Abstract
Global mature microRNA (miRNA) expression is downregulated in cancers, and impaired miRNA processing enhances cancer cell proliferation. These findings indicate that the miRNA system generally serves as a negative regulator during cancer progression. In this study, we investigated the role of the miRNA system in cancer cell invasion by determining the effect of damaging miRNA processing on invasion-essential urokinase-type plasminogen activator (uPA) expression in breast cancer cells. Short hairpin RNAs specific for Drosha, DGCR8, and Dicer, key components of miRNA processing machinery, were introduced into 2 breast cancer cell lines with high uPA expression and 2 lines with poor uPA expression. Knockdown of Drosha, DGCR8, or Dicer led to even higher uPA expression in cells with high uPA expression, while it was unable to increase uPA level in cells with poor uPA expression, suggesting that the miRNA system most likely impacts uPA expression as a facilitator. In cells with high uPA expression, knockdown of Drosha, DGCR8, or Dicer substantially increased in vitro invasion, and depleting uPA abrogated enhanced invasion. These results thus link the augmented invasion conferred by impaired miRNA processing to upregulated uPA expression. uPA mRNA was a direct target of miR-193a/b and miR-181a, and a higher uPA level in cells with impaired miRNA processing resulted from less mature miR-193a/b and miR-181a processed from their respective primary miRNAs. Importantly, the levels of mature miR-193a, miR-193b, and miR-181a, but not their respective primary miRNAs, were lower in high uPA-expressing cells compared to cells with low uPA expression, and this apparently attributed to lower Drosha/DGCR8 expression in high uPA-expressing cells. This study suggests that less efficient miRNA processing can be a mechanism responsible for reduced levels of mature forms of tumor-suppressive miRNAs frequently detected in cancers.
Keywords: miRNA, Drosha, DGCR8, invasion
Introduction
MicroRNAs (miRNAs) are a class of small RNAs that suppress gene expression posttranscriptionally by base pairing with the 3′-untranslated regions (UTRs) of target mRNA.1 miRNAs are initially generated in the nucleus as long primary transcripts (pri-miRNAs) and subsequently cleaved by Drosha/DGCR8 processing machinery to produce stem loop–structured precursors (pre-miRNAs) that are exported to the cytoplasm and further processed by Dicer into mature miRNAs.2,3 Apparently, it is reasonable to assume that the levels of miRNA processing components may affect the levels of functional mature miRNAs.
miRNAs are predicted to regulate 20% to 30% of genes within the genome.4 Many of these miRNA-targeted mRNAs encode genes essential for cell proliferation/differentiation, cell survival/apoptosis, and cell migration/invasion, processes often altered during tumor progression.5,6 Recent miRNA profiling analysis has identified a set of miRNAs that are differentially expressed and can be used to distinguish between breast cancer and normal breast tissues.7 Certain miRNAs have been found to correlate with clinicopathological features of breast tumors. For example, the loss of expression in the distinct member of let-7 family reflects PR status (let-7c), positive lymph node status (let-7f-1, let-7a-3, and let-7a-2), and high proliferation index (let-7c and let-7d).7,8 Downregulation of miR-31 but upregulation of miR-10b have been found to be associated with the metastatic status of breast tumors.9,10 These findings implicate that signatures of miRNA expression may be used as biomarkers for both diagnosis and patient risk stratification of breast cancer patients.11 Meanwhile, defining cancer-relevant miRNAs may also allow the development of effective anticancer therapeutic means. Two recent studies highlighted the efficacy of such an approach, as either silencing miR-10b or forcing miR-26a expression led to suppression of tumorigenicity in experiment murine models.12,13
miRNAs may inhibit or promote tumor progression. Interestingly, a large scale of miRNA expression profiling analyses reveals that mature miRNAs are globally downregulated in various tumors including breast tumor.14-17 These findings suggest a notion that the miRNA system may generally act as a negative regulator during tumor progression. This notion is supported by recent studies in which reducing global mature miRNA expression by impairing miRNA processing was shown to enhance cellular transformation and tumorigenesis.18-20 In addition, recent studies show that the levels of miRNA processing components including Dicer and Drosha are decreased in various tumors including ovarian, lung, and gastric cancers.21-23 Mutation in the miRNA processing gene TARBP2 enhances transformation by impairing miRNA production.24 Oncogenic miR-103/107 promotes cancer metastasis by reducing Dicer expression.25 These observations raise a possibility that reduced expression of miRNA processing components can be one of the contributors to global downregulation of mature miRNAs in tumors.
