Abstract
The nucleotide sequence was determined for a 340-kb segment of rice chromosome 2, revealing 56 putative protein-coding genes. This represents a density of one gene per 6.1 kb, which is higher than was reported for a previously sequenced segment of the rice genome. Sixteen of the putative genes were supported by matches to ESTs. The predicted products of 29 of the putative genes showed similarity to known proteins, and a further 17 genes showed similarity only to predicted or hypothetical proteins identified in genome sequence data. The region contains a few transposable elements: one retrotransposon, and one transposon. The segment of the rice genome studied had previously been identified as representing a part of rice chromosome 2 that may be homologous to a segment of Arabidopsis chromosome 4. We confirmed the conservation of gene content and order between the two genome segments. In addition, we identified a further four segments of the Arabidopsis genome that contain conserved gene content and order. In total, 22 of the 56 genes identified in the rice genome segment were represented in this set of Arabidopsis genome segments, with at least five genes present, in conserved order, in each segment. These data are consistent with the hypothesis that the Arabidopsis genome has undergone multiple duplication events. Our results demonstrate that conservation of the genome microstructure can be identified even between monocot and dicot species. However, the frequent occurrence of duplication, and subsequent microstructure divergence, within plant genomes may necessitate the integration of subsets of genes present in multiple redundant segments to deduce evolutionary relationships and identify orthologous genes.
Rice (Oryza sativa) is a widely grown crop, and is the staple food for over one-half of the world's population. Extensive classical and molecular genetic maps have been constructed to assist biological analyses and plant breeding applications (Kinoshita 1995; Kurata et al. 1994). The genome size of rice, ∼440 Mb (Arumuganathan and Earle 1991), is one of the smallest of the cereals. It has been postulated that the genes within the genome of rice, as with the genes of other Gramineae, are clustered in gene-rich regions separated by gene-poor DNA (Barakat et al. 1997). A high degree of conservation of the order of gene-specific markers (conserved synteny) has been observed between the genomes of most cereals, including rice (Moore et al. 1995). The sequences of exons and exon–intron structures of orthologous genes in the sh2/a1-homologous regions of rice and sorghum have been shown to be conserved (Chen et al. 1998). However, more divergence of gene content has been found in the Adh1 regions of the genomes of maize and sorghum (Tikhonov et al. 1999). Nevertheless, rice is being developed as the key model monocot species for molecular genetic investigations, with the expectation that, by exploiting conserved synteny, the identification and functional assignment of genes in rice will lead to the identification of the equivalent genes in other cereal species. These applications are being supported by the Rice Genome Project, which commenced in 1991. The main aim of this project is the determination of the complete nucleotide sequence of the rice genome.
A 340-kb region around the rice Adh1–Adh2 region has been sequenced (Tarchini et al. 2000), and is predicted to contain 33 protein-coding genes. Fourteen of these predicted genes were supported by the identification of corresponding transcripts, and 15 genes were similar in structure to genes with known functions. Nineteen of the 33 genes were members of gene families within the sequenced region, although some copies were predicted to be nonfunctional pseudogenes.
The key model dicot plant species is Arabidopsis thaliana (Arabidopsis). Extensive classical genetic, molecular genetic, and physical maps have been developed, along with numerous genome analysis and gene cloning strategies (Koornneef 1990; Lister and Dean 1993; Feldmann et al. 1989; Giraudat et al. 1992; Bancroft et al. 1993; Schmidt et al. 1995; http://nasc.nott.ac.uk/new_ri_map.html;). The Arabidopsis genome has been completely sequenced (The Arabidopsis Genome Initiative 2000). It is very gene-rich, containing 25,498 genes, with an average density of one gene per 4.5 kb. Conservation of gene order has been observed between segments of the genome of Arabidopsis and those of its closest relatives among crops, the cultivated Brassica species (Kowalski et al. 1994; Cavell at al. 1998; Lagercrantz 1998).
It had been predicted that conserved synteny between the genomes of Arabidopsis and cereals, which diverged ca. 200 million years ago (Wolfe et al. 1989), would be detectable for segments of ∼3 cM (Paterson et al. 1996). Such conservation could lead to the use of positional approaches to integrate functional genomics information from both monocot and dicot species. The results of comparative genetic mapping efforts have provided little evidence for conserved gene organization (Gale and Devos 1998). Although some conserved synteny can be detected between the genomes of rice and Arabidopsis using physical mapping and sequence analysis approaches, the extent of conservation appears low (Devos et al. 1999; Han et al. 1999; van Dodeweerd at al. 1999). In the present study we report the results of a pilot-scale rice genome sequencing project and the use of the data to further study aspects of genome organization in Arabidopsis.
RESULTS
Gene Prediction
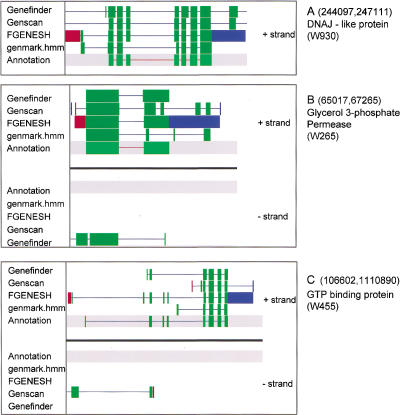
Four overlapping BACs representing the 340-kb region to be sequenced had been identified previously (van Dodeweerd et al. 1999). A shotgun sequencing strategy was used and annotation performed on a 339,972-bp contiguous assembly as submitted to EMBL (accession no. AJ307662). Four gene prediction programs were used for modeling exon structure: Genemark.hmm, FGENESH, Genscan, and GeneFinder. Comparisons of the outputs from these programs with gene structures determined using EST matches and protein homologies for three genes are shown in Figure 1. Although all programs correctly predicted the presence of a gene, none of the predictions accurately identified the exon–intron structures of the genes.
Figure 1.
Comparison of gene models predicted by Genemark.hmm, FGENESH, Genscan, and GeneFinder for predicted genes W930 (A), W265 (B), and W455 (C). Green boxes denote predicted as well as annotated protein coding regions.
