Abstract
This study reports generation of large-scale genomic resources for pigeonpea, a so-called ‘orphan crop species’ of the semi-arid tropic regions. FLX/454 sequencing carried out on a normalized cDNA pool prepared from 31 tissues produced 494 353 short transcript reads (STRs). Cluster analysis of these STRs, together with 10 817 Sanger ESTs, resulted in a pigeonpea trancriptome assembly (CcTA) comprising of 127 754 tentative unique sequences (TUSs). Functional analysis of these TUSs highlights several active pathways and processes in the sampled tissues. Comparison of the CcTA with the soybean genome showed similarity to 10 857 and 16 367 soybean gene models (depending on alignment methods). Additionally, Illumina 1G sequencing was performed on Fusarium wilt (FW)- and sterility mosaic disease (SMD)-challenged root tissues of 10 resistant and susceptible genotypes. More than 160 million sequence tags were used to identify FW- and SMD-responsive genes. Sequence analysis of CcTA and the Illumina tags identified a large new set of markers for use in genetics and breeding, including 8137 simple sequence repeats, 12 141 single-nucleotide polymorphisms and 5845 intron-spanning regions. Genomic resources developed in this study should be useful for basic and applied research, not only for pigeonpea improvement but also for other related, agronomically important legumes.
Keywords: Cajanus cajan L., next generation sequencing, transcriptome assembly, molecular markers and gene discovery
1. Introduction
Pigeonpea (Cajanus cajan L.) is one of the major pulse crops of the tropics and subtropics. The crop is grown on over 4.8 Mha across the world, with an annual production of 4.1 Mt (http://faostat.fao.org). It is a major food legume crop in South Asia and East Africa, with India as the largest producer (3.07 Mha) followed by Myanmar (0.54 Mha) and Kenya (0.20 Mha). It is the only cultivated food crop of the Cajaninae subtribe and has a diploid genome with 11 pairs of chromosomes (2n = 2x = 22) and an estimated genome size of 858 Mb.1 Pigeonpea is a rich source of protein and vitamin B, and as a leguminous plant, it contributes as much as 40 kg nitrogen per hectare to the soil.
Despite its economic and ecological importance, this crop has gained less attention in terms of improvement in production. As a result, production has reached a plateau. As demonstrated in other crops, modern genomics approaches can facilitate breeding, leading to enhanced crop productivity. Integration of genomic approaches in breeding programmes has been referred to as ‘genomics-assisted breeding’.2 Availability of genomic resources like molecular markers, genetic maps, transcriptomic or genome sequence data, and metabolome analyses are the pre-requisites for undertaking genomics-assisted breeding. While these platforms are available in many cereal and major legume crops like soybean, cowpea and common bean,3 pigeonpea has not had the development and application of this genomic revolution. In this crop, only a few hundred (172) simple sequence repeat (SSR) markers4–7 and a recent genetic map8 have become available.
Without a genome sequence, transcriptome sequencing is an effective approach for gene discovery and identifying transcripts involved in specific biological processes. Expressed sequence tag (EST) studies have provided insight into genomic architecture and helped to elucidate genes involved in biological processes [grapevine, Populus, Arabidopsis].9–11 As of October 2010, ∼10 817 pigeonpea EST sequences were available at the NCBI GenBank (www.ncbi.nlm.nih.gov/sites/gquery?term=pigeonpea). This small set of ESTs highlights the need for a larger collection of sequence information before employing effective functional genomics studies in pigeonpea research.
In recent years, the advent of next generation sequencing (NGS) technologies has made it possible to inexpensively and quickly generate large-scale transcript sequence data.12 With an objective of characterizing functionally important genes in pigeonpea, transcriptome analysis was undertaken by sampling a large number of reads from normalized cDNA libraries prepared from different tissues, developmental conditions and physiological stages. The initial part of the present study was aimed at discovering and characterizing genes from the pigeonpea variety Pusa Ageti by developing and analysing the transcriptome assembly of pigeonpea based on FLX/454 sequencing. Secondly, the Illumina 1G, RNA-seq approach was employed on mRNA of 10 parental genotypes of five mapping populations that segregate for Fusarium wilt (FW) and/or sterility mosaic disease (SMD). Alignment of Illumina sequence data of these parental lines with the transcriptome assembly provided a framework for quantitative measurement of gene expression as well as single-nucleotide polymorphism (SNP) detection in these parental lines.
This may be the first report in pigeonpea of large-scale transcriptome data generation and the corresponding comprehensive analysis to understand transcriptome and evolutionary genome dynamics. Based on digital expression analyses, we have also identified candidate FW- and SMD-responsive genes. Finally, this study provides a resource of both SSR and SNP markers identified from these sequences.
2. Materials and methods
2.1. Plant material and library construction
Pusa Ageti (ICP 28), an early maturing pigeonpea variety, was selected for library construction and transcriptome analysis. Seeds were sown in pots (three seeds per pot), and maintained in a glass-house. In order to maximize the diversity of expressed genes in pigeonpea, tissues from different developmental stages were targeted for collection and construction of cDNA libraries. These tissue samples included embryo, cotyledon, root and shoot primordia, apical meristem, leaves, senescing leaves, flowers, stamen and roots, harvested from several individual glass-house grown pigeonpea plants at different time intervals. This was done to analyse gene expression associated with the developmental process. Tissues were washed briefly with 0.1% DEPC water and then were frozen in liquid nitrogen. Total RNA was extracted from all the harvested tissues using modified hot-acid phenol method.13 The integrity and purity of all the samples were assessed both on 1.2% formaldehyde agarose gel and UV Spectrophotometer at A260:A280. An equal amount of each appropriate RNA sample was pooled to form a composite collection of total RNA sample for each tissue. Ten cDNA libraries were constructed to characterize specific stages of gene expression.
In order to minimize differences among the abundance of different transcripts (i.e. genes expressed at different levels), amplified cDNA was normalized employing the Smart cloning methodology14 using the services of Evrogen (www.evrogen.com) and Sfi IA/B primers/adapters that permit directional cloning. The detailed methodology of library construction and normalization has been described in Cheung et al.15
2.2. Sequence data assembly and clustering
Sequence analyses and assembly was conducted using publicly available software and custom Perl scripts. Quality trimming of the sequences involved trimming adapter sequences, removing short sequences (<50 nucleotides) for the assembly process as this will lead to false joining of reads, and chimeras that were sequenced hence reducing the quality of unique sequences. The vector-trimmed high-quality sequences were selected for further clustering and alignment to form transcript assemblies (TAs) using the CAP3 program.16 The following parameters were used for all CAP3 assemblies: –p 95–o 50–g 3–y 50–t 1000. These parameters were chosen to satisfy three primary goals: (i) to maximize contig length, (ii) to minimize production of contigs with highly variable read coverage, as these tend to be spurious assemblies and (iii) increasing the value of the ‘–t’ parameter improves the quality of the assembly at the cost of using additional memory on the assembly server; the value of 1000 was chosen as it was higher than the default but remained within the memory constraints of the assembly server. The assembly included the publicly available 10 817 ESTs of pigeonpea along with the FLX/454 reads.
