Abstract
We have constructed NheI and XhoI optical maps of Escherichia coli O157:H7 solely from genomic DNA molecules to provide a uniquely valuable scaffold for contig closure and sequence validation. E. coli O157:H7 is a common pathogen found in contaminated food and water. Our approach obviated the need for the analysis of clones, PCR products, and hybridizations, because maps were constructed from ensembles of single DNA molecules. Shotgun sequencing of bacterial genomes remains labor-intensive, despite advances in sequencing technology. This is partly due to manual intervention required during the last stages of finishing. The applicability of optical mapping to this problem was enhanced by advances in machine vision techniques that improved mapping throughput and created a path to full automation of mapping. Comparisons were made between maps and sequence data that characterized sequence gaps and guided nascent assemblies.
Modern approaches to understanding the detailed molecular mechanisms that underlie microbial biological systems often start with whole genome sequencing and annotation (Ruepp et al. 2000; Shigenobu et al. 2000; Stover et al. 2000). Since the first microbe was fully sequenced a mere six years ago (Fleischmann et al. 1995), a large number of microbial genomes have been sequenced and an even larger number are slated to be completed over the coming year. Although new sequencing technologies (Dovichi 1997; Dolnik 1999; Endo et al. 1999; Pang et al. 1999; Wei and Yeung 2000) have to some extent ameliorated the daunting task of amassing the large number of sequence reads required to assemble a completed genome sequence, significant progress has not been made in new approaches to finish and validate such data. Whole genome shotgun sequencing techniques are widely used to eliminate the need for time-consuming mapping. The situation, however, is more complex. We think that shotgun sequencing approaches have not totally eliminated the requirement for maps but have instead developed the need for new types of maps in order to fully complement these high-throughput approaches.
Optical mapping is now a proven system for the construction of whole genome maps from genomic DNA molecules directly extracted from both bacteria and unicellular parasites (Lai et al. 1999a; Lin et al. 1999). The system creates ordered restriction maps using randomly selected individual DNA molecules mounted on specially prepared surfaces (Aston et al. 1999; Jing et al. 1999; Lai et al. 1999; Lin et al. 1999), without the use of electrophoresis, hybridization, PCR, or clones. Ordered restriction maps of an entire genome form a useful scaffold for guiding sequence assembly and for validating finished sequence. Because such maps are directly linked with the genome, they do not suffer from clone- or PCR-based artifacts, making them ideal for cross-checking sequencing efforts. Previous whole genome optical maps have indeed served in this capacity to aid large-scale sequencing efforts (Lai et al. 1999; Lin et al. 1999).
Pathogenic microbes are numerous and clinically important, but are often lacking well-developed genomic resources such as genetic markers, simple physical maps, and definitively characterized genome structural features. Such organisms are a challenge to genomicists engaged in large-scale sequencing projects, since simple facts regarding accurate genome size and chromosome number are obscure. Variation in pathogenicity observed between related bacterial strains can sometimes be associated with significant alterations to genome structure (Karaolis et al. 1994; Sokurenko et al. 1998). The bacterial genome we have optically mapped here, Escherichia coli O157:H7 EDL933, produces a Shiga toxin. E. coli expressing this toxin cause over 100,000 cases of human illness annually in the United States alone and pose a significant threat to public health worldwide. Most cases are associated with strains of the serotype O157:H7, and 85% of these are linked to contaminated food (Mead et al. 1999).
To sequence and annotate this virulent bacterium, the Blattner laboratory has adopted a strategy of using the E. coli K-12 genome (Blattner et al. 1997) as a backbone for new sequence assembly and annotation. This strategy was designed to quickly highlight a subset of additional candidate genes for further characterization by comparison of the O157:H7 sequence to that of the nonpathogenic E. coli K-12. The O157:H7 genome was expected to be considerably larger than that of K-12 based on the sizes of fragments generated by digestion of genomic DNA with a rare cutting restriction enzyme (Bergthorsson and Ochman 1998). However, those regions common to both genomes were expected to be nearly identical (Whittam et al. 1998). Genome sequencing has now confirmed that there are extensive differences between the two genomes that are distributed throughout a backbone of highly conserved and basically colinear shared genes (Blattner et al. 1997; Perna et al. 2001). A strategy employed in the O157:H7 genome project was to capitalize on this backbone by using sequences similar to regions of the K-12 genome as an indicator of contig order and to direct gap closure. The optical maps presented here were undertaken to provide a unique scaffold for assembly of the O157:H7 genome, but they also proved invaluable in providing an early indication of a major genomic rearrangement that simplified gap closure efforts.