Urokinase-type plasminogen activator (uPA) is a serine protease that initiates the activation of metalloproteinases as well as the conversion of plasminogen to plasmin, thus conferring cancer cells with the capability to degrade surrounding extracellular proteins.26 The interaction of uPA with its receptor also triggers cellular responses leading to cell migration, proliferation, and expression of specific genes.27 Elevated uPA expression is detected in most metastatic tumors including breast cancer28 and may be regulated at both levels of transcription29,30 and posttranscription.31,32 Recent studies also report that miR-23b and miR-193b can regulate uPA expression in human hepatocellular carcinomas and breast cancer cells, respectively.33,34
The objective of this study was to investigate the effect of impaired miRNA processing on uPA expression and in vitro invasion of breast cancer cells. We show that knockdown of Drosha, DGCR8, or Dicer leads to an even higher uPA level in high uPA-expressing cells, but it was unable to enhance uPA expression in cells with low uPA expression, indicating that the miRNA system is most likely to play a regulatory rather than decisive role in uPA expression. Similarly, knockdown of Drosha, DGCR8, and Dicer was only able to substantially enhance in vitro invasion of high uPA-expressing cells. As depleting uPA abrogated in vitro invasion of Drosha, DGCR8, and Dicer knockdown cells, it indicates that the enhanced invasion conferred by impaired miRNA processing is functionally linked to upregulated uPA expression. Moreover, we show that uPA mRNA is a direct target of miR-193a/b and miR-181a and that the damaged processing of these 3 miRNAs in Drosha, DGCR8, and Dicer knockdown cells is responsible for upregulated uPA expression. As Drosha and DGCR8 levels are relatively lower in high uPA-expressing cells than cells with low uPA expression, this may explain lower levels of mature miR-193a/b and miR-181a in high uPA-expressing cells. In fact, forced Drosha/DGCR8 expression elevated the levels of these uPA mRNA-targeted miRNAs and inhibited uPA expression. Our studies indicate that low abundance of Drosha/DGCR8 can contribute to less efficient processing of uPA mRNA-targeted miRNAs, leading to upregulated uPA expression and augmented in vitro invasion in breast cancer cells.
Results
miRNA-193a, miRNA-193b, and miR-181a effectively inhibit uPA expression in breast cancer cells
miR-23b and miR-193b have recently been shown to regulate uPA expression in human hepatocellular carcinomas and breast cancer cells, respectively,33,34 suggesting the possibility that the miRNA system can regulate uPA expression in breast cancer cells. To test this possibility, we initially analyzed potential miRNA target sites in 3′-UTR of uPA mRNA with a web-based miRNA target prediction program TargetScanHuman 5.1.35,36 There are 2 miR-181 target sites and 1 target site each for miR-143, miR-193, and miR-23 in 3′-UTR of human uPA mRNA (Fig. 1A). To determine the effect of these miRNAs on uPA expression, synthesized, mature miRNA mimics were introduced into MDA-MB-231 and MDA-MB-436 cells that were known to express high levels of uPA.37 Immunoblotting with anti-uPA mAb showed that, among those tested, miR-193a, miR-193b, and miR-181a mimics significantly downregulate uPA expression in both lines (Fig. 1B). The inhibitory effect of these mimics on uPA expression was clearly specific because the respective miRNA inhibitors (inhibitory antisense molecules for miRNAs) largely abolished their inhibitory effect on uPA expression in MDA-MB-231 cells (Fig. 1C).
Figure 1.
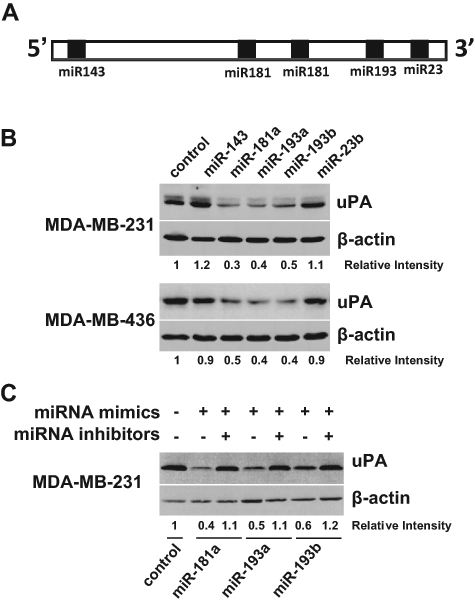
miR-193a, miR-193b, and miR-181a effectively inhibit uPA expression in breast cancer cells. (A) Diagram of potential miRNA target sites in 3′-UTR of human uPA mRNA. The solid box denotes miRNA target site. (B) MDA-MB-231 and MDA-MB-436 cells were transfected with negative control or indicated miRNA mimics for 3 days. Cells were lysed, and cell lysates were subjected to immunoblotting to detect uPA with uPA mAb. The membrane was stripped and reprobed for β-actin to ensure equal protein loading. (C) MDA-MB-231 cells were transfected with a mixture of miRNA mimic and its respective inhibitor for 3 days and then analyzed for uPA expression by immunoblotting.