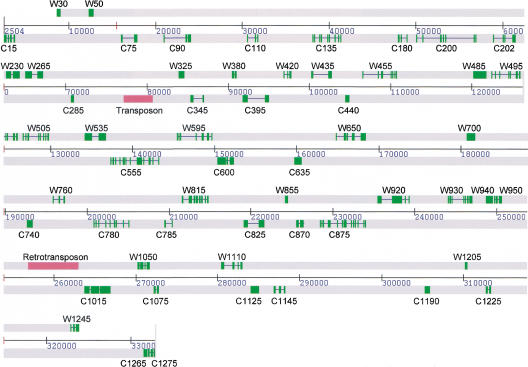
Gene prediction in rice is complicated by the fact that a rice species setting is available only for Genemark.hmm. Nevertheless, our data suggest that even Genemark.hmm output is not reliable enough to perform an in silico whole-genome analysis in rice. Further adjustment and refinement of gene prediction programs is necessary for large-scale automated genome analysis. Similarities of genomic sequences with EST sequences, matches of predicted protein products with known proteins, and matches with transposable elements were also used to derive the final gene modeling, as shown in Figure 2. In total, 56 potential protein-coding genes were identified, along with a region containing a retrotransposon and a region showing homology to transposon Tnr1. Two tRNAs were also identified. A summary of the positions of the identified genes and other features is presented in Table 1.
Figure 2.
Positions of predicted genes (predicted protein coding regions denoted by green boxes) and transposable elements (red boxes).
Table 1.
Features Identified in Rice Sequence Data
| Feature | Position | Protein similarity/EST matches |
|---|---|---|
| C15 | C/2504-3864 | |
| W30 | W/8619-9131 | hypothetical protein F30H5.2, Caenorhabditis elegans, 1.6e-05 |
| W50 | W/12306-12944 | glycine-rich protein GRP22, rape, PIR:S31415, 1.6e-36 |
| C75 | C/16037-17967 | PREG like protein, Arabidopsis thaliana, EMBL:AB012239, 5.2e-38 |
| C90 | C/20929-24200 | DNA-binding protein ABF1—wild oat, PIR:S61413, 5.0e-28 |
| C110 | C/30589-31850 | |
| C135 | C/38132-41466 | predicted protein, Arabidopsis thaliana, PIR:T49221, 8.7e-133/D25127 |
| C180 | C/47976-49075 | predicted protein, Oryza sativa, EMBL:AC026815, 1.9e-23 |
| C200 | C/50060-56958 | 27k vesicle-associated membrane protein-associated protein (VAMP), curled-leaved tobacco, EMBL:AB018117, 4.8e-84 |
| C202 | C/58901-61517 | plastid ribosomal protein L19 precursor, Spinacia oleracea, EMBL:AF250384, 5.3e-59,/C74047 |
| W230 | W/62697-64415 | extensin-like protein—maize, PIR:S49915, 1.2e-06,/AU030763, AU070696, AU030762 |
| tRNA | C/64886-65017 | tRNA predict as a tRNA- Arg : anticodon acg |
| tRNA | C/64886-64957 | tRNA predict as a tRNA- Pro : anticodon ggg |
| W265 | W/65017-67265 | cAMP inducible 2 protein, Mus musculus, EMBL:AF121081, 1.5e-123 |
| C285 | C/70640-71035 | |
| transposon | C/77159-80730 | transposon |
| W325 | W/83990-84706 | glycine-rich protein 2, Arabidopsis thaliana, PIR:T05494, 7.5e-09 |
| C345 | C/85408-87116 | uclacyanin 3, Arabidopsis thaliana, PIR:T49223, 3.3e-36/BE040770, BE040904, AU071152, AU071239, AU070738, AU071112, AU071195, AU070659, AU070774, AU071224, AU095618, AU163454, AU071095, AU070835, AU071193, AU070840, C25085 |
| W380 | W/90519-91174 | |
| C395 | C/91818-95108 | predicted protein, Arabidopsis thaliana, PIR:T48310, 3.7e-28 |
| W420 | W/96931-97865 | |
| W435 | W/100319-102879 | predicted protein, Oryza sativa, EMBL:AB026295, 8.5e-12 |
| C440 | C/104495-104992 | |
| W455 | W/106602-110890 | GTP-binding protein—maize, PIR:B38202, 2.7e-136/BE039980, AU092678, AU092609, AU092889, AU030259, AU064907, AU064033, D49344, C93488, D2387, C25096, AU066159, AU066160, AU030258, C28045, C25059, C27780, D22939, AU057614, C28341 |
| W485 | W/120269-121903 | probable triacylglycerol lipase, Arabidopsis thaliana, PIR:E71435, 2.3e-110/AU101257, AU064146, AU064563, AU064925, AU065141, AU065117, AU094966 |
| W495 | W/122503-126090 | probable enoyl-CoA hydratase, Arabidopsis thaliana, PIR:C71435, 4.6e-60 |
| W505 | W/126711-129864 | probable enoyl-CoA hydratase, Arabidopsis thaliana, PIR:C71435, 5.7e-64 |
| W535 | W/134248-136831 | protein kinase (EC 2.7.1.37) 5, Arabidopsis thaliana, PIR:JN0505, 4.8e-138 |
| C555 | C/137352-143243 | predicted protein, Arabidopsis thaliana, EMBL:AB008267, 3.1e-81 |
| W595 | W/145530-149814 | ubiquitin thiolesterase (EC 3.1.2.15) L3, human, PIR:A40085, 3.3e-52/AU058175, C25250 |
| C600 | C/150409-152412 | bZIP protein, Arabidopsis thaliana, PIR:T49227, 5.4e-73,/AU163243 |
| C635 | C/159754-160665 | CDBP Mesembryanthemum crystallinum AP2-related transcription factor, EMBL:AF245119, 7.9e-51/ AU057740, AU083516 |