2.3. Identification of paralogoues
Identification of paralogous genes was conducted using both the contig consensus sequences and the singletons following assembly. The longest open reading frame was identified using EMBOSS: getorf (http://emboss.open-bio.org/wiki/Appdocs) to identify all open reading frames and a custom Perl script to retain only the longest. Clustering of these sequences followed using a virtual suffix tree generation with six frame translation using Vmatch.17 Gene families of size 2–6 were clustered with the following parameters, i.e. subject per cent match of 85, query per cent match of 70, a minimum length of 20 amino acids and an exdrop of 30. Pair-wise alignments were obtained using ClustalW18 and synonymous distances (Ks values) calculated using the method of Goldman and Yang19 as implemented in PAML.20
2.4. Alignment of CcTA to the soybean genome
Alignments of CcTA with the soybean genome were made with GMAP, requiring 80% identity and 80% coverage, maximum intron length of 10 000 bp, and maximum of 10 introns per gene fragment. The highest scoring alignment satisfying the stringency criteria was taken as the best hit. Alignments within 1% identity and 1% coverage of each other were retained as multiple equally good matches.
2.5. Functional annotation and similarity search
Functional annotations of 127 754 TUSs were made using BLASTX comparisons against the UniRef non-redundant protein database. Sequence similarity was considered at a bit-score >50 and a significant e-value ≤ 1E−08. Each TUS was assigned a putative cellular function based on the significant database hit with the lowest E-value. Subsequently, TUSs that showed a significant BLASTX hit were used for functional annotation based on Gene Ontology (GO) categories from the UniProt database (UniProt-GO). TUSs were thus assigned to primary and sub-GO functional categories.
2.6. Gene expression analysis
cDNA pools from disease-stressed tissues of 10 pigeonpea genotypes were sequenced using Illumina 1G sequencing. These 10 genotypes are responsive to SMD and FW and represent parental genotypes of five mapping populations and segregate for the given stresses. FW stress was induced in four genotypes: ICPL 87119 and ICPL 99050 (resistant), ICPL 87091 and ICPB 2049 (susceptible). SMD stress was induced in six genotypes: ICPL 20096, ICPL 7035 and BSMR 736 (resistant), ICPL 332, TTB 7 and TAT 10 (susceptible). Stress was imposed on 15th day after sowing, using two methods: (i) root dipping method for FW infection and (ii) leaf stapling method for sterility mosaic virus infection. The tissues were harvested after 10 days of infection. Total RNA was extracted from all the harvested tissues using modified hot-acid phenol method.13 cDNA libraries were constructed and subjected to Illumina 1G sequencing. Illumina tags were aligned to the CcTA and counts were assigned to each TUS for all 10 genotypes. These expression values were used for estimation of expression patterns of these TUSs in the parental combination of each cross. Differentially expressed genes/TUSs between pairs of SMD- and FW-responsive genotypes were identified. The expression values of TUSs were transformed to log 2 scale. Expression values of each gene were used to compare respective libraries of susceptible and resistant genotypes in each cross. Expression differences between TUSs from susceptible and resistant lines were considered with a minimum of a two-fold difference in log 2 expression. The set of TUSs with high differential expression ≥5 were considered for functional annotation using BLASTX analysis as described above.
2.7. Identification of microsatellite/SSRs
SSR mining of 127 754 TUSs was carried out using the MIcroSAtellite (MISA)21 (http://pgrc.ipk-gatersleben.de/misa/) program, with the following parameters: at least 10 repeats for mono-, 6 repeats for di- and 5 repeats for tri-, tetra-, penta- and hexa-nucleotide for simple SSRs. Both perfect (i.e. SSRs containing a single repeat motif such as ‘AGG’) and compound (i.e. composed of two or more SSRs separated by ≤100 bp) SSRs were identified. The Primer3 program22 was used for designing the primer pairs for identified SSRs based on the following criteria: (i) annealing temperature (Tm) between 50–65°C with 60°C as optimum; (ii) product size ranging from 100 to 350 bp; (iii) primer length ranging from 18 to 24 bp with an optimum of 20 bp; and (iv) GC% content in the range of 40–60%.
2.8. Identification of SNPs
Identification of SNPs from Illumina data was carried out using the Alpheus software system.23 SNPs were identified on the basis of alignment of Illumina reads generated from each of the genotypes against a reference—in this case, the CcTA and respective counter genotype, allowing not more than two mismatches. Based on alignment results, variants at a particular nucleotide position were identified. Significant variants were selected based on two criteria, allele frequency between two genotypes >0.8, and number of tags aligned to the reference >5.
2.9. Identification of intron-spanning region (ISR) markers
Using alignments of paralogous genes (Section 2.4), primers were selected to span introns in predicted genes. The Primer3 program was used to design primers with default parameters, except for the requirement of spanning one predicted intron.
3. Results and discussion
This is the most comprehensive study of pigeonpea transcriptomic data to date. Pusa Ageti (ICP 28), a leading pigeonpea variety in India, was chosen for developing these genomic/transcriptomic resources, based on its phenology and the utility of this genotype in breeding programmes. The sequence data generated have been analysed to understand the transcriptome architecture and genome organization with respect to potential duplication, identification of candidate genes for FW and SMD based on digital gene expression profiling and the development of genetic markers.
3.1. Clustering and assembly of transcript reads
Until recently, only 10 817 ESTs were available, of which >90% were developed during last 2 years. With the objective of generating a comprehensive transcriptomic resource, deep sequencing was undertaken on cDNAs pools of 31 different developmental stages from early vegetative growth through the reproductive organs (Supplementary Fig. S1). Following cDNA synthesis, these libraries were pooled and normalized. FLX/454 sequencing of this normalized cDNA pool generated 494 353 short transcript reads (STRs), with an average length of 171 bp. At the time of data analysis, 10 817 Sanger ESTs, with average read length 527 bp, were available in the public domain via NCBI. These two data sets were analysed separately and in combination. Clustering of 354 131 STRs alone yielded 52 827 contigs, with an average length of 262 bp including 4308 high confidence singletons (that contain only two reads with zero read coverage) and 140 222 singletons. Clustering of Sanger ESTs yielded 746 contigs, with an average length 637 bp, and 5553 singletons. In order to develop a transcriptome reference in pigeonpea, 505 170 FLX/454 STRs and Sanger ESTs were assembled in combination to yield a pigeonpea transcriptome assembly (CcTA) comprising of a total 127 754 tentative unique sequences (TUSs). The sequence data from this study have been submitted to the Legume Information System (LIS) (http://comparative-legumes.org). The sequence data can be accessed at http://cajca.comparative-legumes.org/data/2011/1e0b2bbdb4a3dca874759a9c7d23d46b/transcript_contigs.fa.gz.
