Abstract
Here we report a novel design of linker primer that allows one to differentially amplify long tracts (average 3.0 kb with size ranges of 1–7 kb) or short DNAs (average 1.5 kb with size ranges of 0.5–3 kb) from a complex mixture. The method allows one to generate cDNA libraries enriched for long transcripts without size selection of insert DNAs. One representative library from newborn kidney includes 70% of clones bearing ATG start codons. A comparable library has been generated from 20 mouse blastocysts, containing only ∼40 ng of total RNA. This universal PCR amplification scheme can provide a route to isolate very large cDNAs, even if they are expressed at very low levels.
[The sequence data described in this paper have been submitted to the GenBank data library under accession numbers BG060207–BG062928.]
A catalog of genes in the form of cDNA clones is the complement to the sequence of genomes, providing not only the confirmation of predicted gene structures, but also the materials for cDNA microarrays and for functional analyses or proteomics (Takahashi and Ko 1994; Lennon et al. 1996; Marra et al. 1999; Strausberg et al. 1999; Tanaka et al. 2000). Because many genes apparently are expressed only at limited times and places during embryogenesis (Ko et al. 2000), a complete cohort requires cDNA libraries from microdissected tissues and small numbers of cells from developing embryos. Because they require large amounts of RNA, conventional cDNA construction methods are precluded for such purposes. The problem is especially acute for full-length cDNA libraries, which require 10–50 μg of polyA+ RNA, equivalent to milligram amounts of total RNA (Maruyama and Sugano 1994; Edery et al. 1995; Carninci et al. 1996, 1997, 2000; Suzuki et al. 1997, 2000).
PCR amplification can permit the use of smaller amounts of RNA (Belyavsky et al. 1989), but presents the technical dilemma that it customarily yields a shorter average insert size (e.g., ∼0.5 kb [Peterson et al. 1998] and ∼1.5 kb [Ko et al. 2000]), too short to recover many full-length cDNAs. While working on various linker designs and PCR conditions, we serendipitously found a condition that overcomes size limitations. Here, we report a novel design of linker primer that allows one to differentially amplify long cDNAs or short cDNAs from a complex mixture.
RESULTS AND DISCUSSION
To recover novel genes from mouse embryos, we have been using a PCR-amplification method to construct cDNA libraries. In the original protocol (Takahashi and Ko 1994), double-stranded cDNA mixtures were ligated with a lone linker (LL-Sal3 [Ko et al. 1990]; Fig. 1A) and amplified by Taq polymerase (Fig. 1B). The resultant cDNA libraries have relatively short inserts (average 0.8 kb, range 0.5–2.5 kb). Subsequent introduction of new enzymes and long PCR methods (Barnes 1994; Cheng et al. 1994) dramatically increased the size of recovered DNA inserts (average 1.5 kb, range 0.5–3.0 kb), and such libraries have been used for a large-scale expressed sequence tag (EST) project (Ko et al. 2000). However, the inserts were not long enough to recover full-length cDNAs of long transcripts.
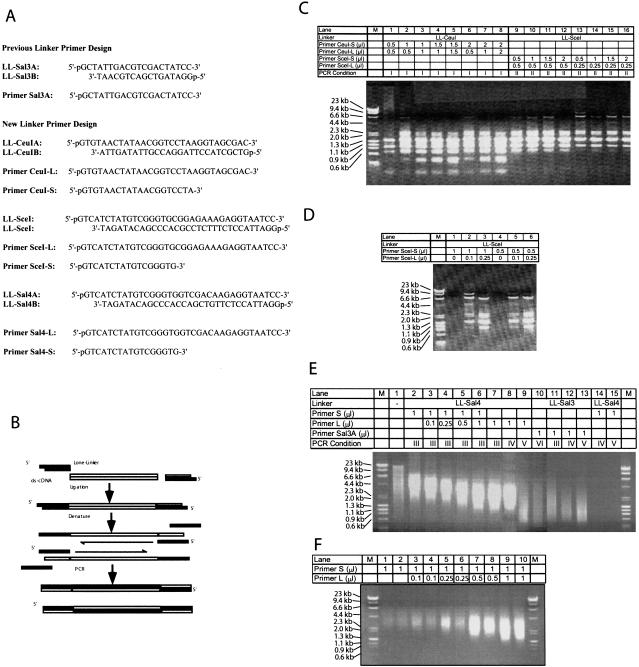
Figure 1.