| C650 | W/164858-168510 | predicted protein, Arabidopsis thaliana, PIR:T00408, 2.0e-77 |
| W700 | W/180768-181805 | ethylene responsive element binding factor 5, Arabidopsis thaliana, PIR:T52020, 1.1e-35 |
| C740 | C/192640-193413 | 3-isopropylmalate dehydratase-like protein (small subunit), Arabidopsis thaliana, PIR:T47781, 4.5e-64 /AU172969, AU163547, AU096007, AU096006, D22329, AU101178, AU162746, AU094753, AU101179 |
| W760 | W/195850-197387 | predicted protein, Arabidopsis thaliana, 1.8e-10 |
| C780 | C/200773-205219 | predicted protein, Arabidopsis thaliana, EMBL:AB023044 |
| C785 | C/209473-210472 | |
| W815 | W/211666-217666 | predicted protein, Arabidopsis thaliana, EMBL:AC024174, 1.6e-244 |
| C825 | C/219153-221701 | kinase-like transmembrane protein TMKL1 precursor, Arabidopsis thaliana, PIR:S39476, 1.8e-49,/ AU056887 |
| W855 | W/224272-224625 | |
| C870 | C/225607-226540 | predicted protein, Arabidopsis thaliana, EMBL:ATH288958, 9.9e-13 |
| C875 | C/228468-234134 | predicted protein, Arabidopsis thaliana, EMBL:AB023044, 1.5e-98 |
| W920 | W/235513-239385 | CDC23, human, EMBL:AB011472, 7.0e-138,/C28583 |
| W930 | W/244097-247111 | LDJ2 (DnaJ) protein, leek, PIR:S42031, 8.0e-226/AU031322, C26206, AU030551, AU092294, AU101483, C28691, AU031323, AU070856, AU070832, D38865, D38877 |
| W940 | W/248749-249612 | transcription factor TINY, Arabidopsis thaliana, PIR:T01076, 7.4e-39 |
| W950 | W/249771-250715 | probable tonoplast aquaporin—maize, PIR:T01648, 9.7e-117/D23748, AU173162, AU163445, AU083016, D24190, D24763, AU031632, AU065449, AU173163, D24895 |
| Retrotransposon | W/256930-263084 | Ty3/Gypsy-like retrotransposon |
| C1015 | C/263711-266955 | predicted protein, Oryza sativa, EMBL:AP000399, 0.0 |
| W1050 | W/270285-271770 | hypothetical protein, Arabidopsis thaliana, PIR:G71442, 1.7e-80 |
| C1075 | C/272186-272851 | predicted protein, Oryza sativa, EMBL:AC051633, 3.5e-23 |
| W1110 | W/280506-283120 | predicted protein, Arabidopsis thaliana, PIR:T05538, 8.3e-24/BE230101, BE230297 |
| C1125 | C/284043-285083 | acetylglutamate kinase-like protein, Arabidopsis thaliana, PIR:T46192, 8.0e-113 |
| C1145 | C/286907-288305 | |
| C1190 | C/305291-305968 | AP2 domain transcription factor, Arabidopsis thaliana, EMBL:AC006068, 1.9e-40 |
| W1205 | W/310298-310597 | predicted protein, Oryza sativa, EMBL:AP002094, 8.3e-37 |
| C1225 | C/312769-313464 | fibroin-2, Araneus diadematus, EMBL:AD47854, 1.5e-13 |
| W1245 | W/322960-323955 | |
| C1265 | C/331539-332006 | transcription factor TINY—Arabidopsis thaliana, PIR:T01076, 6.4e-17 |
| C1275 | C/332068-332877 | tonoplast intrinsic protein 1 (TIP1), Hordeum vulgare, EMBL:AF254799, 4.9e-90/AU173162, D23748, AU163445, AU083016, D24190, D24763, AU065449, D24895 |
Identification of Homologous ESTs and Proteins
The rice genomic nucleotide sequence was used to query a rice EST database to identify ESTs corresponding to modeled genes. A threshold of at least 90% sequence identity over at least 150 bp was applied. Each predicted gene was used to query all available nucleotide and protein databases for homologous identified or predicted protein sequences. The results of both analyses are summarized in Table 1. Sixteen of the 56 modeled genes match ESTs, supporting the prediction of the presence of a gene. The predicted proteins of 29 putative genes (52%) match known proteins, and the predicted proteins of 17 putative genes (30%) match proteins predicted from genome sequence data. The predicted proteins of the 10 remaining putative genes (18%) show no similarity to known proteins, so may either represent new types of proteins or be the result of false gene predictions.
The predicted proteins for all 56 putative genes were analyzed for the presence of functional domains using Interpro (The InterPro Consortium 2000). The results are shown in Table 2. Characterized functional domains of protein products were identified for 33 of the 56 putative genes (59%). These allowed us to identify a four-member gene family (C635, W700, W940, and C1190) encoding protein products with AP2 domain/ethylene responsive element binding protein functional domains, which was the largest gene family we identified in the sequenced region.
Table 2.