To determine whether uPA mRNA is a direct target of miR-193a/b and miR-181a, we linked 3′-UTR of uPA mRNA to downstream of the luciferase gene in pMIR reporter plasmid. Cotransfection experiments showed that miR-193a, miR-193b, and miR-181a mimics, but not the control, inhibited luciferase activity in MDA-MB-231 and MDA-MB-436 cells (Fig. 2A). To confirm that these miRNAs target uPA mRNA through their predicted pairing sites in 3′-UTR of uPA mRNA, we introduced G/C→C/G and A/U→U/A mutation in these regions in order to disrupt miRNA/mRNA interaction (Fig. 2B). Mutation in the predicted miR-193 target site diminished the ability of miR-193a or miR-193b mimic, but not miR-181a mimic, to inhibit luciferase activity (Fig. 2C), while mutation in the predicted miR-181 target site only prevented miR-181a mimic–caused reduction in luciferase activity (Fig. 2C). These results confirm that uPA mRNA is a direct target of miR-193a/b and miR-181a in breast cancer cells.
Figure 2.
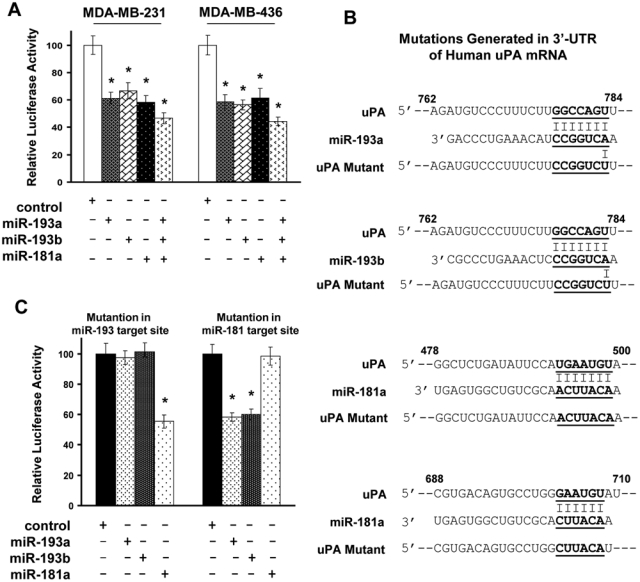
miR-193a, miR-193b, and miR-181a directly target uPA mRNA. (A) pMIR containing uPA 3′-UTR was cotransfected into MDA-MB-231 and MDA-MB-436 cells with 5 µM negative control, miR-193a, miR-193b, or miR-181a precursor for 2 days. Cells were lysed, and cell lysates were analyzed for luciferase activity. Data are mean ± SE (n = 3). *P < 0.01 versus negative control. (B) Diagram of mutation generated in 3′-UTR of human uPA mRNA. Alphabetical numbers are the relative nucleotide position in 3′-UTR of human uPA mRNA. (C) pMIR containing uPA 3′-UTR with mutation in a particular miRNA target site was cotransfected into MDA-MB-231 cells with 5 µM miR-193a, miR-193b, or miR-181a precursors for 2 days, followed by the analysis of luciferase activity. Data are mean ± SE (n = 3). *P < 0.01 versus negative control.
The levels of mature miR-193a/b and miR-181a, but not the levels of their respective pri-miRNAs, are inversely correlated to uPA expression in breast cancer cells
To investigate whether miR-193a/b and miR-181a participated in the regulation of uPA expression in breast cancer cells, we first performed qRT-PCR to quantitate the primary, precursor, and mature forms of these miRNAs in both high uPA-expressing (MDA-MB-231 and MDA-MB-436) and low uPA-expressing (MCF-7 and T47D) cells (Fig. 3B). The levels of the primary forms of these miRNAs were similar in all 4 lines (Fig. 3A). In contrast, the levels of both mature and precursor forms of miR-193a, miR-193b, and miR-181a were lower in MDA-MB-231 and MDA-MB-436 cells than those in MCF7 and T47D cells (Fig. 3A). As mature miRNA is generated through the consecutive steps of pri- to pre-miRNA and pre- to mature miRNA processing,2,3 these results indicate that the processing of primary to precursor uPA mRNA-targeted miRNAs is inefficient in high uPA-expressing breast cancer cells.
Figure 3.