(A) Linkers and primers used in this paper. (B) Experimental procedures of lone-linker PCR. (C, D) Amplification of blunt-ended DNA size markers, λ-HindIII and φ×174-HaeIII, by various PCR conditions. Size distributions of amplified DNAs were analyzed on 0.7% agarose gels. M, DNA size marker. (E) Amplification of mouse genomic DNAs by various PCR conditions. Size distributions of amplified DNAs were analyzed on 0.7% agarose gels. M, DNA size marker; Lane 1, PvuII-digested mouse genomic DNAs (C57BL/10J); Lanes 2–15, PCR products amplified from PvuII-digested mouse genomic DNA by a different combination of linkers, primers, and PCR condition. See Methods section for PCR conditions. (F) Amplification of mouse newborn kidney cDNAs by various PCR conditions. Size distributions of amplified cDNAs in duplicate were analyzed on 0.7% agarose gels.
Our initial goal was to incorporate rare-cutter restriction enzyme sites into the linker so that cDNA inserts would not be truncated. For example, the previous lone linker, LL-Sal3 (Fig. 1A), has a SalI restriction enzyme site and will be cut by SalI to clone into a plasmid vector. If, by chance, the cDNA inserts contain a SalI site, the cDNAs are cut into two pieces, preventing the cloning of full-length cDNAs. Two relatively new enzymes, PI-SceI and I-CeuI, called homing endonucleases, have long recognition sites and have been used as extremely rare-cutting enzymes (Belfort and Roberts 1997). By replacing the SalI site of LL-Sal3 by I-CeuI and PI-SceI recognition sequences, we designed and synthesized LL-CeuI and LL-SceI (Fig. 1A). As we did for the LL-Sal3 system, a longer strand of linker was used as a PCR primer, CeuI-L (30 mer) and SceI-L (35 mer), respectively (Fig. 1A). To lower the melting temperature of primers, shorter primers, CeuI-S (20 mer) and SceI-S (17 mer), also were designed (Fig. 1A).
The linkers first were ligated to the blunt-ended DNA size markers, λ-HindIII and φx174-HaeIII, and amplified by PCR with various concentrations and combinations of primers. To our surprise, longer DNA fragments up to 9.4 kb were amplified efficiently among other shorter DNA fragments (Fig. 1C). Such long DNA fragments never were amplified as a part of complex mixtures of DNA fragments using the previous LL-Sal3 system. The results also showed that for both LL-CeuI and LL-SceI, shorter primers (CeuI-S and SceI-S) contributed the amplification of longer DNA fragments (6.6 and 9.4 kb). Furthermore, the LL-SceI system tended to amplify longer DNA fragments than LL-CeuI, because the 0.6 kb DNA fragments were not amplified in LL-SceI system. Additional experiments using LL-SceI consistently showed that the shorter primer SceI-S (17 mer) contributed the amplification of longer DNA fragments, whereas the longer primer SceI-L (35 mer) contributed the amplification of shorter DNA fragments (Fig. 1D). Based on these preliminary analyses, we tentatively concluded that the combination of longer linkers such as LL-SceI and shorter primers such as SceI-S could amplify longer DNA fragments, whereas a long primer such as SceI-L amplifies only short DNA fragments.
Unfortunately, we later realized that commercially available PI-SceI and I-CeuI enzymes did not work efficiently, failing to digest linkers completely (data not shown). Because of this undesirable feature, we modified the experimental design. To incorporate the principles of the above findings into a usable lone-linker system, a part of the LL-SceI sequence was modified and replaced by a SalI restriction enzyme site (Fig. 1A: LL-Sal4). We also designed a long primer, Sal4-L and short primer, Sal4-S (Fig. 1A). To test the new linker primers, PvuII-digested mouse genomic DNA was ligated to the LL-Sal4 linker and amplified with either long or short primer, or with a mixture of the two (Fig. 1E). The short primer (S) alone amplified DNA fragments in the 2.0- to 8.0-kb size range (Fig. 1E, lane 2). In contrast, the long primer (L) alone amplified DNA fragments of 0.5–3.0 kb (Fig. 1E, lane 9). By changing the ratio of primers S to L, DNAs with intermediate size ranges were obtained (Fig. 1E; lanes 3–6). A striking feature of this method is the essentially complete lack of shorter DNA fragments when primer S is used (see below for verification of the sizes of cloned species). This is in sharp contrast to other linker and PCR conditions that have been used, which typically only amplify short DNA fragments (Fig. 1E, lanes 10–13). The differences are attributable to the new linker-primer system, with no significant difference in PCR conditions (Fig. 1E, all lanes, particularly lanes 14 and 15).