Functional Domains of Predicted Rice Proteins
| Predicted gene | Interpro domains |
|---|---|
| C15 | IPR000104 Type I antifreeze protein |
| W50 | IPR001687 ATP/GTP-binding site motif A (P-loop) |
| IPR000817 Prion protein | |
| IPR002952 Eggshell protein | |
| C90 | IPR001472 Bipartite nuclear localization signal |
| IPR000104 Type I antifreeze protein | |
| C200 | IPR000535 Major sperm protein (MSP) domain |
| C202 | IPR001857 Ribosomal protein L19 |
| W230 | IPR002965 Proline rich extensin |
| W265 | IPR000694 Proline-rich region |
| W325 | IPR001878 Zn-finger CCHC type |
| C345 | IPR002965 Proline rich extensin |
| IPR001402 Probable G protein-coupled receptor GPR10 | |
| W455 | IPR001687 ATP/GTP-binding site motif A (P-loop) |
| IPR002078 Sigma-54 factor interaction protein family | |
| IPR001806 Ras family | |
| W485 | IPR000734 Lipase |
| W495 | IPR001753 Enoyl-CoA hydratase/isomerase |
| W505 | IPR001753 Enoyl-CoA hydratase/isomerase |
| W535 | IPR002290 Serine/Threonine protein kinases active-site |
| IPR000379 Esterase/lipase/thioesterase active site | |
| IPR000719 Eukaryotic protein kinase | |
| W595 | IPR001578 Ubiquitin carboxyl-terminal hydrolases family 1 |
| IPR000694 Proline-rich region | |
| C600 | IPR000694 Proline-rich region |
| C635 | IPR001471 AP2 domain/ethylene responsive element binding protein |
| IPR000104 Type I antifreeze protein | |
| W700 | IPR001471 AP2 domain/ethylene responsive element binding protein |
| W760 | IPR002293 Permease for amino acids and related compounds, family I |
| C780 | IPR000379 Esterase/lipase/thioesterase active site |
| W815 | IPR001865 Ribosomal protein S2 |
| C825 | IPR001245 Tyrosine kinase catalytic domain |
| C875 | IPR001472 Bipartite nuclear localization signal |
| IPR000719 Eukaryotic protein kinase | |
| W920 | IPR001440 TPR repeat |
| W930 | IPR001623 DnaJ N-terminal domain |
| IPR001305 DnaJ central domain (CXXCXGXG) | |
| IPR003095 DNAJ heat shock protein | |
| IPR001472 Bipartite nuclear localization signal | |
| W940 | IPR001778 Pollen allergen Poa pI signature |
| IPR001471 AP2 domain/ethylene responsive element binding protein | |
| IPR000104 Type I antifreeze protein | |
| W950 | IPR001472 Bipartite nuclear localization signal |
| IPR000425 MIP family | |
| W1050 | IPR001778 Pollen allergen Poa pI signature |
| W1110 | IPR000173 Glyceraldehyde 3-phosphate dehydrogenase |
| C1125 | IPR001472 Bipartite nuclear localization signal |
| IPR001057 Glutamate 5-kinase | |
| C1145 | IPR001472 Bipartite nuclear localization signal |
| C1190 | IPR001471 AP2 domain/ethylene responsive element binding protein |
| IPR000104 Type I antifreeze protein | |
| C1225 | IPR002086 Aldehyde dehydrogenase family |
Analysis of Genome Organization in Rice and Arabidopsis
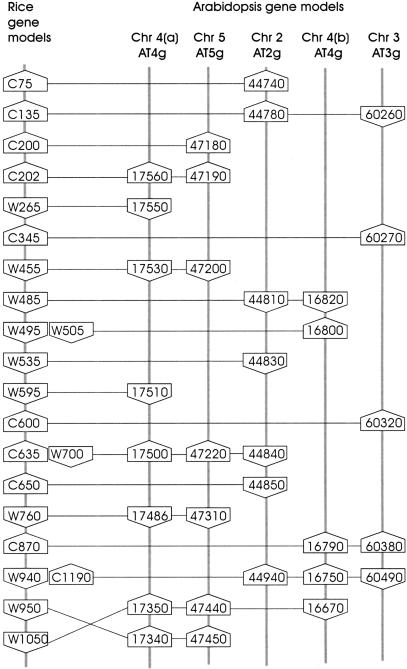
The extent of conservation of both the presence and the position of genes in the sequenced segment of the rice genome and the corresponding segments of the genome of Arabidopsis was analyzed. BLASTP analyses were performed using the extracted amino acid sequences of the annotated rice genes to query a database of all predicted Arabidopsis protein sequences, using a P-value of ≤e−5 as a cutoff. The results were then filtered to remove adjacent matches (indicative of tandem duplications) and clusters of three or more nearby matches recorded. The results for this analysis of the 340-kb region are summarized in Table 3. The relative coding strand for each gene model is denoted by W or C. Five segments of the Arabidopsis genome contained conserved subsets of the rice genes, as shown in Figure 3. These segments represented regions of the Arabidopsis genome containing approximately 22, 27, 20, 15, and 23 genes, for the chromosome 4(a), 5, 2, 4(b), and 3 segments, respectively, shown in Figure 3. Overall, 22 of the 56 rice genes are represented in the five Arabidopsis chromosome segments, counting both copies of three pairs of related genes (W495/W505, C635/W700, and W940/C1190) that show homology to common Arabidopsis genes. The most highly conserved segment, chromosome 4(a), contains eight conserved genes, with one pair (W950-AT4g17340 and W1050-AT4g17350) reversed. The relative coding strand orientation of the genes is also conserved, except for the reversal of the final pair, which is consistent with the inversion of the segment containing the genes. This region of the Arabidopsis genome had been shown previously to be related to the sequenced segment of the rice genome (van Dodeweerd at al. 1999). The Arabidopsis chromosome 5 segment contains seven conserved genes. These are also in conserved order and orientation, except the same reversed pair of genes. This region of the Arabidopsis genome had been shown previously to be related to the chromosome 4(a) segment (Bancroft 2000). The remaining segments contain seven, five, and five conserved genes for the chromosome 2, 4(b), and 3 segments, respectively, all in conserved order. However, the orientation of several of the individual genes is reversed, indicating possible small-scale inversion events.
Table 3.