The CcTA includes 48 726 (38.1%) contigs (average length 273 bp, maximum length 2067 bp) and 79 028 (61.9%) singletons (average length 198 bp, maximum length 1720 bp). A total of 3021 contigs (6.1%) were longer than 500 bp. Details about length distribution and read depth of contigs are given in Table 1 and Fig. 1, respectively. The overall redundancy of the library was calculated at 25.2%, suggesting that the normalization process was effective and that the libraries generated have the potential to uncover many more unique transcripts. These results support that FLX/454-based gene discovery represents a viable and perhaps favourable alternative to Sanger-based sequencing of EST libraries, when a diverse sampling of genes is more important than obtaining full transcript-length contigs.15,24
Table 1.
Sequence length distribution before and after assembly of FLX/454 STRs and Sanger ESTs
| Range of nucleotide length (bp) | Raw 454 STRs | Raw Sanger ESTs | Assembled 454 STRs | Assembled Sanger ESTs | Assembled 454 STRs + Sanger ESTs |
|---|---|---|---|---|---|
| 50 | 31 876 (6.4%) | 44 (0.4%) | 0 | 0 | 0 |
| 51–100 | 61 172 (12.3%) | 180 (1.6%) | 2282 (4.7%) | 5 (0.6%) | 2253 (4.6%) |
| 101–150 | 84 878 (17.1%) | 420 (3.8%) | 4854 (10.0%) | 4 (0.5%) | 4829 (9.9%) |
| 151–200 | 88 806 (17.9%) | 449 (4.1%) | 5934 (12.2%) | 17 (2.2%) | 5874 (12.0%) |
| 201–250 | 185 863 (37.5%) | 658 (6.0%) | 12 780 (26.3%) | 24 (3.2%) | 12 561 (25.7%) |
| 251–300 | 41 758 (8.4%) | 630 (5.8%) | 9224 (19.0%) | 20 (2.6%) | 9015 (18.5%) |
| 301–350 | 401 (3.7%) | 4960(10.2%) | 21 (2.8%) | 4821 (9.8%) | |
| 351–400 | 603 (5.5%) | 3415 (7.0%) | 42 (5.6%) | 3349 (6.8%) | |
| 401–450 | 666 (6.1%) | 1901 (3.9%) | 37 (4.9%) | 1879 (3.8%) | |
| 451–500 | 573 (5.2%) | 3169 (6.5%) | 57 (7.6%) | 1124 (2.3%) | |
| 501–550 | 740 (6.8%) | 58 (7.7%) | 3021 (6.1%) | ||
| 551–600 | 575 (5.3%) | 65 (8.7%) | |||
| 601–650 | 621 (5.7%) | 51 (6.8%) | |||
| 651–700 | 887 (8.2%) | 45 (6.0%) | |||
| 701–750 | 1590 (14.6%) | 79 (10.5%) | |||
| 751–800 | 682 (6.3%) | 42 (5.6%) | |||
| 801–850 | 1098 (10.1%) | 74 (9.9%) | |||
| 851–900 | 24 (3.2%) | ||||
| 901–950 | 18 (2.4%) | ||||
| 951–1000 | 12 (1.6%) | ||||
| 1001–1050 | 11 (1.4%) | ||||
| 1051–1100 | 40 (5.3%) | ||||
| Total reads | 494 353 | 10 817 | 48 519 | 746 | 48 726 |
Figure 1.
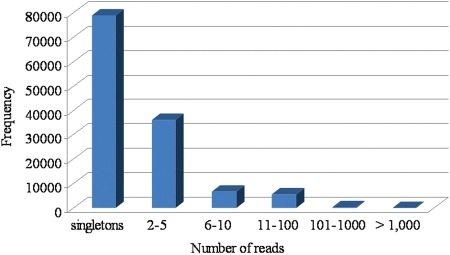
Distribution and read depth of sequence tags in the contigs. Number of 454 STRs aligning to form a contig ranged from 2 to >1000. A total of 36 152 contigs showed a read depth ranging from 2 to 5 tags, followed by 6755 contigs with read depth 6–10 tags, 5517 contigs with read depth 11–100 tags and 290 contigs with read depth ranging from 100 to 1000 tags. A maximum of 12 contigs showed a read depth of >1000 tags.
3.2. Evaluation of paralogous genes
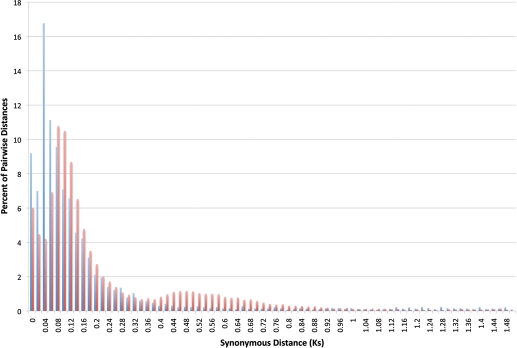
To evaluate characteristics of the CcTA sequences and to identify potential signatures of genome duplication in pigeonpea, the transcriptome assembly was analysed by comparing all TUSs against one another. Of the 127 754 TUSs sequences, 9.8% (12 515) could be clustered into families of similar genes (requiring subject per cent match of 85, query per cent match of 70, and a minimum alignment length of 60 nt). Of these, 3098 occurred in clusters of size 2, 537 of size 3, 181 of size 4, 89 of size 5 and 68 of size 6. The modal per cent identity among paralogues was 98.5, with the proportion of paralogues dropping to less than half the modal value at ∼96.5 per cent identity. Stated differently, the paralogue pairs are dominated by nearly identical or highly similar sequences. These alignments can also be used to find pair-wise synonymous distance measures, and tallied for Ks ranges for comparison with published Ks values for soybean (Fig. 2). There is a sharp Ks peak for Cajanus at roughly 0.04—contrasting with the soybean peak at ∼0.12.25 While this might be indicative of a recent whole-genome (or other large-scale segmental) duplication in Cajanus, a likely cause is incompletely collapsed contigs. The chromosome number of pigeonpea (2n = 22) is the same as in the other phaseoloids such as common bean (Phaseolus vulgaris) and cowpea (Vigna unguiculata), which are not known to have experienced recent polyploidy. Examination of 40 random near-identical paralogous alignments (data not shown) shows that the majority (>60%) of the differences are indels in the vicinity of homopolymer runs—which very likely derive from the large proportion of 454 STRs used in the TA construction.