Essentially equivalent observations were made for the amplification of anonymous cDNA mixtures (Fig. 1F). The mechanism underlying the differential amplification of DNA fragments with different sizes is not clear, and in fact, preferential amplification of longer DNA fragments with no shorter DNA fragments is counterintuitive. Nevertheless, this empirical protocol has worked reproducibly in the construction of all seven consecutive libraries attempted.
For example, to apply this method, 1 μg of total RNA extracted from newborn mouse kidney was subjected to double-stranded cDNA synthesis in a standard manner with an oligo(dT)-NotI primer. The resultant mixture of cDNAs was divided into two portions: one was ligated to a “traditional” linker (LL-Sal3) and the other to the new linker system (LL-Sal4). Both cDNA mixtures were amplified by 25 cycles of PCR. LL-Sal3 was able to amplify cDNAs of 0.5- to 4.0-kb range (average 1.5 kb), in keeping with standard experience (Ko et al. 2000). Similarly, LL-Sal4 with primer L was able to amplify cDNAs of 0.5- to 4.0-kb range. In contrast, LL-Sal4 with primer S was able to amplify cDNAs of 1.0- to 7.0-kb range (Fig. 2A). The PCR products with LL-Sal4 and primer L showed the same size distribution observed with LL-Sal3. This indicates that amplification of short cDNA fragments with LL-Sal3 (Fig. 2A, lane 2) is equivalent to results with LL-Sal4 and primer L (Fig. 2A, lane 4).
Figure 2.
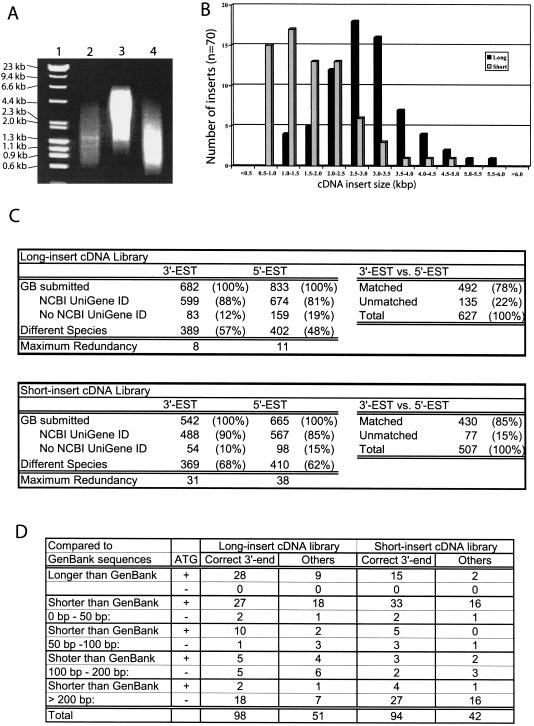
Application of the new method to the construction of newborn mouse kidney DNA libraries. (A) Size distributions of amplified cDNAs were analyzed on 0.7% agarose gels. Lane 1, DNA size marker. Lane 2, previous method. The cDNAs were tagged by LL-Sal3, and amplified by the PCR condition VI with 1 μL of LL-Sal3A primer. Lane 3, new method. The cDNAs were tagged with LL-Sal4 and amplified by the PCR condition III with 1 μL of primer S. Lane 4, new method. The cDNAs were tagged with LL-Sal4 and amplified by the PCR condition V with 1 μL of primer L. (B) Comparison of size distributions of cDNA inserts between the short and long cDNA libraries. (C) Analysis of the complexity of cDNA libraries based on NCBI Mouse UniGene Index. (D) Sequence analyses of the long- and short-insert cDNA libraries. “Correct 3′-end” means that the 3′ end of cDNA sequence shows exact match to the 3′-end sequence of GenBank entry. “Others” means that the 3′ end of cDNA sequences does not match to the 3′ end sequence of GenBank entry. This can happen when either correct 3′-end sequences are not reported in the GenBank or our cDNA clones are truncated at 3′ ends. Although most cDNA clones have polyA sequence at the 3′ end, possible 3′-end truncation by Oligo(dT) mispriming cannot be excluded.