Homology Scores for Conserved Arabidopsis and Rice Genes: Rice Chromosome 2 Segment
| Rice gene | Arabidopsis genes (P value) |
|---|---|
| C75 | AT2g44740 (8.1e-37) C |
| C135 | AT2g44780 (1.2e-83) C; AT3g60260 (1.5e-83) W |
| C200 | AT5g47180 (4.3e-54) C |
| C202 | AT4g17560 (1.0e-22) W; At5g47190 (2.3e-39) C |
| W265 | AT4g17550 (2.1e-143) C |
| C345 | AT3g60270 (1.1e-25) C |
| W455 | AT4g17530 (6.0e-96) C; AT5g47200 (1.1e-71) W |
| W485 | AT2g44810 (3.7e-70) W; AT4g16820 (1.3e-105) W |
| W495 | AT4g16800 (3.7e-54) C |
| W505 | AT4g16800 (2.0e-58) C |
| W535 | AT2g44830 (1.4e-170) W |
| W595 | AT4g17510 (3.8e-71) C |
| C600 | AT3g60320 (9.6e-74) C |
| C635 | AT4g17500 (6.6e-35) W; AT5g47220 (4.7e-31) C; AT2g44840 (4.1e-30) W |
| C650 | AT2g44850 (8.9e-63) W |
| W700 | AT4g17490 (3.1e-21) C; AT5g47230 (9.2e-27) W |
| W760 | AT4g17486 (2.7e-44) C; AT5g47310 (1.1e-50) W |
| C870 | AT4g16790 (5.5e-7) C; AT3g60380 (4.4e-9) C |
| W940 | AT3g44940 (1.7e-36) C; AT4g16750 (1.3e-31) C |
| W950 | AT4g17340 (6.7e-97) W; AT5g47450 (4.1e-97) C |
| W1050 | AT4g17350 (8.0e-46) W; AT5g47440 (1.8e-46) C; AT4g16670 (1.3e-36) W |
| C1190 | AT2g44940 (1.6e-31) C; AT3g60490 (3.7e-31) W |
Figure 3.
Comparison of the organization of conserved putative genes in the 333-kb rice DNA sequence and five segments of the Arabidopsis genome sequence. The relative coding strands of the genes are indicated by up- or down-pointing polygons. Duplicated genes in the rice sequence that detected homology to common Arabidopsis genes are indicated next to each other at the position of the gene with the lower reference number.
Analysis of Additional Regions of the Rice Genome
To assess the generality of our findings of gene density in the rice genome and the conservation of microstructure with the genome of Arabidopsis, we selected for analysis two further BACs that had been sequenced and submitted to public databases. One of these, P0436E04 (accession no. ap002818), was selected as the sequenced clone nearest to a telomere (map position 0.3 cM on chromosome 1). The other, P0406H10 (accession no. ap002524), was near the middle of a chromosome arm (20.2 cM on chromosome 1). We implemented our annotation protocols using these data and compared the putative genes derived with those accompanying the database submission. For P0436E04, 26 genes and three transposons were identified in 145 kb of sequence, compared with 24 genes and two transposons recorded with the submission. For P0406H10, 25 genes and one transposon were identified in 156 kb of sequence, compared with 26 genes and one transposon recorded with the submission. The densities of putative genes identified, one per 5.6 kb and one per 6.2 kb for P0436E04 and P0406H10, respectively, are very similar to the density found in the 340-kb region analyzed (one per 6.1 kb). Although the annotation accompanying database submissions of rice genome sequence suggested significantly different gene structures to those predicted by our protocols, the overall gene density predicted is very similar. The gene densities of these clones are typical of those accompanying the rice BAC sequences presently in the public databases. These results suggest that a typical gene density for the rice genome is around one gene per 6 kb.
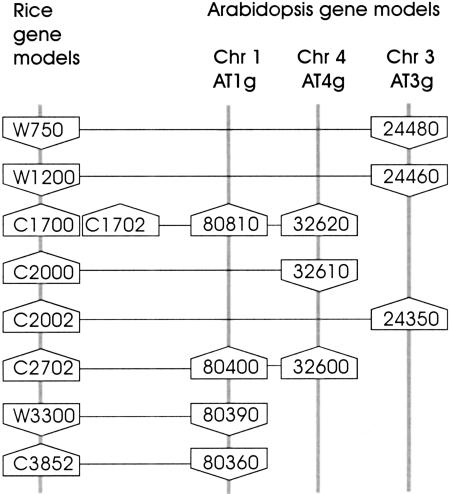
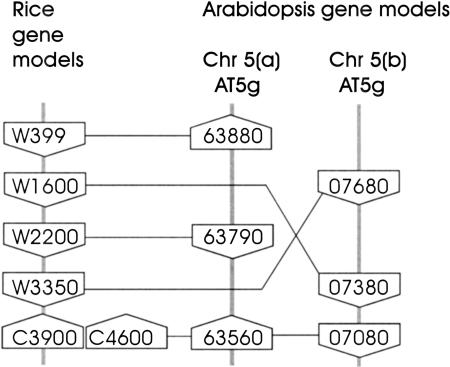
Searches were conducted for segments of the Arabidopsis genome that contain conserved gene content and order for each of BAC clones P0436E04 and P0406H10. The same methods and recording criteria were used. The results are summarized in Table 4 and Figure 4 for P0406H10, and Table 5 and Figure 5 for P0436E04. In both cases multiple conserved segments were identified. Only three or four conserved genes were identified in each segment; there was one reversal of gene order (W1600-AT5g07380 and W3350-AT5g07690), and the strand orientation of several of the genes was not conserved (i.e., C3852-AT1g80360, C2000-AT4g32610, W399-AT5g63880, C3900-AT5g07080). However, these results indicate that it may be feasible to align much of the rice genome with duplicated segments of the genome of Arabidopsis.
Table 4.
Homology Scores for Conserved Arabidopsis and Rice Genes: BAC P0406H10
| Rice gene | Arabidopsis genes (P value) |
|---|---|
| W750 | AT3g24480 (1.5e-106) C |
| W1200 | AT3g24460 (2.9e-32) C |
| C1700 | AT4g32620 (8.1e-100) W |
| C1702 | AT1g80810 (8.1e-08) W; AT4g32620 (3.7e-32) W |
| C2000 | AT4g32610 (4.1e-09) C |
| C2002 | AT3g24350 (4.8e-94) W |
| C2702 | AT1g80400 (3.7e-96) W; AT4g32600 (8.9e-102) W |
| W3300 | AT1g80390 (7.7e-24) C |
| C3852 | AT1g80360 (7.3e-68) C |
Figure 4.