Low abundance of Drosha/DGCR8 leads to inefficient processing of uPA mRNA-targeted miRNAs in breast cancer cells. (A) Total RNA was isolated from MCF-7, MDA-MB-231, MDA-MB-436, and T47D cells and then subjected to qRT-PCR to measure the levels of primary, precursor, and mature forms of miR-193a, miR-193b, and miR-181a. The levels of GAPDH were also determined and serve as an internal control for standardization. Data are mean ± SE (n = 3). (B) Overnight-cultured MCF-7, MDA-MB-231, MDA-MB-436, and T47D cells were lysed, and cell lysates were subjected to immunoblotting to detect uPA, Drosha, DGCR8, Dicer, and β-actin with the respective antibodies. (C) MDA-MB-231 cells were transfected with empty vector, Drosha, and DGCR8 expressing vectors together or Dicer expression vector for 2 days. Total RNA was isolated from these cells, and qRT-PCR was performed to measure the levels of primary and mature forms of miR-193a, miR-193b, and miR-181a. The levels of GAPDH were also determined and serve as an internal control for standardization. Data are mean ± SE (n = 3). *P < 0.001 versus control. (D) Drosha/DGCR8 together or Dicer was forced to be expressed in MDA-MB-231 cells for 3 days. Cells were lysed, and cell lysates were analyzed for uPA protein levels by immunoblotting with uPA mAb.
The processing of pri- to pre-miRNA and pre- to mature miRNA is respectively carried out by Drosha/DGCR8 and Dicer complexes.2,3 Immunoblotting with specific antibodies showed that Drosha and DGCR8 protein levels were significantly lower in cells with high uPA expression compared to cells with low uPA expression (Fig. 3B). In contrast, the level of Dicer was similar in all 4 lines (Fig. 3B). Together with the observation that both mature and precursor miR-193a/b and miR-181a levels were lower in high uPA-expressing cells (Fig. 3A), these results indicate the possibility that lower Drosha/DGCR8 expression may cause inefficient processing of primary to precursor uPA mRNA-targeted miRNAs, leading to upregulated uPA levels in MDA-MB-231 and MDA-MB-436 cells.
To further investigate the link between miRNA processing complexes and uPA expression, we simultaneously introduced Drosha and DGCR8 genes into MDA-MB-231 cells. qRT-PCR showed that enforced Drosha/DGCR8 expression led to a significant increase in mature miR-193a/b and miR-181a levels while displaying little effect on the levels of respective primary miRNAs (Fig. 3C). Immunoblotting also showed that enforced Drosha/DGCR8 expression greatly decreased uPA levels in MDA-MB-231 cells (Fig. 3D). In parallel, we introduced Dicer gene into MDA-MB-231 cells. However, forced Dicer expression exhibited little effect on either uPA mRNA-targeted miRNA levels or uPA expression (Fig. 3C and 3D). These results support the notion that low abundance of Drosha/DGCR8, rather than Dicer, contributes to low levels of mature miR-193a/b and miR-181a levels as well as high uPA expression in high uPA-expressing cells.
Impaired miRNA processing inhibits the production of mature miR-193a/b and miR-181a while upregulating uPA expression in breast cancer cells
To further establish the functional link between inefficient uPA mRNA-targeted miRNA processing and high uPA expression in breast cancer cells, we examined the effect of silencing Drosha, DGCR8, or Dicer on miR-193a/b and miR-181a levels in breast cancer cells. Cells were transduced with lentivirus containing control (scrambled sequence), or shRNAs specific for Drosha, DGCR8, or Dicer populations of the transduced cells were confirmed for gene-specific inhibition (Fig. 4A), followed by qRT-PCR to determine the levels of uPA mRNA-targeted miRNAs. While Drosha, DGCR8, or Dicer knockdown cells showed similar levels of pri–miR-193a, pri–miR-193b, or pri–miR-181a to those in the control, the respective mature miRNA levels were significantly less in knockdown cells than in the control (Fig. 4B). These results confirm that the processing of primary to mature miR-193a/b and miR-181a requires the presence of Drosha/DGCR8 and Dicer. Interestingly, immunoblotting showed that silencing miRNA processing components only greatly elevated uPA levels in high uPA-expressing cells, but it was unable to enhance uPA expression in low uPA-expressing cells (Fig. 5A). These results indicate that the miRNA system is most likely to act as a facilitator to regulate uPA expression.
Figure 4.

Primary miR-193a/b and miR-181a are not efficiently processed to their respective mature miRNAs in Drosha, DGCR8, and Dicer knockdown cells. (A) Cells were transduced with control lentiviral vector or vector encoding Drosha, DGCR8, or Dicer shRNA for 4 days. A portion of transduced cells was lysed for immunoblotting to detect Drosha, DGCR8, or Dicer with the respective antibodies. The membrane was stripped and reprobed for β-actin to ensure equal protein loading. (B) Total RNA was isolated from control, Drosha, DGCR8, and Dicer knockdown cells and then subjected to qRT-PCR to measure the levels of primary and mature forms of miR-193a, miR-193b, and miR-181a. The levels of GAPDH were also determined and serve as an internal control for standardization. Data are mean ± SE (n = 3). *P < 0.01 versus control.