This approach allows us to make two cDNA libraries with different insert size ranges from the cDNA mixtures tagged with LL-Sal4. Each of these two cDNA mixtures (Fig. 2A, lanes 3,4) was double digested with SalI and NotI and cloned into a plasmid vector pSPORT1. From each library, 70 cDNA clones were randomly selected and digested with SalI and NotI. The distributions of insert sizes then were monitored by agarose gel electrophoresis (Fig. 2B). The two libraries clearly differ; the long-insert library indeed contains cDNA clones between 1.0 and 6.0 kb (average 2.5 kb); the short-insert library has inserts between 0.5 and 2.5 kb (average 1.5 kb).
To begin to assess the gene content of the library, ∼1000 cDNA clones from each library were selected randomly and subjected to single-pass sequencing from both 5′ and 3′ ends. The sequences were deposited in the National Center for Biotechnology Information's (NCBI) database of expressed sequence tags (dbEST) as ESTs (GenBank accession numbers BG060207–BG062928). Because one of the major concerns about PCR-amplified cDNA libraries is the possibility of biased gene representation, we first evaluated the complexity of the cDNA libraries. Because the majority of these ESTs already were incorporated into the NCBI Mouse UniGene Index (Boguski and Schuler 1995), the UniGene ID can be used to group the ESTs into distinctive genes. In the long-insert cDNA library, 682 3′ ESTs represented 389 (57%) different genes, while 833 5′ ESTs represented 402 (48%) different genes (Fig. 2C). The most abundant gene, eukaryotic translation elongation factor 1 alpha 1, was represented only eight times in 3′ ESTs and 11 times in 5′ ESTs (“Maximum Redundancy” in Fig. 2C). Thus, we did not observe any significant PCR-based bias in the amplification of specific cDNA species. The overall redundancy of cDNAs is only about twofold, consistent with high complexity of the library. Similar results were obtained for the short-insert cDNA library (Fig. 2C). The most abundant gene in this library also was eukaryotic translation elongation factor 1 alpha 1.
UniGene ID clusters also were used to estimate the number of undesirable chimeric cDNA clones. When both 3′ EST and 5′ EST for each cDNA clone have the same UniGene ID (“Matched” in Fig. 2C), the cDNA clone most likely is not chimeric. The long-insert library contains 492 (78%) such cDNA clones, and the short-insert library contains 430 (85%) such cDNA clones (Fig. 2C). This indicates that the majority of cDNA clones are not chimeric. Although there are slightly more “unmatched” cDNA clones in the long-insert cDNA library than the short-insert one (Fig. 2C), this does not necessarily indicate the presence of chimeric cDNA clones. In fact, among the “EST-unmatched” cDNA clones, we have seen no clear example that 3′ EST and 5′ EST are matched to different known genes.
For the long-insert cDNA library, 627 cDNA clones yielded both 5′ and 3′ ESTs. For the short-insert cDNA library, 507 clones yielded both 5′ and 3′ ESTs. BLASTN searches of these sequences against NCBI's nonredundant database revealed that 5′-end sequences of 149 cDNA clones from the long-insert library and 136 cDNA clones from the short-insert library exactly matched sequences of known genes. To know which region of mRNAs reported in the GenBank was included in our cDNA clones, sequence alignments were inspected carefully (Fig. 2D). Sequence coverage of clones could be inferred because most mRNA sequences reported in GenBank cover complete protein-coding regions (although they are not necessarily full length). From the long-insert cDNA library, 37 (25%) of 149 clones contain sequences longer than those reported in the GenBank and 106 (71%) cDNA clones have the ATG start codon. In contrast, in the short-insert cDNA library, 17 (12%) of 140 clones contain sequences longer than those reported in the GenBank and 81 (60%) clones contain the ATG start codon (Fig. 2D). Therefore, the long-insert cDNA library gives better coverage of complete open reading frames than does the short-insert cDNA library. Considering the average size of mRNAs reported in GenBank, longer transcripts (average 2.73 kb, with a range between 598 and 8233 bp) were represented in the long cDNA library, whereas shorter transcripts (average 1.91 kb, with a range between 490 and 6266 bp) were represented in the short cDNA library. This indicates that the two cDNA libraries represent substantially different sets of genes.