Comparison of the organization of conserved putative genes in rice BAC P0406H10 and three segments of the Arabidopsis genome sequence. Representation is as shown in Figure 3.
Table 5.
Homology Scores for Conserved Arabidopsis and Rice Genes: BAC P0436E04
| Rice gene | Arabidopsis genes (P value) |
|---|---|
| W399 | AT5g63880 (5.5e-08) W |
| W1600 | AT5g07380 (8.5e-55) W |
| W2200 | AT5g63790 (5.3e-53) C |
| W3350 | AT5g07680 (1.9e-60) W |
| C3900 | AT5g63560 (1.4e-46) W; ATg07080 (1.2e-30) W |
| C4600 | AT5g63560 (1.7e-08) W; AT5g07080 (2.1e-06) W |
Figure 5.
Comparison of the organization of conserved putative genes in rice BAC P0436E04 and two segments of the Arabidopsis genome sequence. Representation is as shown in Figure 3.
DISCUSSION
Using a combination of approaches, 56 genes were predicted in the 340 kb of rice genome sequence data we generated and analyzed, indicating a density of one gene per 6.1 kb. This density is close to that found for the genome of Arabidopsis; that is, one gene per 4.76 kb (Bancroft 2000), but higher than that found near the ADH1 locus of rice, one gene per 10.3 kb (Tarchini et al. 2000). Extrapolation to the 440-Mb genome of rice, using the gene densities of one per 6.1 kb or 10.3 kb, would predict the presence of ∼72,000 or ∼43,000 genes in the rice genome, respectively. However, there is evidence of gene-rich and gene-poor isochores in the rice genome based on bulk sequence composition (Barakat et al. 1997), and both regions analyzed are likely to be characteristic of the gene-rich regions. If we estimate that the rice ESTs presently in dbEST represent ∼10,000 nonredundant genes, our observation that 16 of the 56 predicted genes identified (29%) have EST matches leads us to predict a total gene number for rice of ∼35,000. This would be consistent with a model in which the majority of the rice genes are contained in gene-rich regions comprising 50% of the genomic DNA of rice (220 Mb), with these gene-rich regions typically containing a gene density of one per ∼6 kb, as we have observed.
The composition of the region we have analyzed differs significantly from that near the ADH1 locus (Tarchini et al. 2000). In addition to more predicted genes in a region of almost identical size (56, compared with 33), we identified fewer transposons and retrotransposons (2, compared with 15). There are smaller gene/pseudogene families; for example, the largest gene family we identified contained four members, compared to 13 members. The region around the ADH1 locus contains several genes with homology to genes known to be involved in plant disease resistance. It has a complex structure, including a large family of genes, some of which do not encode a full and functional protein, and several retrotransposons. This resembles the structure of the Arabidopsis ecotype Columbia allele of the RPP5 disease-resistance locus identified on chromosome 4 (Bevan et al. 1998). However, this is an unusual genome organization, and is not representative of the genome structure as a whole (Lin et al. 1999; Mayer et al. 1999).
Sixteen of the 56 modeled genes (29%) match EST sequences, supporting the predicted gene models. Further support for the authenticity of our predicted genes come from the highly significant homology that the predicted products of many of them show to known or predicted proteins in other species. Forty-six of the 56 predicted genes (82%) show such homology. These data suggest that the majority of our gene models correctly indicate the presence of a gene. It also suggests that the EST representation in rice may be relatively low, which in turn might indicate that many of the genes of rice are expressed at low levels generally, only in specific cells or in response to specific conditions. The 10 gene models for which no homology has been identified may be false gene predictions or genes unique to rice.
The framework of conserved genes preserved between segments of the genomes of Arabidopsis and rice suggests that mechanisms of genome evolution have been operating to delete, rearrange, and disperse single or small groups of genes, resulting in extensive genome reshuffling during plant evolution. This is inconsistent with the suggestion that plant genome organization might have evolved primarily by gross rearrangements, permitting the construction of unified genetic maps (Paterson et al. 1996). Mechanisms that might achieve the observed divergence of genome fine structure may involve mobile genetic elements, as has been found to contribute to “exon shuffling” in mammalian systems (Boeke and Pickeral 1999). It is also likely that unequal crossing over contributes to both tandem duplications of genes, and deletion of single or small groups of genes (Bancroft 2001).
Many duplicated regions have been identified within the genome of Arabidopsis (Lin et al. 1999; Mayer et al. 1999; Bancroft 2000). It has been suggested that these may have been the result of an ancestral tetraploidy event (Blanc et al. 2000; The Arabidopsis Genome Initiative 2000), or multiple duplication events (Vision et al. 2000). Our data support the hypothesis that there have been multiple duplication events during the evolution of the genome of Arabidopsis. These duplicated segments appear to have diverged extensively by the loss of different subsets of interspersed genes. The relationships of such highly diverged duplicated segments is revealed most clearly by comparative sequence analysis with relatively distantly related species, such as tomato (Ku et al. 2000) or rice. By integrating the data from multiple duplicated segments of the Arabidopsis genome we have been able to align segments of the rice and Arabidopsis genomes and deduce the ancestral relationships of sets of genes. It is not known whether the 340-kb rice genome segment studied is also the product of genome duplication events during the ancestry of rice. When the rice genome sequence data become available, it should be possible to analyze complex relationships within the rice genome by extensive analysis using the Arabidopsis genome sequence. By taking due account of the mechanisms of the evolution of plant genome structure, it may be possible to make extensive use of comparative genome analysis to integrate structural and functional genomics of dicot and monocot species.
METHODS
Sequencing of BAC Clones
Individual BAC clones were sequenced by standard methods using a shot-gun approach (Bodenteich et al. 1993). Cesium chloride-purified BAC DNA was sheared by nebulization (Roe et al. 1996). After end-filling, DNA fragments were size fractionated and cloned into the SmaI site of pUC18 or HincII site of pUC19 (Amersham Pharmacia Biotech). Clones were sequenced using the ABI PRISM Dye Terminator Cycle Sequencing ready Reaction kit with FS AmpliTaq DNA polymerase (PE Applied Biosystems) and analyzed on ABI 377 (PE Applied Biosystems) sequencing gels. The sequence data were assembled using PHRED/PHRAP software (Green 1996).