RESULTS
Strategy for Mapping
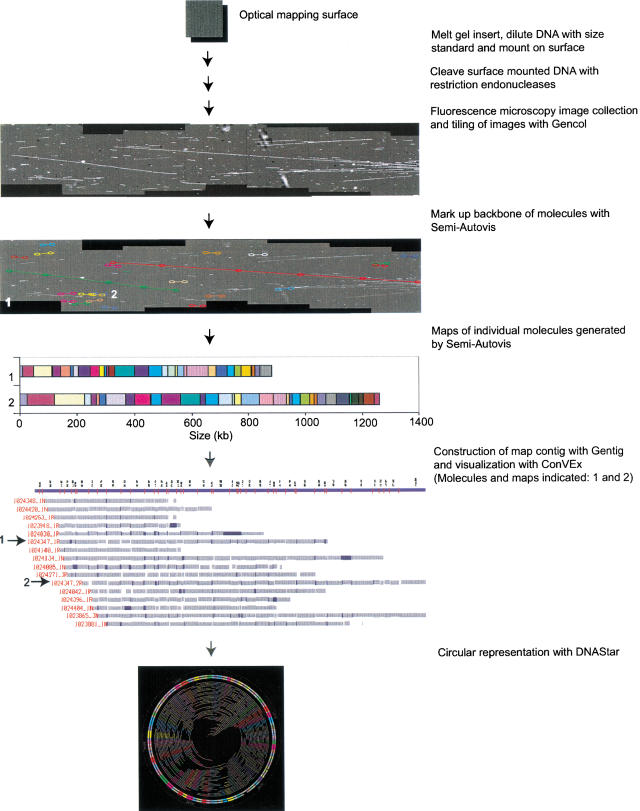
Previously, we developed an approach to mapping entire genomes, termed shotgun optical mapping (Fig. 1; Lai et al. 1999; Lin et al. 1999). Randomly broken DNA molecules that ranged in size from 150–2900 kb were used as the mapping substrate. Molecule breakage was not deliberate, but occurred as a consequence of handling. Surface mounted molecules were digested (on optical mapping surfaces) with restriction endonucleases, and images were collected using Gencol (see Methods). The basis of how shotgun optical mapping assembles whole genome maps is similar in many ways to random clone mapping approaches that assemble tiling paths across chromosomes and entire genomes (Marra et al. 1997; Soderlund et al. 1997; Han et al. 2000). Here, a single molecule optical map corresponds to a clone map discerned by gel electrophoresis. The assembly of maps into complete contigs covering the entire genome was accomplished by software called Gentig (Anantharaman et al. 1997; Lai et al. 1999). The Gentig algorithms were specially created to deal with the types of errors unique to the analysis of single DNA maps. Error processes such as partial digestion, spurious cuts, chimeric molecules (an imaging artifact caused by overlapping molecules), and fragment sizing error were rigorously modeled and integrated into Gentig.
Figure 1.
Scheme for shotgun optical mapping. High-molecular weight DNA is simply extracted from cells and deposited onto an optical mapping surface. After restriction endonuclease digestion and staining with a fluorescent dye, individual molecules are imaged by fluorescence microscopy. Images are collected using Gencol, which accumulates overlapping images in a semiautomated fashion and preserves registration. Semi-Autovis is then used to automatically convert image data into map files after a user selects molecules. Maps are then automatically contiged using Gentig, and the results are displayed using ConVEx. ConVEx allows the user to edit contigs, view statistics, and browse molecular images. Finished maps are visualized as a circular chromosome using software from DNAStar.
Optical Maps
Gentig was used to assemble two separate optical maps of E. coli O157:H7, using XhoI and NheI. The NheI map was first constructed and represents a preliminary map in that final editing was not completed. It became apparent from communications with the group sequencing this genome (F.R. Blattner, pers. comm.) that a second enzyme map was necessary since a difficult and long sequence stretch was not adequately represented in the preliminary NheI map. New in silico analysis of available sequence showed that an XhoI map would be more useful for finishing the sequence data. Additional sequence data and the XhoI map subsequently showed that this difficult stretch (∼ 450 kb) was indeed absent from the preliminary NheI map.
Figure 2a shows a typical molecule and its associated map. A total of 840 molecules were collected and processed for map construction (XhoI: 494 molecules collected, 251 of which went into the final contig; NheI: 346 molecules collected, 220 of which went into the final contig). The two enzymes apparently cleaved the genome to produce random patterns, with no obvious discernment of structural features. However, the average fragment size significantly differed. The XhoI map featured an average restriction fragment size of 25.1 kb versus 32.3 kb calculated for NheI.
Figure 2.
(a) Digital fluorescence micrograph and map of a typical genomic DNA molecule. An E. coli O157:H7 molecule digested with XhoI is shown with its corresponding optical map. Image was constructed by tiling a series of 63× (objective power) images using GenCol. Comounted λ bacteriophage DNA was used as a sizing standard and to estimate enzymatic cutting efficiencies. (b) Whole genome XhoI restriction map of E. coli O157 generated by shotgun optical mapping. The outer circle represents an in silico XhoI digest of the sequence. The second outermost circle shows the consensus optical map. The inner circles represent the individual molecule maps from which the consensus map was generated. XhoI fragment sizes (in kilobases) can be measured from the figure. Colors are arbitrarily assigned to homologous overlapping fragments. The white triangles show discrepancies between the sequence and the optical map. These regions are detailed in Table 1.
Figure 2b shows the finished XhoI map constructed using Gentig with 251 molecules, providing 30× coverage (166 Mb of total DNA analyzed). This map formed a closed circle, with no gaps, and a typical restriction fragment was computed from the average of 20 molecules. Importantly, this depth of coverage ensured confidence in calling restriction cleavage sites and accuracy in fragment sizing. The genome size was calculated to be 5.52 Mb.
Optical Maps versus Sequence
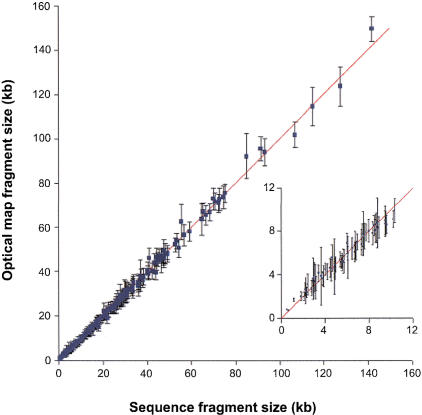
A comprehensive overview of optical mapping accuracy versus sequence is shown in Figure 3. The error bars were calculated as the standard deviation on sets of homologous fragments used to calculate the average consensus map shown in Figure 2b. Overall there was excellent agreement between map fragment sizes and those generated in silico using sequence data. For XhoI, the precision was estimated from the median of the standard deviation determined for all fragments (2.06 kb; for a range in fragment sizes spanning 0.71–149.6 kb). The median of the absolute error (|map-sequence|) was 0.52 kb. Although the average percent relative error ({map − sequence/sequence}*100%) remained somewhat constant at 4.8%, the absolute error expectedly increased with fragment size.
