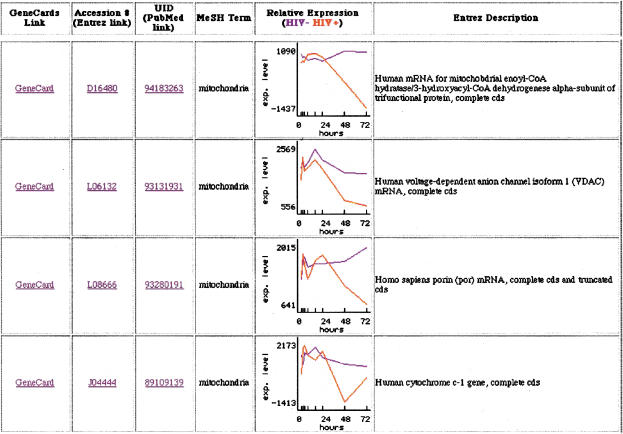
The similarities present in each cluster group based on the enumeration of Medical Subject Heading (MeSH) keywords were determined using 2HAPI. This approach also revealed a dysregulation in gene expression associated with keywords such as mitochondria (52 genes), apoptosis (75 genes), p53 (18 genes), and repair (38 genes). Genes in the mitochondrial category included mitochondrial 3-ketoacyl-CoA thiolase beta-subunit of trifunctional protein, a gene located in the mitochondrial matrix (D16481), human cytochrome c-1, located in the mitochondrial inter membrane space (J04444), ATP synthase alpha subunit found in the mitochondrion inner membrane (D14710), cytochrome bc-1 complex core protein II localized in the matrix side of the inner mitochondrial membrane (J04973), aconitase (U80040), and porin, which forms a channel through the mitochondrial outer membrane (L08666). All were down-regulated significantly in later time points, indicative of mitochondrial dysfunction. A partial list can be found in Fig. 3. Genes involved in DNA repair and apoptosis also were significantly repressed. These include DNA-PK (U35835), a DNA damage-activated enzyme responsible for phosphorylating p53 (Cuddihy et al. 1999), HSP70 (L12723), a heat-shock protein capable of inhibiting apoptosis, and HHR23A (D21235), which has been shown to bind to HIV-1 vpr (Withers-Ward et al. 1997; Gragerov et al. 1998). In contrast, GADD45 (growth arrest and DNA-damage-inducible protein: M60974) was slightly elevated.
Figure 3.
Selected mitochondrial-associated genes modulated by human immunodeficiency virus (HIV) infection. (Blue) control condition; (red) HIV-infected cells. An additional link to GeneCard, Entrez, and PubMed also are displayed with the Entrez description for the gene. Clicking on the graph itself provides the average difference values and call made by the initial GeneChip analysis suite software. Average difference values of <35 reflects that the gene is not present (includes negative values).
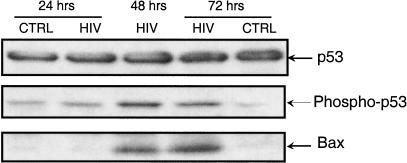
These observations indicated DNA damage or genotoxic stress and prompted an evaluation of the components of DNA repair pathway in the infected cell population at the protein level. p53 was phosphorylated in HIV-1 infected cells (Fig. 4). p53 acts as an inducer of the potent proapoptotic gene Bax (Miyashita et al. 1995; Marzo et al. 1998), which was up-regulated only in HIV-infected cells (Smith et al. 1994). DNA-damage inducible gene Gadd45 also was up-regulated (Zhan et al. 1999). Human mdm2, an inhibitor of p53 function, was down-regulated, possibly leading to an increase in p53 function and GADD45, a repair-sensing protein, was elevated in the infected population (results not shown). This enhanced expression of p53 protein and its stabilization by phosphorylation may be attributable to DNA breaks induced by the integration of the HIV-1 genome into the host genome, the activation of the NFkB pathway (Wu et al. 1994), or the general genotoxic stress induced by HIV-1 infection. The transcription of most genes ultimately was down-regulated by HIV-1 infection. One of the down-regulated genes has been reported previously by Geiss and collaborators (Geiss et al. 2000). The gene prothymosin alpha (M26708) that is expressed to high levels was down-regulated to the same extent in our own experiments. However, we saw a twofold expression enhancement at 30 min after infection. This points out the importance of determining the expression for multiple time points. The general downregulation contrasts with cytomegalovirus infection of primary human fibroblasts (Zhu et al. 1998) for which remarkable amplification of genes resulted from cytomegalovirus infection before the demise of the target cell. Another analysis of the process of HIV infection was performed using differential display by Ryo et al. (2000). Interestingly, they also suggest that HIV exerts a strong cytopathic effect on the host.