Figure 2.
Histogram plot of pair-wise synonymous distances of pigeonpea duplicated genes when compared with soybean. Histogram plot of percentage pair-wise distance to the synonymous distance value (Ks) showing a peak in pigeonpea at 0.06 which gives a divergence estimate of ∼4.9 Mya.
3.3. Characterization of the pigeonpea transcriptome
3.3.1. Comparison with the soybean genome
As an effort to validate gene structures in the newly developed assembly, the 127 754 TUSs were aligned to soybean using GMAP, with identity and coverage thresholds of 90% and 80%, respectively.26 Of these TUSs, 33 874 had matches at these thresholds, corresponding to 10 857 soybean genes. The TUSs were distributed similarly across the 20 chromosomes of soybean, with an average of ∼1693 loci on each chromosome, with the exception of chromosome 13, which had 4162 matches. The excess on chromosome 13 is primarily due to many matches across a long ribosomal array on this chromosome. The alignment results are available as a track in the SoyBase genome browser (http://soybase.org/gbrowse/cgi-bin/gbrowse/gmax1.01/).
3.3.2. Functional annotation and GO categorization
Putative assignments of 127 754 TUSs into functional categories resulted in the assignment of 32 719 TUSs (25.6%) with similarity to the UniRef non-redundant protein database (BLASTX bit-score >50 and a significant e-value ≤ 1E−08), while 8949 sequences (7.0%) had low similarity and 86 086 (67.3%) sequences had no significant matches (Supplementary Table S1). The TUSs could be placed into GO categories: biological process (5455), cellular component (3958) and molecular function (6491). Enzyme IDs retrieved from the UniProt database were distributed in one of the six major enzyme classes: transferases 31% (474), followed by hydrolases 28% (443), oxido-reductases 25% (389) ligases 6% (98), lyases 5% (79) and isomerases 5% (79). Further details about the GO categories and enzyme IDs of the TUSs are shown in Supplementary Table S2. A noteworthy aspect of this analysis is that the majority of the transcripts were involved in metabolic and cellular process—as expected since most libraries were derived from developing tissues.27
3.3.3. Identification of disease-responsive genes
SMD and FW are two serious diseases that adversely affect pigeonpea production. With an objective of identifying candidate genes for these diseases, Illumina 1G sequencing was used on the transcriptomes of FW-challenged root tissues and SMD-challenged leaf tissues of five each resistant (ICPL 87119, ICPB 2049, ICPL 20096, BSMR 736 and ICPL 7035) and susceptible genotypes (ICPL 87091, ICPL 99050, TAT 10, ICPL 332 and TTB 7). The number of Illumina tags (36 bp long) ranged from 18 644 113 (ICPL 87119) to 14 514 194 (TAT 10) for the 10 genotypes. The sequence data of these Illumina tags have been submitted to the National Center for Biotechnology Information (NCBI). The data can be accessed at (http://www.ncbi.nlm.nih.gov/) and accession numbers are: SRA030523.1 to SRP005971.1. These tags were aligned to the CcTA (Supplementary Table S3). As a result, ∼35 million Illumina tags could be aligned to 54 426 TUSs. Numerical comparison of these tags between a pair of resistant and susceptible genotypes for a disease (usually the parents of a mapping population) was used to identify differentially expressed genes for a disease.
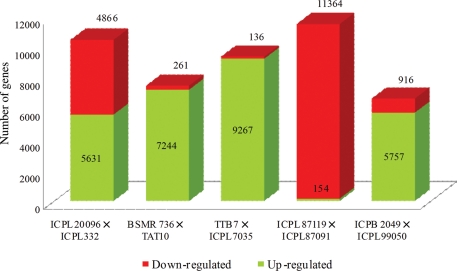
Since the numbers of Illumina tags mapped to the transcriptome assembly varied among genotypes, the data were normalized per million reads. For the SMD study, a numerical comparison of SMD-responsive reads generated from three resistant (ICPL 20096, BSMR 736 and ICPL 7035) and three susceptible (ICPL 332, TAT 10 and TTB 7) genotypes representing three mapping populations was conducted. The Log 2 threshold for this analysis was taken as −2 to +2. The number of TUSs showing expression differences at these cutoffs ranged from 7505 (BSMR 736 × TAT 10) to 10 497 (ICPL 20096 × ICPL 332). In the case of the TTB 7 × ICPL 7035 combination, the number of differentially expressed genes was 9402. Similarly, in the FW study, a comparison was made between the specific parental combinations used to develop two different mapping populations (with the same thresholds) to find TUSs with differential expression. The number of TUSs with significant differentially expressed genes ranged from 6673 (ICPB 2049 × ICPL 99050) to 11 518 (ICPL 87119 × ICPL 87091) (Fig. 3).
Figure 3.
Distribution of differentially expressed genes in SMD- and FW-responsive genotypes. Differential expression was calculated based on Log 2 value, with a threshold of less than −2 to greater than +2 number of differentially expressed gene was calculated for three SMD- and two FW-parental combinations. Comparison of expression values from susceptible parent to the resistant gave an estimate of up- and down-regulated genes in each cross.
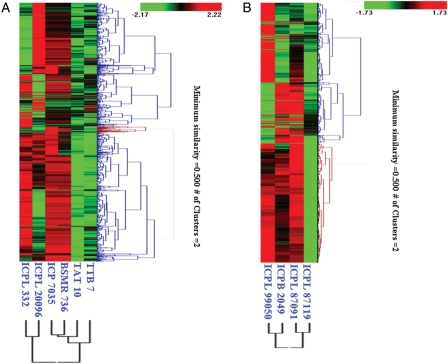
Based on the expression values for differentially expressed genes in SMD- and FW-responsive genotypes, hierarchical clustering was done for SMD- and FW-responsive genes separately to compare the pattern of gene expression. These clusters show the pattern of co-regulated genes for the SMD-responsive genotypes (Fig. 4A) and for the FW-responsive genotypes (Fig. 4B).
Figure 4.