Figure 5.

Impaired miRNA processing–induced uPA expression is functionally linked to inefficient miR-193a/b and miR-181a miRNA processing. (A) Cells were transduced with control lentiviral vector or vector containing Drosha, DGCR8, or Dicer shRNA for 4 days. Cells were lysed, and cell lysates were subjected to immunoblotting to detect uPA with uPA mAb. The membranes were stripped and reprobed for β-actin to ensure equal protein loading. (B) Drosha and DGCR8 knockdown MDA-MB-231 cells were transfected with negative control, miR-23a, miR-193a, miR-193b, or miR-181a mimics for 3 days. Cells were lysed, and cell lysates were subjected to immunoblotting to detect uPA with uPA mAb. The membranes were stripped and reprobed for β-actin to ensure equal protein loading.
To further confirm that the elevated uPA expression induced by the depletion of Drosha/DGCR8 was indeed caused by inefficient processing of uPA mRNA-targeted miRNAs, we introduced miR-193a, miR-196b, and miR-181a mimics into Drosha or DGCR8 knockdown MDA-MB-231 cells. Immunoblotting showed that each of these mimics alone was able to inhibit uPA expression (Fig. 5B). In contrast, treating these knockdown cells with miR-23a mimic did not alter uPA expression (Fig. 5B). These results suggest that upregulated uPA expression in cells with impaired miRNA processing is caused by reduced processing of uPA mRNA-targeted miRNAs.
In vitro invasion augmented by impaired miRNA processing is functionally linked to upregulated uPA expression
The well-established role of uPA in breast cancer cell invasion32,38 prompted us to investigate the consequence of the disruption of miRNA processing on breast cancer cell invasion. Matrigel (BD Biosciences, Franklin Lakes, NJ) in vitro invasion assay showed that MDA-MB-231 and MDA-MB-436 cells were able to invade Matrigel (BD Biosciences), while MCF7 and T47D cells were poorly invasive (Fig. 6A). Knockdown of Drosha, DGCR8, or Dicer rendered MDA-MB-231 and MDA-MB-436 cells significantly more invasive, while it displayed little effect on the invasiveness of MCF7 and T47D cells (Fig. 6A). These results corroborate with the observation that depleting miRNA processing components only upregulated uPA expression in MDA-MB-231 and MDA-MB-436 cells (Fig. 5A). Similar results were also obtained with high uPA-expressing BT549 cells and low uPA-expressing ZR75-1 cells (data not shown). To test whether upregulated uPA expression is necessary for enhanced invasion observed with Drosha, DGCR8, or Dicer knockdown cells, the control and these knockdown cells were treated with uPA siRNA pool or scrambled RNA, followed by the analysis of in vitro invasion. Matrigel (BD Biosciences) invasion assay showed that uPA siRNAs, but not the scrambled RNA, abrogated in vitro invasion of these cells (Fig. 6B). These results suggest that impaired miRNA processing–induced invasion is at least partially contributed by upregulated uPA expression.
Figure 6.
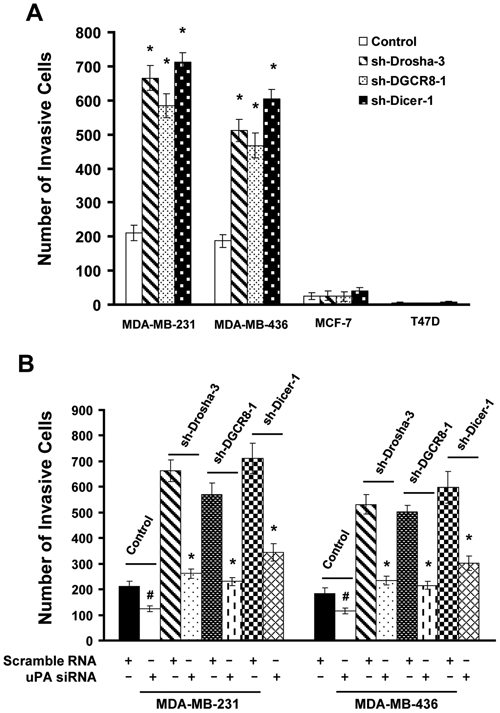
Impaired miRNA processing–enhanced in vitro invasion requires upregulated uPA expression. (A) Cells were transduced with control lentiviral vector or vector containing Drosha, DGCR8, or Dicer shRNA for 4 days. The population of the transduced cells was detached and analyzed for their ability to invade Matrigel. The invading cells on the undersurface of invasion chambers were stained and counted under a phase-contrast microscope. Data are mean ± SE (n = 3). *P < 0.005 versus control. (B) Scrambled RNA or uPA siRNA pool (5 µM) was transfected into control, Drosha, DGCR8, or Dicer knockdown cells for 3 days and then analyzed for in vitro invasion. Data are mean ± SE (n = 3). #P < 0.05 versus scrambled RNA. *P < 0.005 versus scrambled RNA.