To improve the coverage of full-length cDNA clones, we tried to adopt the well-established 5′-end capping method of Maruyama and Sugano (1994) and Suzuki et al. (2000). One drawback of that method has been the use of a PCR step, which again makes it difficult to clone full-length cDNAs of long transcripts (i.e., the libraries tend to be enriched for full-length cDNA clones of relatively short transcripts). For example, in the latest report of 1010 cDNA clones, about 80% are shorter than 3 kb, with the most frequent clone size clustered around 2 kb (Suzuki et al. 2000). The PCR amplification method that we have developed here may help to alleviate this problem. However, in first experiments with a small amount of total RNA, it rather appeared to reduce the diversity of cDNA species in the library (data not shown). It remains to be seen if the method performs better with a large amount of polyA+ RNAs. Interestingly, published information indicates that even when the 5′-end capping method was used, only 59% of cDNAs contained the ATG start codon (Suzuki et al. 2000). This number appears comparable to that in the short-insert cDNA library reported here, whereas the long-insert cDNA library shows better coverage (70%). This is still less than the 80%–100% full-length clones by another reported method (Carninci et al. 1996, 2000), but provides a simple route to large clones with coverage of ATG start codon.
The method reported in this paper has several advantages over previous protocols. First, it makes possible for the first time the construction of long-transcript–enriched, unidirectional cDNA libraries from small amounts of total RNAs. We have thus far successfully made such cDNA libraries from as few as 20 mouse blastocyst-stage embryos (∼1000 cells containing ∼40 ng of total RNA). This PCR-amplified cDNA library shows comparable or better size distributions than full-length cDNA libraries made without PCR amplification using large amounts of polyA+ RNAs (e.g., compared to Carninci et al. 1997; Suzuki et al. 2000). The representation of complete open reading frames in the inserts of the new libraries, indicated by the presence of ATG start codons, is comparable to current “full-length” cDNA libraries. In addition, clear separation of long inserts from short inserts can largely alleviate problems that are inherent to cloning procedures, particularly biased cloning of short inserts. That recurrent problem has imposed a need for cumbersome agarose gel electrophoresis to eliminate short-insert cDNAs, and has inhibited the construction of full-length cDNA libraries from even moderate amounts of total RNAs. The new method thus may facilitate the ongoing assembly of the complete catalog of mammalian genes with full open reading frames (Strausberg et al. 1999). Large-scale sequencing of such cDNA libraries is being conducted (NIA Mouse cDNA Project; http://lgsun.grc.nia.nih.gov/cDNA/cDNA.html) and will correspondingly contribute to genomics and proteomics. The method also can be applied to the amplification of a mixture of variably sized genomic DNA fragments, which could help to preserve genomic DNA from human populations, rare animal species, and archaeological materials in an indefinitely amplifiable form (Weiss et al. 1994).
The general idea of the method is to use a 35-bp long lone-linker sequence (LL-Sal4, Fig. 1A), but with only part of the linker as the PCR primer. Starting from the same DNA mixtures, a longer primer (35 bp) then amplifies only short cDNA fragments, whereas a shorter primer (17 bp) unexpectedly amplifies only long cDNA fragments. The basis for these PCR effects remains unknown, but the length of the linker and primer seem to be more important than their sequence. For example, both LL-SceI and LL-Sal4, which have the same nucleotide length, work in the same manner, though their sequences are different. A slightly shorter linker, LL-CeuI (30 bp) and longer primer, CeuI-S (20 mer) work less efficiently to amplify longer DNA fragments, and the shortest linker, LL-Sal3 (20 bp) and primer, Sal3A (20 mer) are even less efficient. In addition, the higher G/C-content in the 3′ end of primers also may contribute the amplification of longer DNA fragments. For example, the Sal4-S and SceI-S have six G/C in the eight nucleotides in 3′ end, whereas the CeuI-S has five G/C and Sal3A has only four G/C (Fig. 1A). More systematic testing of different linker/primer designs should help to understand the mechanism, but the results indicate that the interaction of particular polymerases with primer complexes of different lengths can affect product length and even template selection.
