Abstract
One new nucleoside derivative, named 3-acetyl-5-methyl-2′-deoxyuridine (1), along with two known compounds 3,5-dimethyl-2′-deoxyuridine (2) and 3-methyl-2′-deoxyuridine (3), were isolated from the cultures of Streptomyces microflavus. This strain was an associated actinomycete isolated from the marine sponge Hymeniacidon perlevis collected from the coast of Dalian (China). Their structures were elucidated by detailed NMR and MS spectroscopic analysis as well as comparison with literature data.
Keywords: actinomycete, Streptomyces microflavus, Hymeniacidon perlevis, deoxyuridine
1. Introduction
The marine environment, particularly with sponges, is a rich source of new bioactive metabolites—287 novel metabolites were isolated from marine sponges in 2008 [1]. The availability of biomass is a limiting factor for isolating marine natural products. The widespread isolation of typical microbial metabolites from sponges leads to the hypothesis that these metabolites are in fact the products of microbial metabolism [2]. The isolation of secondary metabolite-producing bacteria from sponges and of microbial secondary metabolism gene clusters from the metagenome of sponges has led to the general understanding that these metabolites are, in many cases, the products of microbial symbionts and are not derived from the microbial diet of sponges [3]. Thus, marine organism-associated microbes have been attracting increasing interest as potential sources of marine natural products in order to solve the supply shortage.
A number of reports have been published on the isolation of actinobacteria from marine organisms [4]. Screening bioactive substances from these marine-derived actinobacteria has yielded several new bioactive metabolites [5–8]. In our recent screening for new natural products, one novel deoxyuridine (1), along with two known nucleoside derivatives (2, 3), were isolated from Streptomyces microflavus strain No. HVG29 belonging to actinomycetes. To date, there have been no reports of deoxyuridine structures isolated from this species. We report herein the isolation and structure elucidation of 1–3.
2. Results and Discussion
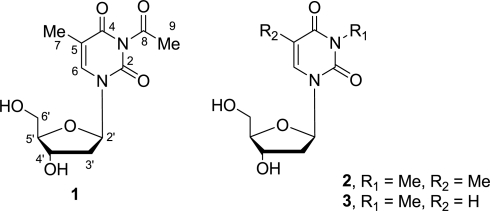
The total ethyl acetate extract from 30 L of fermentation broth was partitioned between hexane and 9:1 MeOH–H2O followed by diluting the aqueous layer to 3:2 MeOH–H2O and solvent partitioning with EtOAc. The EtOAc fraction was subjected to further purification by gel permeation on Sephadex LH-20 to yield compounds 1–3 (Figure 1).
Figure 1.
Structures of compounds 1–3.
Compound 1 was obtained as a white powder, [α]20D +5.4° (c 0.39, MeOH). The IR spectrum possessed absorptions for hydroxyl groups (3394 cm−1) and amide carbonyl groups (1693 cm−1). The EIMS gave a molecular ion peak at m/z 284 [M]+, and the molecular formula C12H16N2O6 was determined by HRESIMS at m/z 307.0905 [M + Na]+ (calcd. for C12H16N2O6Na, 307.0906).
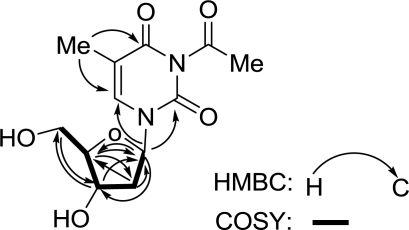
In the 1D NMR spectra, proton signals at δH 6.28 (1H, t, J = 7.0 Hz, H-2′), 2.22 (2H, m, H-3′), 3.90 (1H, q, J = 3.5 Hz, H-4′), 4.40 (1H, dt, J = 3.5, 5.9 Hz, H-5′), 3.79 (1H, dd, J = 3.2, 12.0 Hz, H-6′a), and 3.83 (1H, dd, J = 3.7, 12.0 Hz, H-6′b) and carbon resonances at δC 86.3 (CH, C-2′), 41.2 (CH2, C-3′), 72.2 (CH, C-4′), 88.9 (CH, C-5′), and 62.9 (CH2, C-6′) supported the presence of a pentose moiety in the molecule. The HMBC spectrum showed correlations from H-6′ to C-4′, from H-5′ to C-2′ and C-3′, from H-4′ to C-2′, from H-3′ to C-5′and C-2′, and from H-2′ to C-5′ and C-4′ (Figure 2). In addition, the correlations between H-2′/H2-3′, H2-3′/H-4′, H-4′/H-5′, and H-5′/H2-6′ were observed in the 1H-1H COSY spectrum (Figure 2). The above NMR evidence established the presence of a 2′-deoxyribose moiety, which was supported by the similarity of the 1H and 13C NMR data to those reported in the literatures for such a unit [9,10].