Figure 3.
XhoI restriction endonuclease fragment sizing results for E. coli O157:H7 plotted against sequence data. The diagonal line is for reference. The error bars represent the standard deviation of the fragment sizes. (Inset) Fragment sizes <10 kb.
Comparisons of the NheI map with sequence showed errors similar to the XhoI map, when the missing genomic region was taken into consideration. The average and median relative error values were 5.43%, and 3.32%; respectively.
Table 1 shows a detailed comparison of selected portions of the XhoI optical map with the corresponding restriction map predicted from sequence. These regions of the genome were selected since they show discrepancies between the optical map and sequence. Two discrepancies are readily discerned and are correspondingly noted in the table and in Figure 2b as “O” and “R.” These correspond to regions in the genome where there are phage insertions (CP-933O and CP-933R, Perna et al. 2001). Manual rearrangement of some of their phage sequence here and elsewhere in the genome may result in a sequence map that aligns more closely with the optical map in these regions (B. Mau, pers. comm.). The remaining discrepancies in regions “1,” “2,” “3,” and “V” (in Table 1 and Fig. 2b) have either extra cuts in the sequence or missing cuts in the optical map. The region in V is similar to O and R in that it contains a phage insertion (CP-933V, Perna et al. 2001). The relative error for these discrepancies was calculated by adding the sequence fragments together and comparing them to the corresponding optical map fragments. The following section discusses these remaining discrepancies in more detail, in the context of the composite optical maps (NheI and XhoI).
Table 1.
Comparison of Portions of the XhoI with the Corresponding Restriction Map
| Region with discrepancy | Sequence fragment size (kb) | Optical map fragment size (kb) | Difference (kb) | % relative error | Standard deviation |
|---|---|---|---|---|---|
| 1 | 3.12 | 3.38 | −0.27 | 8.60 | 0.65 |
| 30.82 | 31.53 | −0.71 | 2.32 | 3.60 | |
| 25.00 | 31.82 | −2.12 | 7.14 | 4.64 | |
| 4.70 | |||||
| 7.72 | 7.89 | −0.17 | 2.16 | 1.12 | |
| 18.16 | 18.48 | −0.32 | 1.78 | 2.43 | |
| O | 2.43 | 3.12 | −0.69 | 28.40 | 0.70 |
| 4.02 | 4.50 | −0.48 | 11.94 | 0.73 | |
| 0.31 | 0.31 | ||||
| 7.95 | −7.95 | 0.98 | |||
| 40.90 | 46.25 | −5.35 | 13.08 | 4.45 | |
| 24.44 | 23.54 | 0.90 | 3.68 | 2.00 | |
| R | 8.00 | 8.02 | −0.02 | 0.25 | 1.06 |
| 47.68 | 49.36 | −1.68 | 3.52 | 5.29 | |
| 6.47 | −6.47 | 0.84 | |||
| 21.72 | 19.12 | 2.60 | 11.97 | 2.33 | |
| 8.88 | 8.32 | 0.56 | 6.31 | 1.34 | |
| V | 29.95 | 31.06 | −1.12 | 3.73 | 4.73 |
| 4.48 | 4.61 | −0.13 | 2.91 | 1.02 | |
| 22.42 | 39.70 | 0.60 | 1.48 | 3.05 | |
| 17.88 | |||||
| 17.01 | 17.33 | −0.32 | 1.88 | 2.24 | |
| 3.82 | 3.61 | 0.21 | 5.54 | 0.97 | |
| 2 | 3.17 | 3.21 | −0.04 | 1.23 | 0.75 |
| 25.93 | 25.94 | −0.01 | 0.04 | 2.86 | |
| 13.60 | 39.11 | 1.29 | 3.18 | 2.45 | |
| 14.32 | |||||
| 12.49 | |||||
| 70.73 | 72.05 | −1.32 | 1.87 | 3.99 | |
| 36.14 | 34.70 | 1.44 | 3.99 | 2.25 | |
| 3 | 72.69 | 72.62 | 0.07 | 0.10 | 5.21 |
| 24.39 | 23.58 | 0.82 | 3.36 | 1.98 | |
| 79.67 | 113.50 | 1.61 | 1.40 | 9.42 | |
| 35.44 | |||||
| 28.22 | 27.37 | 0.84 | 2.99 | 2.38 | |
| 20.06 | 22.16 | −2.10 | 10.47 | 1.21 |
Composite Maps
Composite maps constructed from multiple enzymes are more informative than a single enzyme map showing the same average fragment size (Cai et al. 1998). For small clones, the alignment of separate maps derived from different enzymes is laborious, but straightforward. This task becomes difficult when multiple map alignments must be done covering an entire genome. We previously aligned two separate restriction maps spanning an entire chromosome (∼1 Mb) from Plasmodium falciparum (Jing et al. 1999), and our analysis indicated a complex set of errors, which were made apparent by local inversions in the order of closely spaced cleavage sites (between the two maps). Essentially, if one simply aligns several maps at a single end, the registration wanders from one end to the other. Here we were faced with the task of aligning two circular maps covering over 5 Mb.