METHODS
RNA Preparation and cDNA Synthesis
Two pieces of a newborn kidney were collected from C57BL/6J mouse. Total RNAs were extracted from these tissues according to a standard method using the Trizol reagent (Life Technologies). In brief, the tissues were homogenized in 1 mL of Trizol reagent, and extracted with 0.2 mL of chloroform. After extraction, RNA was precipitated from the aqueous phase by adding 0.5 mL of isopropanol. Then, RNA was pelleted by centrifugation at 12,000 rpm for 10 min in a refrigerated centrifuge. The pellet was washed with 75% ethanol. The RNA was dissolved in 9 μL in DEPC-treated water, heated at 70°C for 10 min, and quick chilled on ice. The RNA was mixed with the 9 μL of first-strand buffer and incubated at 37°C for 6 min. Finally, 2 μL of SuperScript II reverse transcriptase (200 units/μL) was added and incubated at 37°C for 1 h. The final concentration of each ingredient in the first-strand synthesis reaction was 50 μg/mL oligo(dT)-NotI primer (0.5 μg/μL: 5′-pGACTAGTTCTAGATCGCGAGCGGCCGC CCTTTTTTTTTTTTTTT-3′ [Life Technologies]), 50 mM Tris-HCI (pH 8.3), 75 mM KCl, 3 mM MgCl2, 10 mM DTT, 500 μM each dATP, dCTP, dGTP, dTTP, and 26 μg total RNA. To help resolve the RNA secondary structure, the reaction mixture was heated at 65°C for 5 min. Then, 2 μL of SuperScript II RT was added to the reaction, followed by the incubation at 37°C for 15 min. This step was repeated two more times.
The second strand was synthesized in a total volume of 150 μL reaction [25 mM Tris-HCI(pH 7.5), 100 mM KCI, 5 mM MgCl2, 10 mM (NH4)2SO4, 0.15 mM β-NAD+, 250 μM each dATP, dCTP, dGTP, dTTP and 1.2 mM DTT, 65 units/mL DNA ligase, 250 units/mL DNA polymerase I, 13 units/mL RNaseH]. After incubating the reaction at 16°C for 2 h, the cDNAs were treated with 2 μL of T4 DNA polymerase (5 units/μL) by incubating at 16°C for 5 min and purified by ethanol precipitation.
PCR Amplification of cDNAs
The double-stranded cDNAs were ligated to two different lone linkers, LL-Sal4 (LL-Sal4A, 5′-pGTCATCTATGTCGGGTGGTC GACAAGAGGTAATCC-3′ LL-Sal4B, 5′-pGGATTACCTCTT GTCGACCACCCGACATAGAT-3′) and LL-Sal3 (LL-Sal3A, 5′-pGCTATTGACGTCGACTATCC-3′ LL-Sal3B, 5′-pGG ATAGTCGACGTCAAT-3′) with 5 μL of T4 DNA ligase (1 unit/μL ) in a total volume of 50 μL reaction at 16°C overnight. The cDNAs were purified by phenol/chloroform and separated from free linkers by Centricon 100 (Amicon). The final volume is around 100 μL.
The 0.5 ∼ 1.0 μL of purified DNAs were amplified by long-range, high-fidelity PCR using 0.25 μL (5 units/μL) of Ex Taq polymerase (Takara) with 20 pmol of PCR primer (Primer L, 5′-GTCATCTATGTCGGGTGGTCGACAAGAGGTAATCC-3′ or Primer S, 5′-GTCATCTATGTCGGGTG-3′) in a total 50 μL reaction. The amplification cycles were 25 cycles at 94°C for 30 sec, at 55°C for 30 sec, and 68°C for 12 min for long insert; 25 cycles at 94°C for 40 sec, and 72°C for 10 min for short insert. Then, the amplified cDNAs were purified by phenol/chloroform and by Centricon 100. The cDNAs were double-digested with SalI/NotI enzymes and purified by phenol/chloroform extraction, followed by ethanol precipitation. The cDNAs were separated from the linkers by a cDNA size fractionation column (Life Technologies) and subjected to the ethanol precipitation. Then, cDNAs were cloned into SalI/NotI site of pSPORT1 plasmid vector in a total volume of 20 μL reaction at room temperature for 3 h. The 100 μL of DH10B Escherichia coli host (Life Technologies) was transformed with the 5 μL ligation mixture by the chemical method according to the manufacturer's instruction. The cultures were spread on LB-agar plate with 100 μg/mL ampicillin and incubated at 37°C overnight. Then, single colonies were randomly picked and arrayed into 96-well microtiter plates and sequenced from both 5′ and 3′ ends by ABI 3700 DNA sequencer. Sequence similarity searches were performed using NCBI/NIH BLASTN server against nonredundant GenBank sequence database.