Synthesized miR-193a, miR-193b, and miR-181a mimics inhibit in vitro invasion of breast cancer cells
As synthesized miR-193a/b and miR-181a mimics were able to inhibit uPA expression (Fig. 1B), we determine whether they were also able to suppress breast cancer cell invasion. MDA-MB-231 and MDA-MB-436 cells were treated with each of these miRNA mimics individually or in combination and subsequently analyzed for in vitro invasion using Matrigel (BD Biosciences) invasion chambers. Treatment of individual miR-193a, miR-193b, and miR-181a mimics, but not the control, resulted in approximately 40% to 50% of reduction in invasion, and combined treatment blocked over 70% of invasion (Fig. 7A). However, adding soluble single-chain uPA (scuPA) to the cells was able to rescue invasion of these cells in a dose-dependent manner (Fig. 6B). Especially at the concentration of 4 nM, scuPA restored over 60% of invasion in cells treated with these miRNA mimics (Fig. 7B). These results suggest that miR-193a/b and miR-181a inhibit in vitro invasion of breast cancer cells mainly by downregulating uPA expression.
Figure 7.
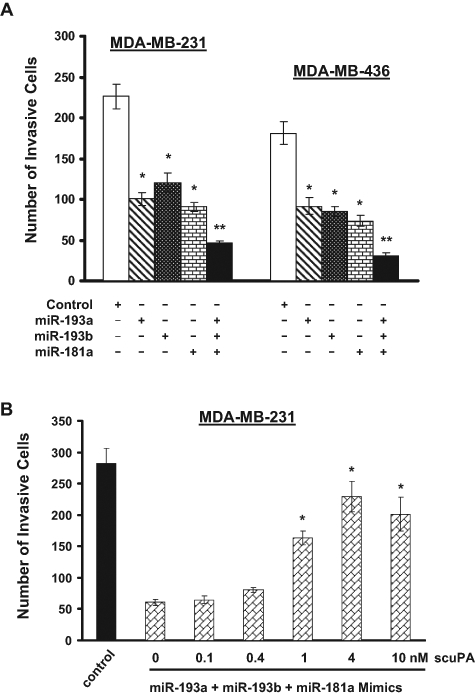
miR-193a, miR-193b, and miR-181a precursors inhibit breast cancer cell invasion. (A) MDA-MB-231 and MDA-MB-436 cells were transfected with 5 µM negative control, miR-193a, miR-193b, or miR-181a either individually or in combination for 3 days. Cells were then detached and added into the invasion chamber for 24 hours to allow invasion. The invading cells on the undersurface of the chamber were stained and counted under a phase-contrast microscope. Data are mean ± SE (n = 3). *P < 0.005 versus negative control. **P < 0.001 versus negative control. (B) MDA-MB-231 cells were treated with a combination of 5 µM miR-193a, miR-193b, and miR-181a precursors for 3 days. Cells were analyzed for in vitro invasion in the presence of various concentrations of scuPA (0.1-10 nM). Data are mean ± SE (n = 3). *P < 0.005 versus 0 nM scuPA.
Discussion
The observation that disrupting miRNA processing enhances carcinogenesis18-20 indicates that a large population of miRNAs may act as negative regulators of tumor progression. This possibility is supported by the findings that 1) global levels of miRNA expression are reduced in various tumor types including breast tumor14-17; 2) the expression of various miRNA processing components is downregulated in various tumor types including ovarian, lung, and gastric cancer21-23; 3) mutation has been found in the miRNA processing gene TARBP224; and 4) certain oncogenic miRNAs target Dicer to promote cancer metastasis.25 In this study, we showed that knockdown of Drosha, DGCR8, and Dicer, components of miRNA processing machinery, upregulated uPA expression and rendered breast cancer cells more invasive (Figs. 5 and 6). Our study provides another evidence that the miRNA system generally serves as a negative regulator of cancer progression.
Recent efforts have identified miRNAs that can suppress tumor growth and metastasis. For example, let-7 inhibits cell growth by inhibiting Ras and HMGA2 expression.39,40 miR-31 blocks breast cancer metastasis by targeting multiple migration-relevant genes including Fzd3, ITGA5, RDX, or RhoA.9 In this study, we showed that miR-193a, miR-193b, and miR-181a effectively inhibited breast cancer cell invasion (Fig. 7) by downregulating uPA expression in breast cancer cells (Fig. 1). The ability of these 3 miRNAs to reduce uPA expression was mediated by directly targeting 3′-UTR of uPA mRNA (Fig. 2). Our results indicate that miR-193a/b and miR-181a may also be classified into the category of currently expanding tumor/metastasis-suppressing miRNAs.