Figure 4.
Western blot for p53, phosphorylated p53, and BAX. p53 concentration remained constant but phosphorylated p53 was markedly increased in HIV 1 at 48 and 72 h, a situation mimicked for BAX.
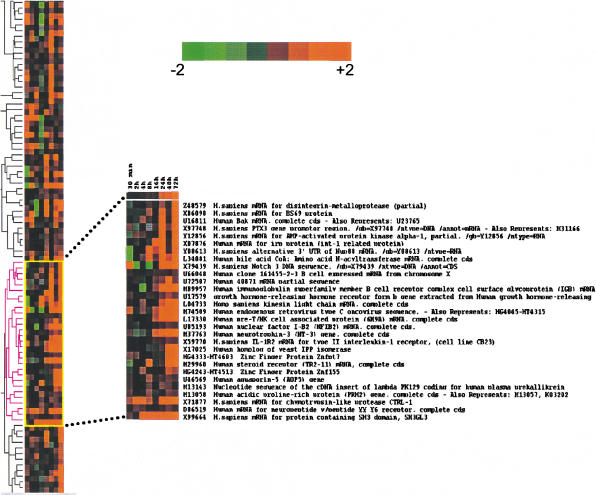
Using the cluster and treeview applications of Eisen et al. (1998), we ordered the genes for similarity of expression over time. The resulting ratios of expression for HIV over control were clustered. A section of the hierarchical tree is shown in Figure 5, representing a subset of genes whose expression levels differed by at least fourfold between HIV and control. We focused on a section where expression was increased in HIV-infected cells at the later time point (16 h to 72 h). A selection of these also is shown in supplemental Figure B in our 2HAPI format. Interestingly, several genes were significantly up-regulated.
Figure 5.
Hierarchical cluster analysis. We selected genes with average difference values >50 for at least four out of the eight time points. Also, the ratio of the average difference of HIV over control had to be ≥2 (422 genes met the criteria). The data then were processed using Cluster. The values were log transformed, and self-organizing map was applied (100,000 iterations) prior to generating the hierarchical clusters. The display software Treeview was used to render the figure. Cross section of the tree and magnification of a subset of the hierarchical cluster showing genes that clustered with NFIB-2. These selected genes were overexpressed as compared to control at the later time points. Intensity of color from bright green to bright red (log scale) is shown. (Green) overexpressed in control; (red) overexpressed in HIV-infected population.
One such gene identified by the 2HAPI overlap function and the hierarchical clustering was Nuclear Factor I (NFIB-2). This gene was highly expressed at all the later time points (16, 24, 48, and 72 h) during the process of HIV-1 infection. NFIB-2 is part of a family of dimeric DNA-binding proteins with very similar, possibly identical, DNA-binding specificity. These proteins are required for cell-specific transcription of many viral and cellular genes as cellular transcription factors, and as replication factors for adenovirus DNA replication (Qian et al. 1995; Chaudhry et al. 1997). NFIB-2 also has been identified as a recurrent translocation partner gene of high-mobility-group IC (HMGIC) in pleomorphic adenomas (Geurts et al. 1998). This increase in NFIB-2 expression was confirmed using real-time RT-PCR; the values were 3.5-fold greater at 24 h, 10-fold greater at 48 h, and 505-fold greater at 72 h when compared to the mock-infected controls. This increase is remarkable in the face of a general down-regulation of host-cell mRNA expression in infected cells. Interestingly, expressing this factor constitutively in a Jurkat cell line resulted in decreased production of virus over time as monitored by p24 expression possibly resulting from a reduction in surface expression of the CD4 receptor (data not shown). The role of this factor remains to be fully elucidated in the context of HIV infection. Additionally, this observation was confirmed in primary CD4+ T lymphocytes infected with HIV-1 with a 55-fold increase of this transcript at 24 h (data not shown). Real-time RT-PCR also confirmed that Bax and GADD45 were up-regulated, 2- and 2.1-fold respectively, compared to control cells at 72 h. Bax also was shown t be significantly up-regulated by Western-blot analysis (Fig. 4). Other genes of interest were: SH3GL3, reported to be involved in selective neuronal cell death (Sittler et al. 1998), the type II receptor for interleukin (IL) 1, characteristic of inflammation processes, CD79B (M89957) associated with the immunoglobulin gene superfamily, human pre-T/NK associated protein (CD73) involved in signal transduction associated with increased purine salvage, and the generation of adenosine (Ado) for Ado-receptor signaling (Peola et al. 1996).