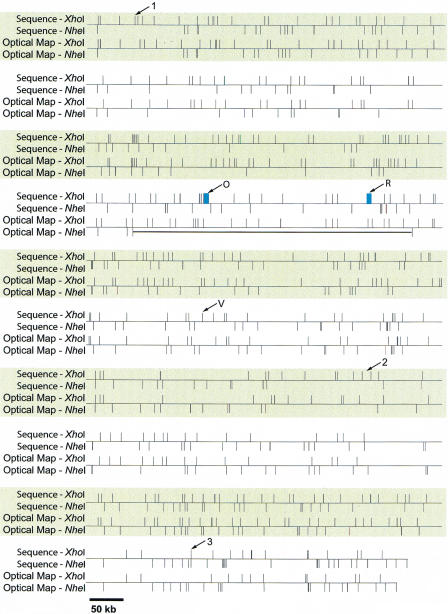
Figure 4 shows the alignment of the nascent NheI map with the finished XhoI results. The alignments were done by first normalizing each map, and then breaking them into discrete ∼ 500 kb sections. Alignments were then locally made by hand using the in silico (sequence) maps as a template. Leftmost alignments were done; however, this simple approach does not optimally fit all restriction sites to the sequence data. Errors in fragment sizing will shift restriction fragments relative to each other, and this becomes apparent when large map sections are simply aligned. Statistical analysis by our laboratory (Jing et al. 1999) predicted that misalignment grows as the square root of the distance from a known alignment (here, left end of alignments in Fig. 4), and that smaller fragments should show more instances of position reversal (i.e., restriction site of enzyme “A” vs. “B”). The data presented here had 197 instances where consecutive restriction sites were NheI followed by XhoI (or vice versa). In 61 of those instances the expected misalignment exceeded the distance between the restriction sites. Only half of all misalignments on average produce reversals of the restriction site order. Hence we can predict about 15–40 reversals. Actual data were observed to have 30 reversals, which is consistent with our prediction. A more appropriate approach we plan to implement will use a set of algorithms to optimize alignments for all fragments, which will rigorously model errors in both map and sequence data. Despite these concerns, the alignments show a high degree of correspondence and serve to flag errors in both sequence assembly and map construction.
Figure 4.
Alignment of map and sequence data. The use of sequence information to link single-enzyme optical maps. The composite optical map was generated by normalizing the single-enzyme maps to be the same size. The resulting multienzyme map was aligned with the map predicted from sequence. The thick black horizontal line denotes a missing region in the NheI optical map. The arrows show discrepancies between sequence and the optical maps. These discrepancies correspond to those in Figure 2b. The blue rectangles denote gaps in the sequence data compared to the XhoI optical map.
Several discrepancies between the optical maps and sequence were detected upon alignment. Notably, the absence of a 450 kb region is immediately evident in the NheI map, which was confirmed in both the XhoI map and sequence data. These data showed that the preliminary NheI map contained an assembly error, which omitted this 450 kb region. A gap in sequence (∼54kb) was also revealed when the composite optical maps were compared to sequence (gap 2, Perna et al. 2001). Since this gap was closed after sequencing new templates derived from fractionated genomic DNA, it is not reported here.
There are two small regions (∼ 7 and ∼ 6 kb fragments) present in the XhoI optical map that are missing from sequence (denoted in Table 1, Fig. 2b, and Fig. 4 as “O” and “R”). Unfortunately, these two regions could not be verified as “missing” using the NheI optical map, because they are located within the 450 kb region that was absent from the NheI optical map. However, these regions in the XhoI optical map each had significant coverage underlying the consensus map (roughly 20 molecules). This discrepancy between the XhoI optical map and sequence may be due to the fact that these regions coincide with phage elements that were difficult to assemble correctly because some sequence reads match the assembly in several different places where related phage are integrated. These phage regions are currently undergoing final sequence assembly (B. Mau, pers. comm.).
There are four regions where the number of fragments from sequence does not exactly match that from the optical map. These regions are denoted in Table 1, Fig. 2b, and Fig. 4 as “1,” “2,” “3,” and “V.” Optical map data in these regions showed the absence of 1–2 restriction enzyme sites. V is another instance of partially completed sequence assembly due to the difficulty of matching sequence reads to the correct phage locus. As an aside, we compared these regions with the recently released sequence (Hayashi et al. 2001), which matched the optical map in regions 1, 2, and V. However, such direct comparisons can only be used as a guide, since a different bacterial strain (RIMD 0509952) with the same O157:H7 serotype was sequenced.