Analysis of miRNA expression profiling has shown that the levels of many tumor-suppressing miRNAs are reduced in tumor or metastatic tissues. For instance, the expression of metastasis-suppressive miR-126 and miR-335 is lost in metastatic breast tumors, and the loss of their expression is associated with poor distal metastasis-free survival.41 Also, miR-17 and miR-20, in which their levels are inversely correlated to cyclin D1, were often reduced or even lost in breast tumors.42 Several recent studies suggest that loss or reduction in tumor-suppressive miRNA expression is associated with CpG island hypermethylation in the respective miRNA promoters.43-46 In this study, we show that the levels of mature miR-193a/b and miR-181a are lower in high uPA-expressing breast cancer cells in comparison with those in low uPA-expressing cells, while the levels of their respective primary miRNAs are very similar between them (Fig. 3). This observation clearly shows that low levels of mature miR-193a/b and miR-181a in high uPA-expressing cells are not due to reduced gene transcription. In fact, the levels of Drosha/DGCR8 are low in high uPA-expressing cells (Fig. 3). Forced expression of Drosha/DGCR8 increases levels of mature miR-193a/b and miR-181a (Fig. 3). Our results suggest that low levels of mature uPA mRNA-targeted miRNAs (miR-193a/b and miR-181a) in invasive breast cancer cells are most likely caused by poor pri- to pre-miRNA processing. Recent studies have shown that downregulation of Drosha and Dicer is associated with a specific subgroup of breast cancer (high grade and high Ki67 index).47 Certain alternative initiation and splicing of Dicer gene observed in breast cancer cells can inhibit the translation of Dicer mRNA to protein.48 Although our results generated from the established breast cancer cell lines argue that reduced Drosha/DGCR8 expression may be the main cause contributing to the lower level of uPA mRNA-targeted miRNAs in breast cancer cells, it is very likely that reduced Dicer can also account for inefficient uPA mRNA-targeted miRNA processing in breast cancer cells, especially as concurrent reduction of Drosha and Dicer was detected in 15% of breast tumor specimens.49 Taken together, these studies implicate that in addition to the reduced primary miRNA transcription, poor miRNA processing can be another mechanism contributing to reduced levels of cancer-inhibitory miRNAs.
uPA is well recognized as a key molecule in the process of cancer invasion and matastasis,50 and its overexpression is detected in various metastatic tumors including breast cancer.28 In the MMTV-PymT transgenic breast cancer model, uPA deficiency was found to reduce metastasis more than 7-fold.51 Knockdown of uPA also exhibited great inhibitory effect on breast cancer cell invasion and metastasis.52 These findings indicate the possibility of circumventing metastatic breast tumors by inhibiting uPA expression. As the delivery of miR-26a has exhibited a promising tumor-suppressing effect,13 we reason that uPA mRNA-targeted miRNAs may also be employed as therapeutic agents against breast cancer invasion/metastasis.
Materials and Methods
Cells, shRNAs, miRNAs, and other reagents
MDA-MB-231 and MDA-MB-436 cells were cultured in DMEM supplemented with 10% FBS. MCF-7 and T47D cells were cultured in DMEM supplemented with 10% FBS plus 10 µg insulin. Antibodies used in immunoblotting include anti-Drosha polyclonal antibody (titer: 1:1,000) (Cat. No. 07-717, Millipore, Billerica, MA), anti-DGCR8 polyclonal antibody (titer: 1:1,000) (Cat. No. 10996-1-AP, Proteintech Group, Chicago, IL), anti-Dicer polyclonal antibody (titer: 1:1,000) (Cat. No. 3363, Cell Signaling Technology, Danvers, MA), anti-uPA mAb (titer: 1:1,000) (Cat. No. 394, American Diagnostica, Stamford, CT), and anti–β-actin mAb (titer: 1:1,000) (Cat. No. sc-47778, Santa Cruz Biotechnology, Santa Cruz, CA). Lentiviral vectors containing scrambled sequence, Drosha, DGCR8, or Dicer shRNAs were obtained from Addgene (Cambridge, MA). Mature miRNA mimics (miScript mimics) were purchased from Qiagen (Venlo, the Netherlands). Synthesized miRNA inhibitors were purchased from Applied Biosystems (Foster City, CA). The control and uPA siRNA pool were obtained from Fisher Scientific (Hampton, NH). Recombinant single-chain uPA was purchased from American Diagnostica.