DISCUSSION
This study shows that HIV-1 infection modulates the transcriptional levels of a large number of host genes. Many genes are either suppressed to undetectable levels or are significantly up-regulated very soon after virus inoculation and before the initiation of virus transcription. The changes involved in the culture system (infected and mock infected) are all part of the process encompassing the response of the target cells to productive HIV infection. The mock-infected cells grew normally and cycled normally as seen in Figure 1A, but the HIV-infected cells diverged significantly from the normal cycling phenotype and displayed extensive apoptosis. Fluctuations in gene expression are expected even in normally cycling cells because of differences in proliferation and generation of different effectors during the course of the experiments. The experiments were done in a cell line expressing the green fluorescent protein, however, we controlled that GFP had little to no impact on the target cell (using cell cycle analysis as in Fig. 1A) compared to the virus by constitutively expressing the protein in the CEM-GFP cell line (results not shown). HIV-1 transcription usurps cellular transcription to such an extent that a marked diminution of host-cell transcription is observed. Changes in transcription include genes that may be essential for mitochondrial function and integrity (Chen et al. 2000) and to DNA repair mechanisms leading directly to the induction of apoptosis of infected cells. This situation applies to productively infected CD4+ T-cell lines and possibly in the acute events following infection in HIV-infected individuals when virus titers are high.
Discovery-driven approaches in genomics using high-density gene expression microarrays have permitted the definition of new avenues of research and the identification of pathways and signal transduction events pertinent to the process of HIV-1 infection. High-density microarray interrogation of gene expression represents a powerful tool to dissect the relationships between an infectious agent and its host.
METHODS
Cell Culture and Viral Infection
The CEM-GFP CD4+ lymphoblastoid T-cell line was developed as a reporter line to monitor productive HIV infection. A plasmid consisting of the HIV-1 LTR driving the expression of the GFP was introduced in the CEM parental cell line. Upon productive infection, cells express GFP, which can be monitored by flow cytometry. Virus stocks of HIV-1LAI were produced and titered in the same cell line and infectivity determined by the method of Karber. CEM-GFP cells were infected at a multiplicity of infection of 0.5 in the presence of polybrene (2 μg/mL) added to both mock and HIV-infected cells. A mock-infected control was processed using supernatant of cultured CEM-GFP cells. All the culture medium (for both mock and infected culture) was removed and replaced with fresh media after the initial 2-h incubation. Aliquots of cells were taken at 0.5, 2, 4, 8, 16, 24, 48, and 72 h after infection for both populations. Cells were washed once with PBS, resuspended in 250 μL of PBS to which 750 μL of Trizol (Gibco-BRL) was added, and stored at −80°C.
CD4+ Lymphocyte Isolation and Infection
CD4+ lymphocytes were isolated from ∼300 × 106 PBMC using the Vario MACS CD4+ Cell Isolation Kit and Depletion Columns (Miltenyi Biotec) according to the manufacturer’s protocol. Purified primary CD4+ T cells were infected with the same strain of HIV-1 at the same multiplicity of infection, two days after stimulation with PHA and addition of IL-2 ( 20 unit/mL). Aliquots were taken at 2, 24, 48, and 72 h after infection. Cellular and viral messages were quantified using real-time RT-PCR quantification.