Figure 2.
HMBC and 1H-1H COSY correlations of compound 1.
The remaining portion of 1 implied the elemental composition C7H7N2O3, and accounted for the remaining 5 degrees of unsaturation. In the 1D NMR spectra, proton signals at δH 7.81 (1H, s, H-6) and 1.88 (3H, s, H-7) and carbon resonances at δC 152.4 (C-2), 166.4 (C-4), 111.5(C-5), 138.2 (C-6), and 12.4 (C-7) evidenced the presence of a thymine moiety in the molecule. An acetyl group was supported by proton signal at δH 1.88 (3H, s, H-9) and carbon signals at δC 23.4 (C-9) and 179.2 (C-8). In combination with the chemical shifts of the corresponding carbons and the elemental composition, the acetyl group was assigned to N-3 of the thymine moiety, because only N-3 had a vacancy in the molecule. Meanwhile, HMBC long-range correlations from H-2′ to C-2 and C-6 (Figure 2), clearly located the 2′-desoxyribose moiety to N-1 of the 3-acetyl-5-methylthymine unit. Therefore, the structure of 1 was determined as 3-acetyl-5-methyl-2′-deoxyuridine.
The structures of known compounds 2 and 3 were confirmed by detailed NMR data comparison with those in literatures [11]. To the best of our knowledge, compound 1 is the first example of acetyl deoxyuridine from marine-derived actinomycetes to be isolated from the marine sponge Hymeniacidon perlevis.
3. Experimental Section
3.1. General
NMR spectra were recorded at 500 and 125 MHz for 1H and 13C, respectively, on a Bruker Avance 500 NMR spectrometer in acetone-d6 with TMS as internal standard. Low and high resolution mass spectra were determined on Autospec Premier P776 and VG AutoSpec 3000 mass spectrometers. IR spectra were obtained on a JASCO FT/IR-4100 fourier transform infrared spectrometer. Column chromatography was performed with silica gel (200–300 mesh, Qingdao Haiyang Chemical Co., Qingdao, China) and Sephadex LH-20 (Pharmacia). TLC was carried out with precoated silica gel plates (GF-254, Qingdao Haiyang Chemical Co., Qingdao, China). All solvents were of analytical grade.
3.2. Microorganism Material
The sponge-associated actinobacterium Streptomyces microflavus was isolated from the inner tissue of the marine sponge Hymeniacidon perlevis collected from the inter-tidal beach of the Yellow Sea at Dalian, China (38°52′N, 121°41′E) in March 2003. Sponge specimens were placed in plastic bags containing seawater and immediately transported to the laboratory. Actinobacteria identification was carried out by the method reported by Zhang et al. [12]. The sequence data derived from the strain have been submitted and deposited at GenBank with accession number EU554304. BLAST search result showed that the sequence was similar (99%) to the sequence of S. microflavus (compared to EU273548). The strain is preserved within the biological technology department, Dalian Institute of Chemical Physics, Chinese Academy of Sciences.
3.3. Extraction and Isolation
For chemical investigations, the actinobacterium strain was cultivated statically in special media, which was reported by Xin et al. [13]. Mycelium and culture broth of S. microflavus (10 L) were homogenized and exhaustively extracted with MeOH and EtOAc, resp. Since the TLC and HPLC profiles of the two extracts were nearly identical, they were combined before further separation. The combined extracts (20 g) were subjected to column chromatography (CC) over SiO2 (200–300 mesh) and eluted with different solvents of increasing polarity to yield 11 fractions (Fr. 1–Fr. 11) on the basis of TLC analysis. Fr. 8 was further fractionated by CC on Sephadex LH-20 (CHCl3/MeOH, 2:1) and then purified by CC on Sephadex LH-20 (MeOH) to give compound 1 (4.0 mg). Fr. 11 was subjected to CC over SiO2 and then further purified by CC on Sephadex LH-20 (CHCl3/MeOH, 2:1) to yield compounds 2 (10.8 mg) and 3 (7.0 mg).