PCR Conditions
Seven different PCR conditions were used in this study.
PCR Condition I
Initial denaturation at 94°C for 20 sec, followed by 25 cycles of denaturation at 94°C for 10 sec, annealing at 83°C for 30 sec and at 62°C for 30 sec, and extension at 72°C for 12 min, and final extension at 72°C for 10 min.
PCR Condition II
Initial denaturation at 94°C for 20 sec, followed by 25 cycles of denaturation at 94°C for 30 sec, annealing at 62°C for 30 sec, and extension at 72°C for 12 min, and final extension at 72°C for 10 min.
PCR Condition III
Initial denaturation at 94°C for 30 sec, followed by 25 cycles of denaturation at 94°C for 30 sec, annealing at 55°C for 30 sec, and extension at 68°C for 12 min, and final extension at 72°C for 10 min.
PCR Condition IV
Initial denaturation at 94°C for 45 sec, followed by 25 cycles of denaturation at 94°C for 30 sec, annealing/extension at 68°C for 12 min, and final extension at 72°C for 10 min.
PCR Condition V
Initial denaturation at 94°C for 20 sec, followed by 25 cycles of denaturation at 94°C for 40 sec, annealing/extension at 72°C for 10 min, and final extension at 72°C for 10 min.
PCR Condition VI
Initial denaturation at 94°C for 20 sec, followed by 25 cycles of denaturation at 94°C for 10 sec, annealing/extension at 68°C for 12 min, and final extension at 72°C for 10 min.
Generation and Analysis of EST
Sequencing of cDNA clones and the standard analysis of their sequences were done as described (Ko et al. 2000).
Acknowledgments
We thank George J. Kargul, Dawood B. Dudekula, and Yong Qian for helping DNA sequence analyses, Xiaohong Wang for the initial optimization of the protocol, Tetsuya S. Tanaka, Carole A. Stagg, and Serafino Pantano for providing mouse embryos, and David Schlessinger for discussion and critical reading of the manuscript.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL kom@grc.nia.nih.gov; FAX (410) 558-8331.
Article and publication are at http://www.genome.org/cgi/doi/10.1101/gr.185501.
REFERENCES
- Barnes WM. PCR amplification of up to 35-kb DNA with high fidelity and high yield from λ bacteriophage templates. Proc Natl Acad Sci. 1994;91:2216–2220. doi: 10.1073/pnas.91.6.2216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belfort M, Roberts RJ. Homing endonucleases: Keeping the house in order. Nucleic Acids Res. 1997;25:3379–3388. doi: 10.1093/nar/25.17.3379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belyavsky A, Vinogradova T, Rajewsky K. PCR-based cDNA library construction: General cDNA libraries at the level of a few cells. Nucleic Acids Res. 1989;17:2919–2932. doi: 10.1093/nar/17.8.2919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boguski MS, Schuler GD. ESTablishing a human transcript map. Nat Genet. 1995;10:369–371. doi: 10.1038/ng0895-369. [DOI] [PubMed] [Google Scholar]
- Carninci P, Kvam C, Kitamura A, Ohsumi T, Okazaki Y, Itoh M, Kamiya M, Shibata K, Sasaki N, Izawa M, et al. High-efficiency full-length cDNA cloning by biotinylated CAP trapper. Genomics. 1996;37:327–336. doi: 10.1006/geno.1996.0567. [DOI] [PubMed] [Google Scholar]
- Carninci P, Shibata Y, Hayatsu N, Sugahara Y, Shibata K, Itoh M, Konno H, Okazaki Y, Muramatsu M, Hayashizaki Y. Normalization and subtraction of cap-trapper-selected cDNAs to prepare full-length cDNA libraries for rapid discovery of new genes. Genome Res. 2000;10:1617–1630. doi: 10.1101/gr.145100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carninci P, Westover A, Nishiyama Y, Ohsumi T, Itoh M, Nagaoka S, Sasaki N, Okazaki Y, Muramatsu M, Schneider C, et al. High efficiency selection of full-length cDNA by improved biotinylated cap trapper. DNA Res. 1997;4:61–66. doi: 10.1093/dnares/4.1.61. [DOI] [PubMed] [Google Scholar]