Cell Cycle, Apoptosis, and Mitochondrial Membrane Potential Assays
DNA Fragmentation and Cell-Cycle Analysis
Cells (2 × 106) were washed in PBS, resuspended in 30% ethanol in PBS, and kept at 4°C until analyzed. Cells then were treated with RNAse A (100 μg/mL) and stained with 50 μg/mL of propidium iodide for 1 h at 37°C. Cells were analyzed by flow cytometry as previously described (Telford et al. 1992).
Mitochondrial Transmembrane Potential
Cell pellets were resuspended in 100 nM of orange-fluorescent tetramethylrosamine (CMTMRos, Molecular Probes) and incubated at 37°C for 30 min under the normal growth conditions. Cells were washed and fixed in 1% formaldehyde in PBS and analyzed by flow cytometry (Poot et al. 1999).
Caspase Activity
Cells were resuspended in caspase buffer (50 mM Hepes, pH 7.4, 100 mM NaCl, 1 mM EDTA, 0.1% Chaps, and 5 mM dithiothreitol) for 10 min at 4°C. Lysates (10–20 μg of total protein) were mixed with 50 μL of HEB buffer (PIPES 50 mM, KCl 20 mM, EGTA 5 mM, MgCl2 2 mM, and DTT 1 mM, pH 7), and reactions were initiated by addition of 100 μM of the specific substrate. After 1 h incubation at 37°C, caspase-3-like protease activity was measured by monitoring the digestion of its substrate Ac-DEVD-AMC, caspase-9-like activity using Ac-LEHD-AFC, caspase-6-like protease activity with the substrate Ac-VEID-AMC, caspase-8-like activity with Ac-IETD-AFC, and caspase-2-like activity with Ac-VDVAD-AFC (Calbiochem). Activity was quantified by the release of 7-amino-4-trifluoromethyl-coumarin (AFC) or 7-amino-4-methyl-cumarin (AMC) monitoring fluorescence at excitation and emission wavelengths of 400 and 505 nm, and 380 and 460 nm, respectively, using a fluorimeter (Cytofluor 2000, Millipore).
Immunoblots
Immunoblot analysis was performed on total-cell lysates of 5 × 106 cells in 100 μL RIPA buffer (50 mM TrisHCl, pH 7.4, 50 mM NaCl, 1 mM EGTA, 20 mM glycerophosphate, 0.5% (v/v) Nonidet P-40, 1 mM Na3VO4, 1 mM NaF, 1 μg/mL Aprotinin, 1 μg/mL leupeptin and 1 mM phenylmethanesulfonyl fluoride (PMSF) at the indicated time points (24, 48, and 72 h). Cell extracts (30 μg per lane) were resolved by SDS-PAGE on 8%–20% gels and transferred on PVDF membranes. The indicated antibodies, p53 and phosphospecific p53(S15) (New England Biolabs, and BAX (N20) (Santa Cruz Biotechnology) were visualized by alkaline phosphatase-based enhanced chemiluminescence.
Expression Analysis
The preparation of labeled RNA (cRNA) was performed as described by Affymetrix . The procedure to prepare material for hybridization to the chips involved multiple steps (for more details see our web site at: http://genomics.ucsd.edu).We used 10 million cells per time point and condition. Equal amounts of labeled cRNA (15 μg) were used per GeneChip array. Two chips were used for every time point and condition and the average used for all subsequent analysis. In the case of the HIV-1-infected sample, the viral mRNA contributes significantly to the total RNA loaded into the arrays; however, global normalization and scaling takes account of this contribution. Average difference (intensity of fluorescence) was plotted for duplicate experiments. The variability was within twofold. This was used as our reference for calling a modulation significant (more than twofold). The level of gene expression was normalized for all conditions. The global normalization is a computational technique in which the output of the experimental array is multiplied by a factor (normalization factor) to make the average intensity equivalent to the average intensity of all the other arrays used in the multiple chip experiment. This is based on all probe sets present on the chip. The output yields an intensity of expression and with this and other parameters, a determination of the relative number of copies for a specific mRNA transcript for which an absolute call was made. The data were processed using GeneChip analysis software suite 3.1 (Affymetrix) and then imported into 2HAPI for further analysis (http://array.sdsc.edu).