Hierarchical clustering of differentially expressed TAs within SMD- and FW-responsive genotypes. Hierarchial clustering of the gene involved in SMD- and FW-stress responses was done using HCE version 2.0 beta web tool. These two dendrograms illustrate the co-regulation of genes in resistant and susceptible genotypes under stress, (A) clustering of 5106 genes showing expression variation across six SMD-responsive genotypes (three resistant, ICPL 20096, ICPL 7035, BSMR 736, and three susceptible, ICPL 332, TTB 7, TAT 10). (B) Clustering of 3384 genes showing expression variation across four FW-responsive genotypes (two resistant, ICPL 87119, ICPL 99050, and two susceptible, ICPB 2049, ICPL 87091). Colour scale (from green to red) represents the range of expression level.
In order to study the gene expression pattern between two parental genotypes of a mapping population, the numbers of up-regulated and down-regulated TUSs were calculated with respect to the resistant parent. The numbers of up-regulated TUSs remained high in all the crosses studied, with an exception of ICPL 87119 × ICPL 87091, which had more down-regulated TUSs (11 364) when compared with up-regulated (154). Log 2 fold differences between the parental combinations are shown in Table 2.
Table 2.
Summary of differentially expressed TUSs across five parental combinations
| Fold difference | ICPL 20096 × ICPL 332 | BSMR 736 × TAT 10 | TTB 7 × ICPL 7035 | ICPL 87119 × ICPL 87091 | ICPB 2049 × ICPL 99050 |
|---|---|---|---|---|---|
| Exclusively expressed in resistant | 9149 | 8867 | 9455 | 243 | 10 675 |
| Exclusively expressed in susceptible | 1964 | 269 | 200 | 9784 | 2098 |
| 2 to <3 fold | 3385 | 2663 | 3024 | 3715 | 2638 |
| 3 to <4 fold | 2782 | 2111 | 3082 | 3859 | 1779 |
| 4 to <5 fold | 2108 | 1499 | 2180 | 2618 | 1167 |
| 5 to <6 fold | 1544 | 807 | 856 | 1053 | 674 |
| ≥6 fold | 998 | 385 | 260 | 273 | 415 |
Functional annotations of differentially expressed TUSs are described next, with the additional requirement of ≥5 fold differences across all the parental combinations. The annotation analysis was conducted in three ways: (i) TUSs differentially expressed across all the 10 genotypes, (ii) TUSs differentially expressed in 6 SMD-responsive genotypes separately and in 4 FW-responsive genotypes separately and (iii) the common set of TUSs that is differentially expressed in both FW- and SMD-responsive genotypes. Based on these analyses, in the first category, 6107 TUSs (with fold difference ≥5) were selected for functional classification. Considering an E-value cutoff of ≤1E−08 and a bit-score value of ≥50, functional annotation for 3698 TUSs showed significant similarity with the UniRef non-redundant protein database. No significant matches were found for 2409 TUSs. For 3698 TUSs with functional classes, we found that in addition to basic housekeeping genes, these TUSs also showed homology to genes involved in stress response, such as proline-rich protein, Syringolide-induced protein, desiccation protective protein of soybean, ABA-responsive protein, and leucine zipper protein (Supplementary Table S4).
Among these 3698 TUSs, 2106 could be assigned into three major categories: (i) molecular function, (ii) biological process and (iii) cellular component. These categories were further subcategorized, i.e. under molecular function category, the subcategory ‘binding’ accounted for highest percentage of TUSs (594), followed by ‘catalytic activity’ (513), ‘transporter’ (58), ‘structural molecule’ (52) and rest of the subcategories accounting for 75 TUSs. Similarly, under ‘biological process’ category, the highest number of TUSs were assigned to the subcategory ‘metabolic process’ (571) followed by ‘cellular process’ (542), response to ‘stimuli’ (152), ‘biological regulation’ (118), ‘establishment of localization’ (108) and 363 TUSs accounted for rest of the subcategories. Under ‘cellular component’ category, the highest percentage of TUSs was assigned to the subcategory ‘cell part’ (562) followed by ‘organelle’ (381), ‘organelle part’ (202), ‘macro molecule complex’ (158) and 75 TUSs were assigned to rest of the subcategories.
Differentially expressed genes included those encoding proline-rich proteins, zinc finger proteins, leucoanthocyanidin dioxygenase and RAS-related protein. There were seven TUSs that correspond to proline-rich protein. This protein forms a component of glutamate pathway and has multiple developmental and stress-related functions. High expression of this protein in leaves has been reported to play major role in the early stage of virus infection in soybean.28 The glutamate pathway assimilates nitrogen and produces glutamate, which then acts as a starting point for amino acid synthesis. A 5-fold up-regulation of this gene in resistant genotype (ICPL 2049) probably implicates its role in response to this stress. A gene for zinc finger protein showed an average of 5-fold differential expression in both SMD- and FW-responsive genotypes. This protein is a component of mitogen-activated protein kinase (MAPK) pathways which are demonstrated to play an important role in regulating the gene expression in response to various biotic as well as abiotic stress in species such as Arabidopsis.29,30 MAPK pathways transduce a large variety of external signals, leading to a wide range of cellular responses, including growth, differentiation, inflammation and apoptosis. A total of six TUSs had homologies to leucoanthocyanidin dioxygenase, and showed an average of 5-fold differential expression. The up-regulation of this gene (in the flavonoid pathway) is known to play an important role in defence against both biotic and abiotic stress by acting as a passive or inducible barrier against pathogens.31 A total of 21 TUSs showed homology to gene for RAS-related protein ARA-3. These TUSs were up-regulated 6-fold. This gene is involved in the ethylene-mediated signalling pathway, suggesting an important role in stress response.
With an objective of identifying candidate genes for FW and SMD, as mentioned above, the common set of TUSs with ≥5 fold expression difference was identified in SMD- and FW-responsive genotypes, separately. From this, 99 common TUSs were found for FW-responsive genotypes and 13 for SMD-responsive genotypes. Functional characterization of these genes showed function for 51 FW-responsive TUSs and 3 SMD-responsive TUSs (oxygen-evolving enhancer protein, NADH-ubiquinone oxidoreductase and sedoheptulose-1,7-biphosphate). FW responsive TUSs include genes such as mannose-1-phosphate guanyltransferase, prolinedehydrogenase, cellulose synthase, pectinesterase inhibitor, superoxide dismutase [Fe] and vacuolar protein sorting-associated protein.
Among FW-responsive genes were TUSs showing cellulose synthase homology. These genes are essential for secondary cell wall synthesis. Among the SMD-responsive TUSs, one showed homology to oxygen evolving enhancer protein, with an average 5-fold expression difference. These proteins are components of glycine-rich protein 3/wall-associated kinase. One TUS showed homology to a gene coding for NADH-ubiquinone oxidoreductase and was up-regulated in resistant with respect to susceptible genotypes for SMD. This is a common component for energy evolving pathways in the cell.