3-Acetyl-5-methyl-2′-deoxyuridine (1). White powder; [α]20D +5.4° (c 0.39, MeOH); UV (MeOH) λmax 264 (log ε) (3.52), λmax 256 (log ε) (3.46); 1H NMR (acetone-d6, 500 MHz) δ 7.81 (s, H-6), 6.28 (t, J = 7.0 Hz, H-2′), 4.40 (dt, J = 3.5, 5.9 Hz, H-5′), 3.90 (q, J = 3.5 Hz, H-4′), 3.83 (dd, J = 3.7, 12.0 Hz, H-6′b), 3.79 (dd, J = 3.2, 12.0 Hz, H-6a), 2.22 (m, H-3′), 1.88 (s, H-9), 1.88 (s, H-7); 13C NMR (acetone-d6, 125 MHz) δ 179.2 (C-8), 166.4 (C-4), 152.4 (C-2), 138.2 (C-6), 111.5 (C-5), 88.9 (C-5′), 86.3 (C-2′), 72.2 (C-4′), 62.9 (C-6′), 41.2 (C-3′), 23.4 (C-9), 12.4 (C-7). EIMS m/z (relative intensity): m/z 284 (5), 256 (9), 241 (5), 239 (9), 213 (15), 206 (9), 163 (11), 98 (43), 96 (80), 95 (74), 60 (100); HRESIMS m/z: 307.0905 [M + Na]+, calcd. for C12H16N2O6Na, 307.0906.
4. Conclusions
In previous research into metabolites isolated from S. microflavus, many fattiviracin derivatives and actinomycin X2 have been elucidated [14]. The major product of these fattiviracin derivatives was fattiviracin FV-8, which consists of four glucose and two trihydroxy fatty acid residues [15–17]. Actinomycin X2 included a wide range of fatty acids (C10–C22), which have been isolated from Streptomyces nasri [18]. Compounds 1–3 represent the first examples of deoxyuridine structures isolated from S. microflavus which was associated with sponges.
The known compounds (2 and 3) were originally isolated from a marine sponge [11], Geodia barretti, and now, for the first time, from sponge-associated microorganisms. This finding strengthens the hypothesis that marine microbial symbionts are possibly the true producers or take part in the biosynthesis of some bioactive marine natural products isolated from the marine organism hosts.
These nucleosides may have potent biological or physiological effects. A series of nucleoside analogues were synthesized and exhibited potential antiviral activities against duck hepatitis B virus (DHBV) [19], herpes simplex virus type 1 and 2 (HSV-1 and HSV-2), and varicella zoster virus (VZV) [20–22]. All of those synthesized 2′-deoxyuridine analogues have special substituents on position C-5 of uridine moiety. However, there is no evaluation for the compound which has the substituents on position N-3 of uridine residue. Compounds 1 and 2 provide new thoughts of introducing functional groups on nucleoside structures for synthetic medical chemistry.
Acknowledgments
This work was supported by the Major State Basic Research Development Program of China (973 Program) (No. 2009CB724700), the Knowledge Innovation Program of the Chinese Academy of Science (No. KSCX2-YW-G-073).
Footnotes
Samples Availability: Samples of compounds 1–3 are available from the authors.
References
- 1.Blunt JW, Copp BR, Munro MHG, Northcote PT, Prinsep MR. Marine natural products. Nat Prod Rep. 2010;27:165–237. doi: 10.1039/b906091j. [DOI] [PubMed] [Google Scholar]
- 2.Fortman JL, Sherman DH. Utilizing the power of microbial genetics to bridge the gap between the promise and the application of marine natural products. Chembiochem. 2005;6:960–978. doi: 10.1002/cbic.200400428. [DOI] [PubMed] [Google Scholar]
- 3.Kennedy J, Baker P, Piper C, Cotter PD, Walsh M, Mooij MJ, Bourke MB, Rea MC, O’Connor PM, Ross RP, et al. Isolation and analysis of bacteria with antimicrobial activities from the marine sponge Haliclona simulans collected from Irish water. Mar Biotechnol. 2009;11:384–396. doi: 10.1007/s10126-008-9154-1. [DOI] [PubMed] [Google Scholar]
- 4.Li Z. Advances in marine microbial symbionts in the China Sea and related pharmaceutical metabolites. Mar Drugs. 2009;7:113–129. doi: 10.3390/md7020113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lin Z, Antemano RR, Hughen RW, Tianero MDB, Peraud O, Haygood MG, Concepcion GP, Olivera BM, Light A, Schmidt EW. Pulicatins A–E, neuroactive thiazoline metabolites from cone snail-associate bacteria. J Nat Prod. 2010;73:1922–1926. doi: 10.1021/np100588c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Oh DC, Poulsen M, Currie CR, Clardy J. Sceliphrolactam, a polyene macrocyclic lactam from a wasp-associated Streptomyces sp. Org Lett. 2011;13:752–755. doi: 10.1021/ol102991d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Piel J, Butzke D, Fusetani N, Hui D, Platzer M, Wen G, Matsunaga S. Exploring the chemistry of uncultivated bacterial symbionts: Antitumor polyketides of the Pederin family. J Nat Prod. 2005;68:472–479. doi: 10.1021/np049612d. [DOI] [PubMed] [Google Scholar]