Analysis of Sequence Data
The sequence was subjected to a modified analysis procedure based on that established for genome analysis of Arabidopsis thaliana (Mayer et al. 1999). BLAST (Altschul et al. 1997) analysis of the sequence against the EMBL nucleotide database and MIPS in-house databases (a nonredundant protein database, a plant transposon database, a rice EST database, and an all-plant EST database) was performed. Gene predictions were performed using Genscan (Burge and Karlin 1997), GeneFinder (P. Green and L. Hillier, unpubl. software), FGENESH (A.A. Salamov and V.V. Soloyev, unpubl. software; http://genomic.sanger.ac.uk/gf/gf.shtml), and Genemark.hmm (Lukashin and Borodovsky 1998). An Oryza sativa setting is available only for Genemark.hmm. For GeneFinder as well as Genscan the Arabidopsis setting was used. The Zea mays setting available for Genscan yielded less reliable results. Splice-site predictions using Netplantgene2 (Tolstrup et al. 1997) (Arabidopsis setting) gave unreliable results, and was not used for gene modeling.
Gene modeling was performed by combining intrinsic data (gene predictions) with extrinsic data (database matches). Gene models were adjusted to fit EST data from rice and other plants as well as to homologous protein matches where available. For genes not supported by any database matches the FGENESH prediction was generally used.
Protein domain characterization was performed using the InterPro software (The InterPro Consortium 2000), and similarity analysis of extracted proteins was performed by BLASTP comparison to a nonredundant protein database.
Acknowledgments
This work was funded under the BBSRC GAIT Initiative (grant 208/GAT09069) and the EU Arabidopsis Genome Sequencing Project (CT97-0274).
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL ian.bancroft@bbsrc.ac.uk; FAX: 44 1603 259882.
Article published on-line before print: Genome Res., 10.1101/gr.161701.
Article and publication are at http://www.genome.org/cgi/doi/10.1101/gr.161701.
REFERENCES
- Arumuganathan K, Earle ED. Nuclear DNA content of some important plant species. Plant Mol Biol Rep. 1991;9:208–218. [Google Scholar]
- Altschul SF, et al. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bancroft I. Insights into the structural and functional evolution of plant genomes afforded by the nucleotide sequences of chromosomes 2 and 4 of Arabidopsis thaliana. Yeast. 2000;17:1–5. doi: 10.1002/(SICI)1097-0061(200004)17:1<1::AID-YEA3>3.0.CO;2-V. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ————— Duplicate and diverge: The evolution of plant genome microstructure. Trends Genet. 2001;17:89–93. doi: 10.1016/s0168-9525(00)02179-x. [DOI] [PubMed] [Google Scholar]
- Bancroft I, Jones JDG, Dean C. Heterologous transposon tagging of the DRL1 locus in Arabidopsis. Plant Cell. 1993;5:631–638. doi: 10.1105/tpc.5.6.631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barakat A, Carels N, Bernardi G. The distribution of genes in the genomes of Gramineae. Proc Natl Acad Sci. 1997;94:6857–6861. doi: 10.1073/pnas.94.13.6857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bevan M, et al. Analysis of 1.9 Mb of contiguous sequence from chromosome 4 of Arabidopsis thaliana. Nature. 1998;391:485–488. doi: 10.1038/35140. [DOI] [PubMed] [Google Scholar]
- Blanc G, Barakat A, Guyot R, Cooke R, Delseny M. Extensive duplication and reshuffling in the Arabidopsisgenome. Plant Cell. 2000;12:1093–1101. doi: 10.1105/tpc.12.7.1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bodenteich A, Chissoe S, Wang YF, Roe BA. Shot-gun cloning as the strategy of choice to generate templates for high-throughput dideoxynucleotide sequencing. In: Venter JC, editor. Automated DNA sequencing and analysis techniques. London: Academic Press; 1993. pp. 42–50. [Google Scholar]
- Boeke JD, Pickeral OK. Retroshuffling the genomic deck. Nature. 1999;398:108–111. doi: 10.1038/18118. [DOI] [PubMed] [Google Scholar]
- Burge C, Karlin S. Prediction of complete gene structures in human genomic DNA. J Mol Biol. 1997;268:78–94. doi: 10.1006/jmbi.1997.0951. [DOI] [PubMed] [Google Scholar]
- Cavell AC, Lydiate DJ, Parkin IAP, Dean C, Trick M. Collinearity between a 30-centimorgan segment of Arabidopsis thaliana chromosome 4 and duplicated regions within the Brassica napusgenome. Genome. 1998;41:62–69. [PubMed] [Google Scholar]
- Chen M, SanMiguel P, Bennetzen JL. Sequence organization and conservation in sh2/a1-homologous regions of sorghum and rice. Genetics. 1998;148:435–443. doi: 10.1093/genetics/148.1.435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devos KM, Beales J, Nagamura Y, Sasaki T. Arabidopsis-rice: Will colinearity allow gene prediction across the eudicot-monocot divide? Genome Res. 1999;9:825–829. doi: 10.1101/gr.9.9.825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feldmann KA, Marks MD, Christianson ML, Quatrano RS. A dwarf mutant of Arabidopsisgenerated by T-DNA insertional mutagenesis. Science. 1989;243:1351–1354. doi: 10.1126/science.243.4896.1351. [DOI] [PubMed] [Google Scholar]
- Gale MD, Devos KM. Plant comparative genetics. Science. 1998;282:656–659. doi: 10.1126/science.282.5389.656. [DOI] [PubMed] [Google Scholar]