Considering biotic stress responsive genes in common for FW and SMD, no TUS was found common at the threshold of ≥5 fold difference. When this threshold was decreased to ≥2 fold, the number of common TUSs across FW- and SMD-responsive genotypes was found to be 192. Of this set, 99 TUSs were functionally annotated and 93 were uncharacterized proteins. These annotated TUSs showed sequence similarity to several stress responsive genes such as zinc protein, aminocyclopropane carboxylate oxidase, cysteine protease and hexokinase. For example, two TAs showed homology to a gene corresponding to zinc finger protein and expressed with an average of 3.4-fold difference. As mentioned, this gene is a component of MAPK. TUSs with sequence similarity to gene for 1-aminocyclopropane-1-carboxylate oxidase were expressed with an average 3.3 folds. This gene is a component of ethylene-biosynthesis pathway which plays an important role in ethylene biosynthesis at stress conditions.32 TUSs with sequence similarity to gene for synthesis of germination-specific cysteine protease also showed an average of 3-fold difference in expression value, this gene is responsible for cell death hence regulating response to stress. Sequence similarity for another gene encoding for hexokinase-2 was also discovered for one TUS which showed an average-fold difference of 2.9. This gene is known to play a major role in metabolic pathways, e.g. fructose and mannose metabolism, galactose metabolism and glycolysis.
Genes responding in the FW- and SMD-resistant lines will provide a rich basis for further explorations of the mechanisms of disease resistance for these important viral and fungal diseases, and may also be useful in identifying regulatory networks and targets for breeding efforts, As no controls (Illumina tags generated from non-challenged tissues) were used for identification for FW- and SMD-responsive genes, like recent studies in wheat (Triticum aestivum)33 and yam (Dioscorea alata L.)34, it is, therefore suggested to use other techniques like qRT–PCR to validate and establish magnitudes of expression levels of identified genes before they are used for further studies.
3.4. Development of gene-based molecular markers
Genetic markers are important tools for understanding variation, and for identification of gene/QTLs for traits of interest in molecular breeding activities. Until recently, a very limited number of genetic markers have been available for pigeonpea.35 One of the main reasons for the lack of mapping resources in pigeonpea is the low level of polymorphism. An approach to develop genetic markers is the mining of ESTs or transcript sequences for the presence of SSRs and SNPs.36 Although markers developed from ESTs/transcripts are less polymorphic, they have been found useful for assaying the functional diversity in the germplasm collection37,38, trait mapping27 and comparative genomics studies.39
3.4.1. SSR discovery
As microsatellite or SSR markers are the markers of choice for many plant breeding applications, the TUSs were analysed for identification and development of SSR markers. Analysis of 127 754 TUSs with the MISA search program21 identified 50 566 SSRs in 41 899 TUSs (32.7%), with a frequency of one SSR per 570 bp. In terms of abundance, mono-nucleotide repeats were most abundant (33 262, 65.7%) followed by di- (13 204, 26.1%) and tri-nucleotide repeats (3063, 6%). Other type of repeat units occurred at <1% each (Supplementary Table S5). SSRs were divided into perfect (i.e. SSRs containing a single repeat motif such as ‘AAG’) or compound (i.e. composed of two or more SSRs separated by ≤100 bp) SSRs. A total of 6350 (15.1%) compound SSRs were identified. The frequency of SSR repeat motifs was calculated after excluding the mono-nucleotide repeats. Among di-nucleotide repeats motifs, AG/CT was the most abundant with 46.8%; tri-nucleotide repeats motifs were rich in AAG/CTT (32.5%); and among tetra-, penta- and hexa- nucleotide repeat motifs, the most abundant repeats were AAAT/ATTT (24.1%), AAGGT/ATTCC (20.8%) and AAAAAG/CTTTTT (8.4%), respectively.
With an objective of converting the identified SSRs into genetic markers, 9157 primer pairs were designed for all SSRs except mono-nucleotides. Recently, 3312 SSR markers have been designed for pigeonpea4–8,35. An analysis to determine overlap between the 9157 newly designed primer pairs and 3312 published SSR markers (using BLAST) identified 8137 primer pairs as novel SSR markers for pigeonpea (Supplementary Table S6). Validation of these SSR primer pairs, however, is required to determine their potential to amplify and to detect polymorphisms. In several crop species, SNP markers are becoming more popular because of their low cost and potential for automation.40
3.4.2. SNP discovery
In total, 150.8 million Illumina sequence tags were generated from 10 genotypes. For identification of SNPs, tags for two genotypes of a given mapping population were aligned with 127 754 TUSs (the pigeonpea transcriptome assembly), and variants were identified using the Alpheus program of NCGR.23 The number of SNPs in an individual cross ranged from 704 (BSMR 736 × TAT 10) to 6263 (ICPL 87119 × ICPL 87091) (Table 3). In total, 12 141 SNPs were identified; however, only six SNPs were found in common across three populations (ICPL 20096 × ICPL 332, ICPL 7035 × TTB7 and BSMR 736 × TAT 10). The number of common SNPs across any two mapping populations ranged from 8 (ICPL 99050 × ICPL 2049 and BSMR736 × TAT 10) and 39 (ICPL 87119 × ICPL 87091 and ICLP 20096 × ICPL 332). Although a large number of SNP genotyping platforms are available,36 depending on the need and requirements, most suitable platform can be selected for using the SNPs in pigeonpea genetics and breeding applications. For instance, GoldenGate assays of Illumina (www.illumina.com/) will offer high-throughput SNP genotyping while KASPar (www.kbioscience.co.uk) or cleaved amplified polymorphic sequence (CAPS) assays41 will allow low-cost SNP genotyping.
Table 3.