DISCUSSION
Shotgun optical mapping provides a completely independent means to validate sequence assemblies that does not rely on the analysis of clones. This advantage creates a direct route to sequence information that obviates artifacts created by the cloning process, which include underrepresentation of difficult regions and insert rearrangements. Although Southern blotting analysis also directly analyzes genomic DNA, it is cumbersome and difficult to employ for high-resolution whole genome analysis. Map construction can be influenced by the use of sequencing data, so that finished maps would not represent truly independent results. To minimize any bias in sequence assembly, optical maps were constructed without detailed prior knowledge of sequence data. However, preliminary assessment of enzyme site frequencies facilitates the choice of appropriate mapping enzymes. Restriction enzymes that cut too frequently (fragments of <15 kb on the average) or too infrequently (fragments of >55 kb on the average) are not suitable for optical mapping of bacterial genomes. Problems in map assembly arise with frequent cutters because the average fragment size approaches the optical sizing error, while infrequent cutters provide insufficient information per molecule to allow confident map assemblies. To deal with these issues, partial sequence data were used to determine the approximate frequency of restriction enzyme cleavage. We transmitted the preliminary NheI map to the Blattner laboratory while they were in the early stage of sequence finishing and contig closure. At that point we determined that a critical region was not represented by the NheI map. Furthermore, it was not clear whether this region was absent or if the preliminary sequence assemblies were incorrect. Further analysis by the Blattner laboratory indicated that an XhoI map would facilitate sequence assembly efforts in this particular region (subsequently found missing in the NheI map; Fig. 4). More importantly, an NheI map would show insufficient detail to aid closure; hence an XhoI map was constructed. Given these results, future maps might be constructed in two stages; first, a “generic” optical map would be prepared in the absence of significant sequence data, later followed by an additional map (using a different enzyme) to fully leverage preliminary contig closure efforts.
Optical maps can be used to cross-check data — both derived from sequencing and other maps. Composite maps created using different enzymes require good registration to minimize errors in the relative placement of cleavage sites and thus need a way to anchor one map against another. Here, we used sequence information for this purpose, and the resulting composite map revealed discrepancies in both map and sequence data. A previous approach used an infrequent cutter to generate large fragments (in a tube) that were optically mapped (on surfaces) with a frequent cutter (Lin et al. 1999). Generally, when two maps contradict sequencing results in the same region, it is unlikely that the composite map data are incorrect. Overall, since composite maps are more informative than single enzyme maps, genomic structural details become more apparent, and these maps are a better scaffold for sequence assembly. The maps presented here were useful to the Blattner laboratory through the gap closure stages by identifying errors in preliminary assemblies and characterizing contig order and gap sizes. In addition, an accurate measure of genome size is valuable for estimating the quantity of random sequence to collect before starting gap closure.
Clearly, more maps provide more useful information, but the real net utility must be judged in a fiduciary manner as mapping versus sequence finishing costs. This equation will be different for each bacterial genome, and will depend on factors such as map resolution, as well as the nature and scope of sequencing problems. It is worthwhile considering that although the NheI map was missing a genomic region, the rest of the map was quite accurate and did greatly facilitate contig ordering. Development of a much higher throughput optical mapping system is currently underway via increased automation and new software approaches to better link map with sequence data. The XhoI map presented here took two weeks to complete and required the intensive effort of five individuals to prepare surfaces and mounts and edit assemblies. An important step in this direction was the development of new machine vision approaches embodied in Semi-Autovis. Recent, unpublished developments in the optical mapping system use new surface modalities that obviate operator intervention and potentiate the ability of the machine vision to correctly identify objects for the creation of large data files. This combination would allow for a dramatic reduction in costs and would further accelerate sequence finishing efforts, as well as provide a reliable means for validation.
METHODS
Cell Growth and DNA Preparation
The E. coli O157:H7 strain used for the mapping of this organism was the same strain used for sequencing (Perna et al. 2001). E. coli O157:H7 was grown to late log phase in LB broth (per Liter: 10 g tryptone, 5 g yeast extract, 5 g NaCl). Bacteria were washed in TNE buffer (10 mM Tris, pH 7.2, 200 mM NaCl, 100 mM EDTA) and embedded in low-melting, 1% agarose gel (InCert, FMC) to form 20 μL inserts. Bacteria were lysed with lysozyme (1 mg/mL) followed by proteinase K treatment (0.5 mg/mL) in buffer containing EDTA (100 mM, pH 8.0), sodium deoxycholate (0.2%), Brij-58 (polyoxyethylene 20 cetyl ether, 0.5%), and sarcosyl (0.5%). Prior to use, the DNA inserts were washed thoroughly overnight in TE to remove excess EDTA. To extract DNA, washed inserts were melted at 72°C for 7 min. A β-agarase solution (100 μL of TE + 1 μL (1 U) β-agarase, New England Biolabs), prewarmed to 40°C, was added to the melted inserts, and allowed to incubate at 40°C for 2 h. This concentrated DNA sample was equilibrated to room temperature. Then, 10 μL of the DNA sample was added to 490 μL of 30 pg/μL lambda bacteriophage DNA (New England Biolabs). Such samples were mounted onto an optical mapping surface and examined under a fluorescence microscope to check the integrity of the DNA sample, and also to check the concentration of the genomic DNA. If further dilution was needed, 100 μL of 30 pg/μL lambda bacteriophage was added to the sample. The sample was again examined under the microscope. Dilution and examination was iterated until the genomic DNA was dilute enough so that only a few genomic molecules could be seen distinctively in each field of view of the microscope.
Surface Preparation and Calibration
Glass cover slips (18 × 18 mm; Fisher's Finest) were racked in custom-made Teflon racks, and cleaned by boiling in concentrated nitric acid (HNO3) for at least 12 h. The cover slips were rinsed extensively with high-purity, dust-free water until the effluent attained neutral pH. The cleaning procedure was repeated with concentrated hydrochloric acid (HCl), which hydrolyzes the glass surface, preparing it for subsequent derivatization. The cleaned cover slips were rinsed extensively, and any unused cover slips were stored at room temperature under ethanol in polypropylene containers.