- Giraudat J, Hauge BM, Valon C, Smalle J, Parcy F, Goodman HM. Isolation of the ArabidopsisABI3 gene by positional cloning. Plant Cell. 1992;4:1251–1261. doi: 10.1105/tpc.4.10.1251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green P. Towards completely automated sequence assembly. DOE Human Genome Program Contractor-Grantee Workshop V, 157. Washington, DC.: U.S. Department of Energy, Office of Science, Office of Biological and Environmental Research; 1996. [Google Scholar]
- Han F, Kilian A, Chen JP, Kudrna D, Steffenson B, Yamamoto K, Matsumoto T, Sasaki T, Kleinhofs A. Sequence analysis of a rice BAC covering the syntenous barley Rpg1region. Genome. 1999;42:1071–1076. doi: 10.1139/g99-060. [DOI] [PubMed] [Google Scholar]
- Kinoshita T. Report of committee on gene symbolization. Rice Genet Newslett. 1995;12:9–153. [Google Scholar]
- Koornneef M. Arabidopsis thaliana. In: O'Brien S J, editor. Genetic maps. New York: Cold Spring Harbor Laboratory Press; 1990. pp. 6.93–9.96. [Google Scholar]
- Kowalski SP, Lan T-H, Feldmann KA, Paterson AH. Comparative mapping of Arabidopsis thaliana and Brassica oleraceachromosomes reveals islands of conserved organization. Genetics. 1994;138:499–510. doi: 10.1093/genetics/138.2.499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ku H-M, Vision T, Liu S, Tanksley SD. Comparing sequenced segments of the tomato and Arabidopsisgenomes: Large-scale duplication followed by selective gene loss creates a network of synteny. Proc Natl Acad Sci. 2000;97:9121–9126. doi: 10.1073/pnas.160271297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurata N, et al. A 300 kilobase interval genetic map of rice including 883 expressed sequences. Nat Genet. 1994;8:365–372. doi: 10.1038/ng1294-365. [DOI] [PubMed] [Google Scholar]
- Lagercrantz U. Comparative mapping between Arabidopsis thaliana and Brassica napusindicates that brassica genomes have evolved through extensive genome replication accompanied by chromosome fusion and frequent rearrangements. Genetics. 1998;150:1217–1228. doi: 10.1093/genetics/150.3.1217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin X, et al. Sequence and analysis of chromosome 2 of the plant Arabidopsis thaliana. Nature. 1999;402:761–768. doi: 10.1038/45471. [DOI] [PubMed] [Google Scholar]
- Lister C, Dean C. Recombinant inbred lines for mapping RFLP and phenotypic markers in Arabidopsis thaliana. Plant J. 1993;4:745–750. doi: 10.1046/j.1365-313x.1996.10040733.x. [DOI] [PubMed] [Google Scholar]
- Lukashin AV, Borodovsky M. GeneMark.hmm: New solutions for genefinding. Nucleic Acids Res. 1998;26:1107–1115. doi: 10.1093/nar/26.4.1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer K, et al. Sequence and analysis of chromosome 4 of the plant Arabidopsis thaliana. Nature. 1999;402:769–777. doi: 10.1038/47134. [DOI] [PubMed] [Google Scholar]
- Moore G, Devos KM, Wang Z, Gale MD. Grasses, line up and form a circle. Curr Biol. 1995;5:737–739. doi: 10.1016/s0960-9822(95)00148-5. [DOI] [PubMed] [Google Scholar]
- Paterson AH, et al. Towards a unified genetic map of higher plants, transcending the monocot-dicot divergence. Nat Genet. 1996;14:380–382. doi: 10.1038/ng1296-380. [DOI] [PubMed] [Google Scholar]
- Roe BA, Crabtree JS, Khan AS. DNA isolation and sequencing. New York: John Wiley and Sons; 1996. [Google Scholar]
- Schmidt R, West J, Love K, Lenehan Z, Lister C, Thompson H, Bouchez D, Dean C. Physical map and organization of Arabidopsis thalianachromosome 4. Science. 1995;270:480–483. doi: 10.1126/science.270.5235.480. [DOI] [PubMed] [Google Scholar]
- Tarchini R, Biddle P, Wineland R, Tingey S, Rafalski A. The complete sequence of 340 kb of DNA around the rice Adh1–Adh2region reveals interrupted colinearity with maize chromosome 4. Plant Cell. 2000;12:381–391. doi: 10.1105/tpc.12.3.381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The Arabidopsis Genome Initiative. Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature. 2000;408:796–815. doi: 10.1038/35048692. [DOI] [PubMed] [Google Scholar]
- The InterPro Consortium. InterPro—An integrated documentation resource for protein families, domains and functional sites. Bioinformatics. 2000;16:1145–1150. doi: 10.1093/bioinformatics/16.12.1145. [DOI] [PubMed] [Google Scholar]
- Tikhonov AP, SanMiguel PJ, Nakajima Y, Gorenstein NM, Bennetzen JL, Avramov Z. Colinearity and its exceptions in orthologoues adh regions of maize and sorghum. Proc Natl Acad Sci. 1999;97:7409–7414. doi: 10.1073/pnas.96.13.7409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tolstrup N, Rouze P, Brunak S. A branch point consensus from Arabidopsisfound by non-circular analysis allows for better prediction of acceptor sites. Nucleic Acids Res. 1997;25:3159–3163. doi: 10.1093/nar/25.15.3159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Dodeweerd A-M, Hall CR, Bent EG, Johnson SJ, Bevan MW, Bancroft I. Identification and analysis of homoeologous segments of the genomes of rice and Arabidopsis thaliana. Genome. 1999;42:887–892. [PubMed] [Google Scholar]
- Vision TJ, Brown DG, Tanksley SD. The origins of genomic duplications in Arabidopsis. Science. 2000;290:2114–2117. doi: 10.1126/science.290.5499.2114. [DOI] [PubMed] [Google Scholar]
- Wolfe KH, Gouy M, Yang Y-W, Sharp PM, Li W-H. Date of the monocot-dicot divergence estimated from chloroplast DNA sequence data. Proc Natl Acad Sci. 1989;86:6201–6205. doi: 10.1073/pnas.86.16.6201. [DOI] [PMC free article] [PubMed] [Google Scholar]