- Cheng S, Fockler C, Barnes WM, Higuchi R. Effective amplification of long targets from cloned inserts and human genomic DNA. Proc Natl Acad Sci. 1994;91:5695–5699. doi: 10.1073/pnas.91.12.5695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edery I, Chu LL, Sonenberg N, Pelletier J. An efficient strategy to isolate full-length cDNAs based on an mRNA cap retention procedure (CAPture) Mol Cell Biol. 1995;15:3363–3371. doi: 10.1128/mcb.15.6.3363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ko MSH, Kitchen JR, Wang X, Threat TA, Hasegawa A, Sun T, Grahovac MJ, Kargul GJ, Lim MK, Cui Y, et al. Large-scale cDNA analysis reveals phased gene expression patterns during preimplantation mouse development. Development. 2000;127:1737–1749. doi: 10.1242/dev.127.8.1737. [DOI] [PubMed] [Google Scholar]
- Ko MSH, Ko SBH, Takahashi N, Nishiguchi K, Abe K. Unbiased amplification of a highly complex mixture of DNA fragments by ‘lone linker’-tagged PCR. Nucleic Acids Res. 1990;18:4293–4294. doi: 10.1093/nar/18.14.4293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lennon G, Auffray C, Polymeropoulos M, Soares MB. The I.M.A.G.E. Consortium: An integrated molecular analysis of genomes and their expression. Genomics. 1996;33:151–152. doi: 10.1006/geno.1996.0177. [DOI] [PubMed] [Google Scholar]
- Marra M, Hillier L, Kucaba T, Allen M, Barstead R, Beck C, Blistain A, Bonaldo M, Bowers Y, Bowles L, et al. An encyclopedia of mouse genes. Nat Genet. 1999;21:191–194. doi: 10.1038/5976. [DOI] [PubMed] [Google Scholar]
- Maruyama K, Sugano S. Oligo-capping: A simple method to replace the cap structure of eukaryotic mRNAs with oligoribonucleotides. Gene. 1994;138:171–174. doi: 10.1016/0378-1119(94)90802-8. [DOI] [PubMed] [Google Scholar]
- Peterson LA, Brown MR, Carlisle AJ, Kohn EC, Liotta LA, Emmert-Buck MR, Krizman DB. An improved method for construction of directionally cloned cDNA libraries from microdissected cells. Cancer Res. 1998;58:5326–5328. [PubMed] [Google Scholar]
- Strausberg RL, Feingold EA, Klausner RD, Collins FS. The mammalian gene collection. Science. 1999;286:455–457. doi: 10.1126/science.286.5439.455. [DOI] [PubMed] [Google Scholar]
- Suzuki Y, Ishihara D, Sasaki M, Nakagawa H, Hata H, Tsunoda T, Watanabe M, Komatsu T, Ota T, Isogai T, et al. Statistical analysis of the 5′ untranslated region of human mRNA using “Oligo-Capped” cDNA libraries. Genomics. 2000;64:286–297. doi: 10.1006/geno.2000.6076. [DOI] [PubMed] [Google Scholar]
- Suzuki Y, Yoshitomo-Nakagawa K, Maruyama K, Suyama A, Sugano S. Construction and characterization of a full length-enriched and a 5′-end-enriched cDNA library. Gene. 1997;200:149–156. doi: 10.1016/s0378-1119(97)00411-3. [DOI] [PubMed] [Google Scholar]
- Takahashi N, Ko MSH. Toward a whole cDNA catalog: Construction of an equalized cDNA library from mouse embryos. Genomics. 1994;23:202–210. doi: 10.1006/geno.1994.1478. [DOI] [PubMed] [Google Scholar]
- Tanaka TS, Jaradat SA, Lim MK, Kargul GJ, Wang X, Grahovac MJ, Pantano S, Sano Y, Piao Y, Nagaraja R, et al. Genome-wide expression profiling of mid-gestation placenta and embryo using a 15,000 mouse developmental cDNA microarray. Proc Natl Acad Sci. 2000;97:9127–9132. doi: 10.1073/pnas.97.16.9127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss KM, Buchanan AV, Daniel C, Stoneking M. Optimizing utilization of DNA from rare or archival anthropological samples. Hum Biol. 1994;66:789–804. [PubMed] [Google Scholar]