Real-Time RT-PCR Quantification
The level of expression of specific transcripts were determined using the Perkin-Elmer ABI Prism 7700 (TaqMan) and Sequence Detection System software. Total RNA was isolated using the TRIzol Method (Gibco-BRL) and then digested with deoxyribonuclease to remove any contaminating genomic DNA. Five micrograms of total RNA was used to generate cDNA using a T7-poly dT oligodeoxynucleotide primer (GGC CAG TGA ATT GTA ATA CGA CTC ACT ATA GGG AGG AGG-T24) following the protocol for SuperScript II (Gibco-BRL). Equal amounts of cDNA were used in triplicate and amplified with the Taqman Master Mix provided by Perkin-Elmer. Amplification efficiencies were validated and normalized against GAPDH and fold increases were calculated using the Comparative CT Method for quantitation. The primers and probes used follow: NFIB-2 forward-TCT CAC CAA CGA AGG CTA GGA, reverse-GCT TAT AAA ATG GCT GGC TCA TG probe-CGG CGT CAG AGA TGC TGG GTG A; Bax-alpha forward-CTG ATC AGA ACC ATC ATG GGC, reverse-GAG GCC GTC CCA ACC AC, probe-TCC GGG AGC GGC TGT TGG G, and GADD45 forward-TCT GCA GAT CCA CTT CAC CCT, reverse-GCT GAC GCG CAG GAT GTT, probe-TCC AGG CGT TTT GCT GCG AGA AC. The primers and probes are based on the HIV-1NL4–3 sequence: tat forward-GCC TTC ATT GCC AAG TTT GTT T, reverse-GTC GCT GTC TCC GCT TCT TC, probe-CAA GAA AAG GCT TAG GCA TCT CCT ATG GCA; vpr forward-GGC AGG AGT GGA AGC CAT AAT A, reverse-CTC TCC TCT GTC GAA TTA TGC CTA T, probe-AAT TCT GCA ACA ACT GCT GTT TAT TCA TTT CAG AA; gag forward-AAA AGA GAC CAT CAA TGA GGA AGC, reverse-TGG TGC AAT AGG CCC TGC, probe-CAG AAT GGG ATA GAT TGC ATC CAG TGC A.
Acknowledgments
We thank Drs. Joseph Wong and David Looney for comments and suggestions. This work was supported by the National Institute of Allergy and Infectious Diseases (AI46237 and AI47703), the Center for AIDS Research Genomics Core laboratory (AI36214), the Universitywide AIDS Research program and the San Diego Veterans Medical Research Foundation (J.C.) as well as NIH grants AI27670, AI38858, AI43638 and AI29164 (D.D.R.) and the San Diego Veterans Affairs Healthcare System.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL jcorbeil@ucsd.edu; FAX (858) 552-7445.
Article published on-line before print: Genome Res., 10.1101/gr.180201.
Article and publication are at http://www.genome.org/cgi/doi/10.1101/gr.180201.
REFERENCES
- Ameisen JC, Capron A. Cell dysfunction and depletion in AIDS: The programmed cell death hypothesis. Immunol Today. 1991;4:102–105. doi: 10.1016/0167-5699(91)90092-8. [DOI] [PubMed] [Google Scholar]
- Castedo M, Macho A, Zamzami N, Hirsch T, Marchetti P, Uriel J, Kroemer G. Mitochondrial perturbations define lymphocytes undergoing apoptotic depletion in vivo. Eur J Immunol. 1995;25:3277–3284. doi: 10.1002/eji.1830251212. [DOI] [PubMed] [Google Scholar]
- Chaudhry AZ, Lyons GE, Gronostajski RM. Expression patterns of the four nuclear factor I genes during mouse embryogenesis indicate a potential role in development. Dev Dyn. 1997;208:313–325. doi: 10.1002/(SICI)1097-0177(199703)208:3<313::AID-AJA3>3.0.CO;2-L. [DOI] [PubMed] [Google Scholar]
- Chen Q, Gong B, Almasan A. Distinct stages of cytochrome c release from mitochondrial evidence for a feedback amplification loop linking caspase activation to mitochondrial dysfunction in genotoxic stress induced apoptosis. Cell Death Differ. 2000;7:227–233. doi: 10.1038/sj.cdd.4400629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuddihy AR, Wong AH, Tam NW, Li S, Koromilas AE. The double-stranded RNA activated protein kinase PKR physically associates with the tumor suppressor p53 protein and phosphorylates human p53 on serine 392 in vitro. Oncogene. 1999;18:2690–2702. doi: 10.1038/sj.onc.1202620. [DOI] [PubMed] [Google Scholar]