Illumina sequencing based SNP discovery in five parental combinations
| Genotypes | ICPL 87119 | ICPL 87091 | BSMR 736 | TAT 10 | TTB 7 | ICPL 7035 | ICPL 20096 | ICPL 332 | ICPB 2049 | ICPL 99050 |
|---|---|---|---|---|---|---|---|---|---|---|
| Number of reads (in millions) | 18.4 | 16.8 | 16.9 | 14.5 | 15.7 | 17.2 | 15.5 | 14.8 | 15.7 | 17.3 |
| Number of SNPs in parental combination | ||||||||||
| Substitution | 5965 | 573 | 955 | 1580 | 1878 | |||||
| Insertion | 176 | 87 | 28 | 31 | 61 | |||||
| Deletion | 122 | 44 | 44 | 43 | 38 | |||||
| Total SNPs | 6263 | 704 | 1027 | 1654 | 1977 | |||||
3.4.3. Identification of intron-spanning region markers
Identification of potential splice site was undertaken as an extension to the usability of data generated from 127 754 TUSs aligned against soybean genome sequence for development of genetic markers in pigeonpea. Thresholds were set to a minimum alignment length of 70 nucleotides, minimum query coverage of 90% and minimum per cent identity of 80%. A total of 8532 (6.0%) TUSs showed valid alignment with 8491 unique soybean loci, and having not more than 1000 bp gaps between aligned components. Alignment of these 8532 TUSs with one or more splice sites yielded 13 862 putative intron-spanning splice junctions. Alignment results were used for development of intron-spanning region (ISR) markers in silico for pigeonpea. These markers were derived for portions of genes and were designed from low copy sequences (sequences showing single match to the reference). In summary, a total of 5845 ISR primer pairs were designed across the pigeonpea genome (Supplementary Table S7). These potential ISR markers are also available as a GBrowse track at http://soybase.org/gbrowse/cgi-bin/gbrowse/gmax1.01. These ISR markers can be assayed on mutation detection enhancement (MDE) gel for detection of polymorphism.
In summary, large-scale transcriptomic resource has been developed in an under-resourced crop species by deploying two prominent NGS technologies namely FLX/454 and Illumina IG sequencing. These transcript data have been used for both basic and applied aspects in pigeonpea genetics and breeding. Similarly, genes responsive to FW and SMD have been identified, and are ready for further study and validation. Additionally, a large number of molecular markers have been developed that will accelerate molecular mapping and molecular breeding activities for pigeonpea improvement.
Supplementary data
Supplementary Data are available at www.dnaresearch.oxfordjournals.org.
Funding
This study was supported financially by SP2 Leader Discretionary Grant of CGIAR Generation Challenge Programme, Mexico; and Pigeonpea Genomics Initiative (PGI) of Indian Council of Agricultural Research (ICAR) of Government of India.
Supplementary Material
Acknowledgements
The authors are thankful to Dr Pawan Kulwal, Dr Punjabrao Deshmukh Krishi Vishwavidalaya, Akola (India), for providing the seeds of some genotypes used in this study.
References
- 1.Greilhuber J., Obermayer R. Genome size variation in Cajanus cajan (Fabaceae): a reconsideration. Plant Syst. Evol. 1998;212:135–41. [Google Scholar]
- 2.Varshney R.K., Graner A., Sorrells M.E. Genomics-assisted breeding for crop improvement. Trends Plant Sci. 2005;10:621–30. doi: 10.1016/j.tplants.2005.10.004. [DOI] [PubMed] [Google Scholar]
- 3.Hyten D.L., Song Q., Fickus E.W., et al. High-throughput SNP discovery and assay development in common bean. BMC Genomics. 2010;11:475. doi: 10.1186/1471-2164-11-475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Burns M.J., Edwards K.J., Newbury H.J., Ford-Lloyd B.V., Baggott C.D. Development of simple sequence repeat (SSR) markers for the assessment of gene flow and genetic diversity in pigeonpea (Cajanus cajan) Mol. Ecol. Notes. 2001;1:283–5. [Google Scholar]
- 5.Odeny D.A., Jayashree B., Ferguson M., Hoisington D., Crouch J., Gebhardt C. Development, characterization and utilization of microsatellite markers in pigeonpea. Plant Breed. 2007;126:130–6. [Google Scholar]
- 6.Odeny D.A., Jayashree B., Gebhardt C., Crouch J. New microsatellite markers for pigeonpea (Cajanus cajan (L.) Millsp.) BMC Res. Notes. 2009;2:35. doi: 10.1186/1756-0500-2-35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Saxena R.K., Prathima C., Saxena K.B., Hoisington D.A., Singh N.K., Varshney R.K. Novel SSR markers for polymorphism detection in pigeonpea (Cajanus spp.) Plant Breed. 2010;129:142–8. [Google Scholar]
- 8.Bohra A., Dubey A., Saxena R. K., et al. Analysis of BAC-end sequences (BESs) and development of BES-SSR markers for genetic mapping and hybrid purity assessment in pigeonpea. (Cajanus spp.), BMC Plant Biol. 2011;11:56. doi: 10.1186/1471-2229-11-56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rotter A., Camps C., Lohse M., et al. Gene expression profiling in susceptible interaction of grapevine with its fungal pathogen Eutypa lata: extending MapMan ontology for grapevine. BMC Plant Biol. 2009;9:104. doi: 10.1186/1471-2229-9-104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sterky F., Bhalerao R.R., Unneberg P., et al. A Populus EST resource for plant functional genomics. Proc. Natl Acad. Sci. USA. 2004;101:13951–6. doi: 10.1073/pnas.0401641101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Blanc G., Barakat A., Guyot R., Cooke R., Delseny M. Extensive duplication and reshuffling in the Arabidopsis genome. Plant Cell. 2000;12:1093–102. doi: 10.1105/tpc.12.7.1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Varshney R.K., Nayak S.N., May G.D., Jackson S.A. Next generation sequencing technologies and their implications for crop genetics and breeding. Trends Biotechnol. 2009;27:522–30. doi: 10.1016/j.tibtech.2009.05.006. [DOI] [PubMed] [Google Scholar]
- 13.Schmitt M.E., Brown T.A., Trumpower B.L. A rapid and simple method for preparation of RNA from Saccharomyces cerevisiae. Nucleic Acids Res. 1990;18:3091–2. doi: 10.1093/nar/18.10.3091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhu Y.Y., Machleder E.M., Chenchik A., Li R., Siebert P.D. Reverse transcriptase template switching: a SMART approach for full length cDNA library construction. Biotechniques. 2001;4:892–7. doi: 10.2144/01304pf02. [DOI] [PubMed] [Google Scholar]