A stock (2% by weight) solution of 3-aminopropyldiethoxymethylsilane (APDEMS; Gelest), distilled under argon, was prepared by dissolving APDEMS in deionized water and allowed to hydrolyze on a shaker at room temperature for 7.5 h. Thirty-six cleaned cover slips were treated in 4.2 to 5.8 μm hydrolyzed APDEMS in 250 mL distilled ethanol on a 50 rpm shaker at room temperature for 48 h. Any unused derivatized surfaces were stored in the silane solution and were used for up to two weeks. The surfaces were assayed by digesting lambda bacteriophage DNA with 60 units of XhoI enzyme diluted in 100 μL of digestion buffer with 0.02% Triton at 37°C to determine optimal digestion times, which ranged from 9 to 12 min.
Sample Mounting
Capillary action was used to draw DNA solution (5 μL E. coli O157:H7) between a derivatized surface and a glass slide. Two sets of protocols were used for digestion: NheI — The resulting sandwich was allowed to sit at room temperature for a few minutes, then carefully peeled from the slide. Surface mounted DNA was digested with 1.5 μL (15 U) NheI (New England Biolabs) in 50 μL NEB buffer 2 for 8–15 min at 37°C, in a humidity chamber. The buffer was aspirated from the surface to halt digestion, followed by washing (2×) with high-purity water. The mounted sample was dried on a 55°C heating block for one minute. XhoI — Surface mounted DNA was digested with 3.0 μL (60 U) XhoI (New England Biolabs) in 100 μL of 1× NEB Buffer 2 for 9–12 min in a humidity chamber at 37°C. The enzyme solution was carefully pipetted from the surface, and the surface was washed (2×) with excess filtered, high-purity water. The surface was thoroughly dried in a dehumidifying chamber using dessicant (Drierite).
Image Acquisition
Mounted DNA molecules were stained by placing 5 μL 0.1 μM YOYO-1 (in TE containing 20% β-mercaptoethanol; Molecular Probes) on a clean slide. The mounted sample was carefully placed on top of the YOYO-1 solution, avoiding air bubbles. Consecutive microscope images were semiautomatically collected under software control (GenCol software; Lai et al. 1999; Lin et al. 1999) on optical mapping workstations (Aston et al. 1999b) using 63× microscope objectives. Comounted lambda DNA molecules were used to estimate the rate of digestion and to provide a fluorescence standard for sizing (Jing et al. 1999; Lai et al. 1999; Lin et al. 1999).
Image Processing
Images were processed using new software for semiautomatic processing, Semi-Autovis. Fine editing of molecule markups was performed using an image editing program, Visionade (Aston et al. 1999b). Semi-Autovis calculates restriction maps of molecules from an overlapping set of images. User input is limited to identification of the approximate location of suitable molecules, a step we plan to automate in future versions of the software. Semi-Autovis then locates the exact location of the center line (backbone) of all selected molecules as well as any other molecules that are nearby, the most likely locations of restrictions sites on each molecule based on the variation in intensity, and the integrated intensity of each molecule fragment so identified. This is done on each image separately. The results from overlapping images are then combined to merge long molecules, and sizes are translated from intensity units to an absolute scale (kilobases) by identifying nearby size standard molecules in the image whose restriction map and size are known. This produces a physical restriction map for each molecule identified by the user. Additional details are provided below:
A critical feature of Semi-Autovis is that it can automatically deal with crossing molecules, bright spots near molecules, and other object imperfections that can interfere with accurate fragment calling and sizing. Visionade required manual editing to eliminate object noise. Semi-Autovis identifies DNA molecules by looking for long, thin, bright objects that vary slowly in orientation. In the first phase, an algorithm identifies these isolated regions in the image, using both the fluorescence intensity and local directionality properties at each pixel. This is done by first applying a pattern matching filter in the shape of an idealized molecule, which is convolved with the input image in 16 different orientations and produces 16 new images. Each image corresponds to one of 16 different directions, and the value of a pixel in one of these images represents a calculation of the degree to which the pixel appears to lie on a molecule in the particular direction. An image is then constructed which contains, at each pixel, the highest of the 16 values for that pixel. These images are thresholded to remove both the background and small bright objects that do not match molecules in shape. This operation dramatically reduces the number of pixels that remain to be processed. The remaining pixels are clustered into connected regions, each of which may contain one or more DNA fragments; the filter tends to include pixels corresponding to small gaps between fragments, whether in the same molecule or different nearby molecules.
In the next phase, Semi-Autovis identifies the “backbones” (or center- lines) of the DNA fragments by computing the intensity contours at various levels of intensity and identifying “pointed ends” on these contours. The set of all pointed ends represents the end points of fragments thresholded at various levels and collectively define the center lines of the DNA fragments. This formulation has the advantage of only assuming that all objects are thin, without requiring them to be totally straight, and allowing multiple objects to cross each other. In addition, the locus of the thresholded fragment end points can be computed efficiently.
The backbones (DNA center lines) must now be processed to separate out crossed DNA molecules and locate gaps in the DNA molecules corresponding to restriction sites. First, each point on the backbone with more than two continuations (a crossing point) is analyzed by computing the angles of each backbone segment incident at that point and matching backbone segments lying in approximately the same direction. Next, each pair of matched up segments are joined into one DNA molecule. Any unmatched segments at the crossing point are treated as molecule ends. Now each molecule is defined by one or more backbone lines (possibly curved), where each line corresponds to one or more fragments. Within each backbone line the gaps between fragments will be small, since larger gaps would break up the DNA molecule into separate backbone lines. The next step is to locate the smaller gaps by analyzing the intensity profile along the backbone lines. A smooth intensity signal along the backbone is computed; for each position along the backbone, the intensity is calculated by summing the intensities for a set of pixels which are close to the backbone and lying along a line orthogonal to the backbone at that position.