- Eisen MB, Spellman PT, Brown PO, Botstein D. Cluster analysis and display of genome-wide expression patterns. Proc Natl Acad Sci. 1998;95:14863–14868. doi: 10.1073/pnas.95.25.14863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geiss GK, Bumgarner RE, An MC, Agy MB, van 't Wout AB, Hammersmarkm E, Carter VS, Upchurch D, Mullins JI, Katze MG. Large-scale monitoring of host cell gene expression during HIV-1 infection using cDNA microarrays. Virology. 2000;266:8–16. doi: 10.1006/viro.1999.0044. [DOI] [PubMed] [Google Scholar]
- Gervaix A, West D, Leoni LM, Richman DD, Wong-Staal F, Corbeil J. A new reporter cell line to monitor HIV infection and drug susceptibility in vitro. Proc Natl Acad Sci. 1997;94:4653–4658. doi: 10.1073/pnas.94.9.4653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geurts JM, Schoenmakers EF, Roijer E, Astrom AK, Stenman G, van de Ven WJ. Identification of NFIB as recurrent translocation partner gene of HMGIC in pleomorphic adenomas. Oncogene. 1998;16:865–872. doi: 10.1038/sj.onc.1201609. [DOI] [PubMed] [Google Scholar]
- Gragerov A, Kino T, Ilyina-Gragerova G, Chrousos GP, Pavlakis GN. HHR23A, the human homologue of the yeast repair protein RAD23, interacts specifically with Vpr protein and prevents cell cycle arrest but not the transcriptional effects of Vpr. Virology. 1998;245:323–330. doi: 10.1006/viro.1998.9138. [DOI] [PubMed] [Google Scholar]
- Green DR, Reed JC. Mitochondria and apoptosis. Science. 1998;281:1309–1312. doi: 10.1126/science.281.5381.1309. [DOI] [PubMed] [Google Scholar]
- Laurent-Crawford AG, Krust B, Muller S, Rivière Y, Rey-Cuille MA, Bechet JM, Montagnier L, Hovanessian AG. The cytopathic effect of HIV is associated with apoptosis. Virology. 1991;185:829–839. doi: 10.1016/0042-6822(91)90554-o. [DOI] [PubMed] [Google Scholar]
- Marzo I, Brenner C, Zamzami N, Jurgensmeier JM, Susin SA, Vieira HL, Prevost MC, Xie Z, Matsuyama S, Reed JC, et al. Bax and adenine nucleotide translocator cooperate in the mitochondrial control of apoptosis. Science. 1998;281:2027–2031. doi: 10.1126/science.281.5385.2027. [DOI] [PubMed] [Google Scholar]
- Melen K, Keskinen P, Ronni T, Sareneva T, Lounatmaa K, Julkunen I. Human MxB protein, an interferon-alpha-inducible GTPase, contains a nuclear targeting signal and is localized in the heterochromatin region beneath the nuclear envelope. J Biol Chem. 1996;271:23478–23486. doi: 10.1074/jbc.271.38.23478. [DOI] [PubMed] [Google Scholar]
- Miyashita T, Reed JC. Tumor suppressor p53 is a direct transcriptional activator of the human bax gene. Cell. 1995;80:293–299. doi: 10.1016/0092-8674(95)90412-3. [DOI] [PubMed] [Google Scholar]
- Peola S, Borrione P, Matera L, Malavasi F, Pileri A, Massaia M. Selective induction of CD73 expression in human lymphocytes by CD38 ligation: a novel pathway linking signal transducers with ecto-enzyme activities. J Immunol. 1996;157:4354–4362. [PubMed] [Google Scholar]
- Poot M, Pierce RC. Detection of apoptosis and changes in mitochondrial membrane potential with chloromethyl-X-rosamine. Cytometry. 1999;36:359–360. doi: 10.1002/(sici)1097-0320(19990801)36:4<359::aid-cyto12>3.0.co;2-v. [DOI] [PubMed] [Google Scholar]