- 15.Cheung F., Haas B.J., Goldberg M.D., May G.D., Xiao Y., Town C.D. Sequencing Medicago truncatula expressed sequenced tags using 454 life sciences technology. BMC Genomics. 2006;7:272. doi: 10.1186/1471-2164-7-272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Huang X., Madan A. CAP3: a DNA sequence assembly program. Genome Res. 1999;9:868–77. doi: 10.1101/gr.9.9.868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Beckstette M., Homann R., Giegerich R., Kurtz S. Fast index based algorithms and software for matching position specific scoring matrices. BMC Bioinformatics. 2006;7:389. doi: 10.1186/1471-2105-7-389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Thompson J.D., Higgins D.G., Gibson T.J. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–80. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Goldman N., Yang Z. A codon-based model of nucleotide substitution for protein coding DNA sequences. Mol. Biol. Evol. 1994;11:725–36. doi: 10.1093/oxfordjournals.molbev.a040153. [DOI] [PubMed] [Google Scholar]
- 20.Yang Z. PAML: a program package for phylogenetic analysis by maximum likelihood. Comput. Appl. Biosci. 1997;15:555–6. doi: 10.1093/bioinformatics/13.5.555. [DOI] [PubMed] [Google Scholar]
- 21.Thiel T., Michalek W., Varshney R.K., Graner A. Exploiting EST databases for the development and characterization of gene derived SSR-markers in barley (Hordeum vulgare L.) Theor. Appl. Genet. 2003;106:411–22. doi: 10.1007/s00122-002-1031-0. [DOI] [PubMed] [Google Scholar]
- 22.Rozen S., Skaletsky H. In: Bioinformatics Methods and Protocols. Krawetz S., Misener S., editors. Totowa, NJ: Humana Press; 2000. pp. 365–368. http://fokker.wi.mit.edu/primer3/ [Google Scholar]
- 23.Miller N.A., Kingsmore S.F., Farmer A., et al. Management of high-throughput DNA sequencing projects: Alpheus. J. Comput. Sci. Syst. Biol. 2008;1:132–48. doi: 10.4172/jcsb.1000013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Novaes G.J., Drost D.R., Farmerie W.G., et al. High-throughput gene and SNP discovery in Eucalyptus grandis, an uncharacterized genome. BMC Genomics. 2008;9:312. doi: 10.1186/1471-2164-9-312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schmutz J., Cannon S.B., Schlueter J., et al. Genome sequence of the palaeopolyploid soybean. Nature. 2010;463:178–83. doi: 10.1038/nature08670. [DOI] [PubMed] [Google Scholar]
- 26.Wu T.D., Watanabe C.K. GMAP: a genomic mapping and alignment program for mRNA and EST sequences. Bioinformatics. 2005;21:1859–75. doi: 10.1093/bioinformatics/bti310. [DOI] [PubMed] [Google Scholar]
- 27.Zhang W.K., Wang Y.J., Luo G.Z., et al. QTL mapping of ten agronomic traits on the soybean (Glycine max L. Merr.) genetic map and their association with EST markers. Theor. Appl. Genet. 2004;108:1131–9. doi: 10.1007/s00122-003-1527-2. [DOI] [PubMed] [Google Scholar]
- 28.He C-Y., Zhang J-S., Chen S-Y. A soybean gene encoding a proline-rich protein is regulated by salicylic acid, an endogenous circadian rhythm and by various stresses. Theor. Appl. Genet. 2002;104:1125–31. doi: 10.1007/s00122-001-0853-5. [DOI] [PubMed] [Google Scholar]
- 29.Davletova S., Schlauch K., Coutu J., Mittler R. The zinc-finger protein Zat12 plays a central role in reactive oxygen and abiotic stress signaling in Arabidopsis. Plant Physiol. 2005;139:847–56. doi: 10.1104/pp.105.068254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rizhsky L., Davletova S., Liang H., Mittler R. The zinc finger protein Zat12 is required for cytosolic ascorbate peroxidase 1 expression during oxidative stress in Arabidopsis. J. Biol. Chem. 2004;279:11736–43. doi: 10.1074/jbc.M313350200. [DOI] [PubMed] [Google Scholar]
- 31.Dixon R.A., Achnine L., Kota P., Liu C-J., Reddy M.S.S. The phenylpropanoid pathway and plant defense—a genomics perspective. Mol. Plant Pathol. 2002;3:371–90. doi: 10.1046/j.1364-3703.2002.00131.x. [DOI] [PubMed] [Google Scholar]
- 32.Schlagnhaufer C.D., Arteca R.N., Pell E.J. Sequential expression of two 1-aminocyclopropane-1-carboxylate synthase genes in response to biotic and abiotic stresses in potato (Solanum tuberosum L.) leaves. Plant Mol. Biol. 1997;35:683–88. doi: 10.1023/a:1005857717196. [DOI] [PubMed] [Google Scholar]
- 33.Manickavelu A., Kawaura K., Oishi K., et al. Comparative gene expression analysis of susceptible and resistant near-isogenic lines in common wheat infected by. Puccinia triticina, DNA Res. 2010;17:211–222. doi: 10.1093/dnares/dsq009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Narina S. S., Buyyarapu R., Kottapalli K. R., et al. Generation and analysis of expressed sequence tags (ESTs) for marker development in yam. (Dioscorea alata L.), BMC Genomics. 2011;12:100. doi: 10.1186/1471-2164-12-100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Raju N.L., Gnanesh B.N., Lekha P.T., et al. The first set of EST resource for gene discovery and marker development in pigeonpea (Cajanus cajan L.) BMC Plant Biol. 2010;10:45. doi: 10.1186/1471-2229-10-45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Varshney R.K. In: Molecular Techniques in Crop Improvement. Jain S.M., Brar D.S., editors. vol. 2. The Netherlands: Springer; 2010. pp. 119–42. [Google Scholar]
- 37.Eujayl I., Sorrells M.E., Baum M., Wolters P., Powell W. Isolation of EST-derived microsatellite markers for genotyping the A and B genomes of wheat. Theor. Appl. Genet. 2002;104:399–407. doi: 10.1007/s001220100738. [DOI] [PubMed] [Google Scholar]
- 38.Wen M., Wang H., Xia Z., Zou M., Lu C., Wang W. Development of EST-SSR and genomic-SSR markers to assess genetic diversity in Jatropha curcas L. BMC Res. Notes. 2010;3:42. doi: 10.1186/1756-0500-3-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Stein L.D., Bao Z., Blasiar D., et al. The genome sequence of Caenorhabditis briggsae: a platform for comparative genomics. PLoS Biol. 2003;1:e45. doi: 10.1371/journal.pbio.0000045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kota R., Varshney R.K., Prasad M., Zhang H., Stein N., Graner A. EST derived single nucleotide polymorphism markers for assembling genetic and physical maps of the barley genome. Func. Integrat. Genom. 2008;8:223–33. doi: 10.1007/s10142-007-0060-9. [DOI] [PubMed] [Google Scholar]
- 41.Gujaria N., Kumar A., Dauthal P., et al. Development and use of genic molecular markers (GMMs) for construction of a transcript map of chickpea (Cicer arietinum L.) Theor. Appl. Genet. 2011 doi: 10.1007/s00122-011-1556-1. doi:10.1007/s00122-011-1556-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.