Gaps are characterized by intensity dips with a characteristic inverted Gaussian shape. We train the parameters that characterize gaps from hand-marked-up training sets, and the final parameter set is able to find over 95% of the gaps that the human was able to identify with −4% false positives, versus 2.5% for human markups (data not shown).
The backbone section corresponding to each fragment is used to define an area roughly three times as wide as the actual molecule. If two areas overlap, pixels are assigned based on the nearest backbone pixel. The intensity of each fragment's area is integrated and used as an estimate of the mass of the fragment, which is later normalized.
Map Construction
Another software package called Gentig (Anatharaman et al. 1998, 1999; Lai et al. 1999; Lin et al. 1999) takes these single molecule restriction maps and combines them into a genome-wide contig using a Bayesian data error model. This model simultaneously estimates the data error rates while generating a contig map with as little error as possible by using all data redundancy present in the overlapping single-molecule maps. Gentig computes a false-positive probability each time a map overlap is considered, and accept the resulting contig only when we are very sure that the overlap could not be due to chance given the data errors. This way, Gentig avoids the exponential cost of the backtracking that this problem requires to ensure the best possible contig. This does mean that occasionally we may fail to close a gap in the contig when the quantity of data is barely sufficient in theory, but only a very small fraction of extra data is sufficient to allow Gentig to close the gap without exponential backtracking.
Acknowledgments
This work was supported by grants from the National Institutes of Health (HG00225–08, 5U01 A1 44387–05) to D.C.S., (5U01 A1 44387–05), an RMHC to F.R.B., an Alfred P. Sloan/NSF Fellowship in Molecular Evolution to N.T.P., and a Sloan/DOE fellowship to B.M.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL dcschwartz@facstaff.wisc.edu; FAX (608)265-6743.Article published on-line before print: Genome Res., 10.1101/gr. 172101.
Article and publication are at http://www.genome.org/cgi/doi/10.1101/gr.172101.
REFERENCES
- Anantharaman TS, Mishra B, Schwartz DC. Genomics via optical mapping 2. Ordered restriction maps. J Comput Biol. 1997;4:91–118. doi: 10.1089/cmb.1997.4.91. [DOI] [PubMed] [Google Scholar]
- Anantharaman TS, Mishra B, Schwartz DC. Courant Technical Report 760. New York: Courant Institute, New York University; 1998. Genomics via optical mapping III: Contiging genomic DNA and variations. [Google Scholar]
- Anantharaman TS, Mishra B, Schwartz DC. Genomics via optical mapping III: Contiging genomic DNA and variations. The Seventh International Conference on Intelligent Systems for Molecular Biology. 1999;7:18–27. [PubMed] [Google Scholar]
- Aston C, Mishra B, Schwartz DC. Optical mapping and its potential for large-scale sequencing projects. Trends Biotechnol. 1999a;17:297–302. doi: 10.1016/s0167-7799(99)01326-8. [DOI] [PubMed] [Google Scholar]
- Aston C, Hiort C, Schwartz DC. Optical mapping: An approach for fine mapping. Methods Enzymol. 1999b;303:55–73. doi: 10.1016/s0076-6879(99)03006-2. [DOI] [PubMed] [Google Scholar]
- Bergthorsson U, Ochman H. Distribution of chromosome length variation in natural isolates of Escherichia coli. Mol Biol Evol. 1998;15:6–16. doi: 10.1093/oxfordjournals.molbev.a025847. [DOI] [PubMed] [Google Scholar]
- Blattner FR, Plunkett G, III, Bloch CA, Perna NT, Burland V, Riley M, Collado-Vides J, Glasner JD, Rode CK, Mayhew GF, et al. The complete genome sequence of Escherichia coli K-12. Science. 1997;277:1453–1462. doi: 10.1126/science.277.5331.1453. [DOI] [PubMed] [Google Scholar]
- Cai W, Jing J, Irvin B, Ohler L, Rose E, Shizuya H, Kim U, Simon M, Anantharaman T, Mishra B, et al. High-resolution restriction maps of bacterial artificial chromosomes constructed by optical mapping. Proc Natl Acad Sci. 1998;95:3390–3395. doi: 10.1073/pnas.95.7.3390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dolnik V. DNA sequencing by capillary electrophoresis (review) J Biochem Biophys Methods. 1999;41:103–119. doi: 10.1016/s0165-022x(99)00041-x. [DOI] [PubMed] [Google Scholar]
- Dovichi NJ. DNA sequencing by capillary electrophoresis. Electrophoresis. 1997;18:2393–2399. doi: 10.1002/elps.1150181229. [DOI] [PubMed] [Google Scholar]
- Endo Y, Yoshida C, Baba Y. DNA sequencing by capillary array electrophoresis with an electric field strength gradient. J Biochem Biophys Methods. 1999;41:133–141. doi: 10.1016/s0165-022x(99)00043-3. [DOI] [PubMed] [Google Scholar]