- Qian F, Kruse U, Lichter P, Sippel AE. Chromosomal localization of the four genes (NFIA, B, C, and X) for the human transcription factor nuclear factor I by FISH. Genomics. 1995;28:66–73. doi: 10.1006/geno.1995.1107. [DOI] [PubMed] [Google Scholar]
- Roulston A, Marcellus RC, Branton PE. Viruses and apoptosis. Annu Rev Microbiol. 1999;53:577–628. doi: 10.1146/annurev.micro.53.1.577. [DOI] [PubMed] [Google Scholar]
- Ryo A, Suzuki Y, Arai M, Kondoh N, Wakatsuki T, Hada A, Shuda M, Tanaka K, Sato C, Yamamoto M, et al. Identification and characterization of differentially expressed mRNAs in HIV type 1-infected human T cells. AIDS Res Hum Retroviruses. 2000;16:995–1005. doi: 10.1089/08892220050058416. [DOI] [PubMed] [Google Scholar]
- Sittler A, Walter S, Wedemeyer N, Hasenbank R, Scherzinger E, Eickhoff H, Bates GP, Lehrach H, Wanker EE. SH3GL3 associates with the Huntingtin exon 1 protein and promotes the formation of polygln-containing protein aggregates. Mol Cell. 1998;2:427–436. doi: 10.1016/s1097-2765(00)80142-2. [DOI] [PubMed] [Google Scholar]
- Smith ML, Chen IT, Zhan Q, Bae I, Chen CY, Gilmer TM, Kastan MB, O'Connor PM, Fornace AJ., Jr Interaction of the p53-regulated protein Gadd45 with proliferating cell nuclear antigen. Science. 1994;266:1376–1380. doi: 10.1126/science.7973727. [DOI] [PubMed] [Google Scholar]
- Somasundaran M, Robinson HL. Unexpectedly high levels of HIV-1 RNA and protein synthesis in a cytocidal infection. Science. 1988;242:1554–1557. doi: 10.1126/science.3201245. [DOI] [PubMed] [Google Scholar]
- Tamayo P, Slonim D, Mesirov J, Zhu Q, Kitareewan S, Dmitrovsky E, Lander ES, Golub TR. Interpreting patterns of gene expression with self-organizing maps: Methods and application to hematopoietic differentiation. Proc Natl Acad Sci. 1999;96:2907–2912. doi: 10.1073/pnas.96.6.2907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Telford WG, King L E, Fraker P J. Comparative evaluation of several DNA binding dyes in the detection of apoptosis-associated chromatin degradation by flow cytometry. Cytometry. 1992;13:137–142. doi: 10.1002/cyto.990130205. [DOI] [PubMed] [Google Scholar]
- Terai C, Kornbluth RS, Pauza CD, Richman DD, Carson DA. Apoptosis as a mechanism of cell death in cultured T lymphoblasts acutely infected with HIV-1. J Clin Invest. 1991;87:1710–1715. doi: 10.1172/JCI115188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Withers-Ward ES, Jowett JB, Stewart SA, Xie YM, Garfinkel A, Shibagaki Y, Chow SA, Shah N, Hanaoka F, Sawitz DG, et al. Human immunodeficiency virus type 1 Vpr interacts with HHR23A, a cellular protein implicated in nucleotide excision DNA repair. J Virol. 1997;71:9732–9742. doi: 10.1128/jvi.71.12.9732-9742.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H, Lozano G. NF-kappa B activation of p53. A potential mechanism for suppressing cell growth in response to stress. J Biol Chem. 1994;269:20067–20074. [PubMed] [Google Scholar]
- Zhan Q, Antinore MJ, Wang XW, Carrier F, Smith ML, Harris CC, Fornace AJ., Jr Association with Cdc2 and inhibition of Cdc2/Cyclin B1 kinase activity by the p53-regulated protein Gadd45. Oncogene. 1999;18:2892–2900. doi: 10.1038/sj.onc.1202667. [DOI] [PubMed] [Google Scholar]
- Zhu H, Cong JP, Mamtora G, Gingeras T, Shenk T. Cellular gene expression altered by human cytomegalovirus: global monitoring with oligonucleotide arrays. Proc Natl Acad Sci. 1998;95:14470–14475. doi: 10.1073/pnas.95.24.14470. [DOI] [PMC free article] [PubMed] [Google Scholar]