- 8.Izumikawa M, Khan ST, Takagi M, Shin-Ya K. Sponge-derived Streptomyces producing isoprenoids via the mevalonate pathway. J Nat Prod. 2010;73:208–212. doi: 10.1021/np900747t. [DOI] [PubMed] [Google Scholar]
- 9.Ashworth DJ, Chang C. Chemical modification of polynucleotides. Quantitiative studies of polycytidylic acid by nuclear magnetic resonance spectroscopy and secondary-ion mass spectrometry. J Org Chem. 1981;46:4770–4773. [Google Scholar]
- 10.Chang C, Gomes JD, Byrn SR. Chemical modification of deoxyribonucleic acids: A direct study by carbon-13 nuclear magnetic resonance spectroscopy. J Org Chem. 1983;48:5151–5160. [Google Scholar]
- 11.Lidgren G, Bohlin L. Studies of Swedish marine organisms, part X. biologically active compounds from the marine sponge Geodia Baretti. J Nat Prod. 1988;51:1277–1280. [Google Scholar]
- 12.Zhang H, Lee YK, Zhang W, Lee HK. Culturable actinobacteria from the marine sponge Hymeniacidon perleve: Isolation and phylogenetic diversity by 16S rRNA gene-RFLP analysis. Antonie van Leeuwenhoek. 2006;90:159–169. doi: 10.1007/s10482-006-9070-1. [DOI] [PubMed] [Google Scholar]
- 13.Xin Y, Wu P, Deng M, Zhang W. Phylogenetic diversity of the culturable rare actinomycetes in marine sponge Hymeniacidon perlevis by improved isolation media. Acta Microbiol Sin. 2009;49:859–866. [PubMed] [Google Scholar]
- 14.Cao VT, Phan VK, Bui VH. Actinomycin X2, an antibiotic component from the antibiotic biosynthesis fermentation by. Streptomyces microflavus Tap Chi Hoa Hoc. 2010;48:469–474. [Google Scholar]
- 15.Uyeda M, Yokomizo K, Miyamoto Y, Habib E. Fattiviracin A1, a novel antiherpetic agent produced by Streptomyces microflavus strain No. 2445. I. Taxonomy, fermentation, isolation, physicochemical properties and structure elucidation. J Antibiot. 1998;51:823–828. doi: 10.7164/antibiotics.51.823. [DOI] [PubMed] [Google Scholar]
- 16.Yokomizo K, Miyamoto Y, Nagao K, Kumagae E, Suzuki K, Harada S, Uyeda M. Fattiviracin A1, a novel antiviral agent produced by Streptomyces microflavus strain No. 2445. II. Biological properties. J Antibiot. 1998;51:1035–1039. doi: 10.7164/antibiotics.51.1035. [DOI] [PubMed] [Google Scholar]
- 17.Habib E, Yokomizo K, Murata K, Uyeda M. Structures of fattiviracin family, antiviral antibiotics. J Antibiot. 2000;53:1420–1423. doi: 10.7164/antibiotics.53.1420. [DOI] [PubMed] [Google Scholar]
- 18.el-Naggar MY, el-Kersh MA, el-Sharaky AS. Correlation of actinomycin X2 to the lipid profile in static and shaken cultures of Streptomyces nasri strain YG62. Microbios. 1999;100:117–127. [PubMed] [Google Scholar]
- 19.Srivastav NC, Shakya N, Mak M, Agrawal B, Tyrrell DL, Kumar R. Antiviral activity of various 1-(2′-deoxy-β-d-lyxofuranosyl), 1-(2′-fluoro-β-d-xylofuranosyl), 1-(3′-fluoro-β-d-arabinofuranosyl), and 2′-fluoro-2′,3′-didehydro-2′,3′-dideoxyribose pyrimidine nucleoside analogues against duct hepatitis B virus (DHBV) and human hepatitis B virus (HBV) replication. J Med Chem. 2010;53:7156–7166. doi: 10.1021/jm100803c. [DOI] [PubMed] [Google Scholar]
- 20.Chacko AM, Qu W, Kung HF. Synthesis and in vitro evaluation of 5-[18F]fluoroalkyl pyrimidine nucleosides for molecular imaging of herpes simplex virus type 1 thymidine kinase reporter gene expression. J Med Chem. 2008;51:5690–5701. doi: 10.1021/jm800501d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kumar R, Rai D, Sharma SK, Saffran HA, Blush R, Tyrrell DL. Synthesis and antiviral activity of novel 5-(1-cyanamido-2-haloethyl) and 5-(1-hydeoxy (or methoxy)-2-azidoethyl) analogues of uracil nucleosides. J Med Chem. 2001;44:3531–3538. doi: 10.1021/jm010226s. [DOI] [PubMed] [Google Scholar]
- 22.Uyeda M. Fattiviracins, antiviral antibiotics produced by an actinomycete. Actinomycetologica. 2003;17:57–66. [Google Scholar]