- Fleischmann RD, Adams MD, White O, Clayton RA, Kirkness EF, Kerlavage AR, Bult CJ, Tomb J-F, Dougherty BA, Merrick JM, et al. Whole-genome random sequencing and assembly of Haemophilus influenzae Rd. Science. 1995;269:496–512. doi: 10.1126/science.7542800. [DOI] [PubMed] [Google Scholar]
- Han CS, Sutherland RD, Jewett PB, Campbell ML, Meincke LJ, Tesmer JG, Mundt MO, Fawcett JJ, Kim U, Deaven LL, et al. Construction of a BAC contig map of chromosome 16q by two-dimensional overgo hybridization. Genome Res. 2000;10:714–721. doi: 10.1101/gr.10.5.714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi T, Makino K, Ohnishi M, Kurokawa K, Ishii K, Yokoyama K, Han C-G, Ohtsubo E, Nakayama K, Murata T, et al. Complete genome sequence of Enterohemorrhagic Escherichia coli O157:H7 and genomic comparison with a laboratory strain K-12. DNA Res. 2001;8:11–22. doi: 10.1093/dnares/8.1.11. , 47–52. [DOI] [PubMed] [Google Scholar]
- Jing J, Lai Z, Aston C, Lin J, Carucci DJ, Gardner MJ, Mishra B, Anantharaman T, Tettelin H, Cummings LM, et al. Optical mapping of Plasmodium falciparum chromosome 2. Genome Res. 1999;9:175–181. [PMC free article] [PubMed] [Google Scholar]
- Karaolis DK, Lan R, Reeves PR. Sequence variation in Shigella sonnei (soneii), a pathogenic clone of Escherichia coli, over four continents and 41 years. J Clin Microbiol. 1994;32:796–802. doi: 10.1128/jcm.32.3.796-802.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai Z, Jing J, Aston C, Clarke V, Apodaca J, Dimalanta ET, Carucci DJ, Gardner MJ, Mishra B, Anatharaman TS, et al. A shotgun optical map of the entire Plasmodium falciparum genome. Nat Genet. 1999;23:309–313. doi: 10.1038/15484. [DOI] [PubMed] [Google Scholar]
- Lin J, Qi R, Aston C, Jing J, Anatharaman T, Mishra B, White O, Daly MJ, Minton KW, Venter JC, et al. Whole genome shotgun optical mapping of Deinococcus radiodurans. Science. 1999;285:1558–1562. doi: 10.1126/science.285.5433.1558. [DOI] [PubMed] [Google Scholar]
- Marra MA, Kucaba TA, Dietrich NL, Green ED, Brownstein B, Wilson RK, McDonald KM, Hillier LW, McPherson JD, Waterston RH. High throughput fingerprint analysis of large-insert clones. Genome Res. 1997;7:1072–1084. doi: 10.1101/gr.7.11.1072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mead PS, Slutsker L, Dietz V, McCaig LF, Bresee JS, Shapiro C, Griffin PM, Tauxe RV. Food-related illness and death in the United States. Emerg Infect Dis. 1999;5:607–625. doi: 10.3201/eid0505.990502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pang HM, Pavski V, Yeung ES. DNA sequencing using 96-capillary array electrophoresis. J Biochem Biophys Methods. 1999;41:121–132. doi: 10.1016/s0165-022x(99)00042-1. [DOI] [PubMed] [Google Scholar]
- Perna NT, Plunkett G, III, Burland V, Mau B, Glasner JD, Rose DJ, Mayhew GF, Evans PS, Gregor J, Kirpatrick HA, et al. Genome sequence of entrohemorrhagic Escherichia coli O157: H7. Nature. 2001;409:529–533. doi: 10.1038/35054089. [DOI] [PubMed] [Google Scholar]
- Ruepp A, Graml W, Santos-Martinez M, Koretke KK, Volker C, Mewes HW, Frishman D, Stocker S, Lupas AN, Baumeister W. The genome sequence of the thermoacidophilic scavenger Thermoplasma acidophilum. Nature. 2000;407:508–513. doi: 10.1038/35035069. [DOI] [PubMed] [Google Scholar]
- Shigenobu S, Watanabe H, Hattori M, Sakaki Y, Ishikawa H. Genome sequence of the endocellular bacterial symbiont of aphids Buchnera sp. APS. Nature. 2000;407:81–86. doi: 10.1038/35024074. [DOI] [PubMed] [Google Scholar]
- Soderlund C, Longden I, Mott R. FPC: A system for building contigs from restriction fingerprinted clones. Comput Appl Biosci. 1997;13:523–535. doi: 10.1093/bioinformatics/13.5.523. [DOI] [PubMed] [Google Scholar]
- Sokurenko EV, Chesnokova V, Dykhuizen DE, Ofek I, Wu X, Krogfelt KA, Struve C, Schembri MA, Hasty DL. Pathogenic adaptation of Escherichia coli by natural variation of the FimH adhesion. Proc Natl Acad Sci. 1998;95:8922–8926. doi: 10.1073/pnas.95.15.8922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stover CK, Pham XQ, Erwin AL, Mizoguchi SD, Warrener P, Hickey MJ, Brinkman FSL, Hufnagle WO, Kowalik DJ, Lagrou M, et al. Complete genome sequence of Pseudomonas aeruginosa PAO1, an opportunistic pathogen. Nature. 2000;406:959–964. doi: 10.1038/35023079. [DOI] [PubMed] [Google Scholar]
- Wei W, Yeung ES. Improvements in DNA sequencing by capillary electrophoresis at elevated temperature using poly(ethylene oxide) as a sieving matrix. J Chromatogr Biomed Sci Appl. 2000;745:221–230. doi: 10.1016/s0378-4347(00)00069-4. [DOI] [PubMed] [Google Scholar]
- Whittam TS, Reid SD, Selander RK. Mutators and long-term molecular evolution of pathogenic Escherichia coli O157:H7. Emerg Infect Dis. 1998;4:615–617. doi: 10.3201/eid0404.980411. [DOI] [PMC free article] [PubMed] [Google Scholar]