In vitro invasion assay
Cell invasion was performed as previously described.53 Cells were detached with trypsin and then resuspended in serum-free medium at densities of 1.5 × 106 cells/mL, and 300 µL of cell suspension was added into each Matrigel-coated invasion chambers (Cell Biolabs, San Diego, CA). After a 24-hour invasion period, the remaining cells in the chambers were removed by cotton swabs, and the invading cells on the lower surface of the chambers were stained with Diff-Quick staining solution. The number of invading cells was calculated by counting 3 different fields under a phase-contrast microscope. To determine DGCR8, Drosha, or Dicer knockdown effect, cells were infected with lentiviral vectors containing Drosha, DGCR8, or Dicer shRNAs for 4 days before invasion assay. To determine the importance of uPA in invasion, cells were transfected with 5 µM uPA siRNA (siuPA) pool or scrambled RNA using Lipofectamine 2000 (Invitrogen, Carlsbad, CA) for 3 days followed by invasion assay. To determine the effect of exogenous uPA on cell migration, 4 nM recombinant single-chain uPA was added into cells during the invasion period. To determine the effect of miRNA mimics, 5 µM miR-181a, miR193a, and miR193b mimics were transfected into MDA-MB-231 using Lipofectamine 2000 (Invitrogen) for 3 days before the analysis of invasion.
qRT-PCR
Total RNA was extracted using Trizol (Invitrogen) and then treated with DNase I (Fermentas, Burlington, ON, Canada). The levels of mature miR-193a, miR-193b, and miR-181a were analyzed with miR-193a, miR-193b, and miR-181a TaqMan microRNA Assay Kit (Applied Biosystems), respectively. The levels of precursor miRNAs were measured with the respective miScript pre-miRNA assay kit (Qiagen). The levels of primary miRNAs were measured using primer sets specific for each miRNA. The levels of GAPDH mRNA were also measured and used as the internal normalization factor. All primer sequences are included in the supplementary material.
Construction of human uPA 3′-UTR luciferase reporter gene construct
The 3′-UTR of human uPA mRNA was generated by RT-PCR using total RNA isolated from MDA-MB-231 cells, and the PCR fragment was subcloned into pMIR luciferase report vector (Applied Biosystems). Primers for synthesizing 3′-UTR of human uPA mRNA are included in the supplementary material. To destroy the predicted miRNA-binding sites in uPA 3′-UTR, mutagenesis was performed with the aid of a QuikChange kit (Stratagene, La Jolla, CA). The primers used for introducing mutation are included in the supplementary material. To determine miRNA/uPA mRNA interaction, reporter gene constructs were cotransfected into MDA-MB-231 cells with miR193a, miR-193b, or miR-181a mimics for 2 days. Expression vector encoding Renilla luciferase at a 1:50 ratio was included during transfection. A dual luciferase system (Promega, Fitchburg, WI) was used to measure luciferase activity according to the manufacturer’s protocol. The Renilla luciferase activity serves as an internal control for standardization.
Statistical analysis
Statistical analyses of in vitro invasion assays and luciferase activities were performed by the Student t test using Microsoft Excel software (Redmond, WA). P < 0.05 was considered statistically significant.
Supplementary Material
Footnotes
Supplementary material for this article is available on the Genes & Cancer website at http://ganc.sagepub.com/supplemental.
The author(s) declared no potential conflicts of interest with respect to the authorship and/or publication of this article.
This work was supported by the National Institutes of Health [grant numbers CA093926, HL083335].
References
- 1. Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215-33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Ambros V. The functions of animal microRNAs. Nature. 2004;431:350-5 [DOI] [PubMed] [Google Scholar]
- 3. Murchison EP, Hannon GJ. miRNAs on the move: miRNA biogenesis and the RNAi machinery. Curr Opin Cell Biol. 2004;16:223-9 [DOI] [PubMed] [Google Scholar]
- 4. Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15-20 [DOI] [PubMed] [Google Scholar]
- 5. Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281-97 [DOI] [PubMed] [Google Scholar]
- 6. Ventura A, Jacks T. MicroRNAs and cancer: short RNAs go a long way. Cell. 2009;136:586-91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Iorio MV, Ferracin M, Liu CG, et al. MicroRNA gene expression deregulation in human breast cancer. Cancer Res. 2005;65:7065-70 [DOI] [PubMed] [Google Scholar]
- 8. Veeck J, Esteller M. Breast cancer epigenetics: from DNA methylation to microRNAs. J Mammary Gland Biol Neoplasia. 2010;15:5-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Valastyan S, Reinhardt F, Benaich N, et al. A pleiotropically acting microRNA, miR-31, inhibits breast cancer metastasis. Cell. 2009;137:1032-46 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 10. Ma L, Teruya-Feldstein J, Weinberg RA. Tumour invasion and metastasis initiated by microRNA-10b in breast cancer. Nature. 2007;449:682-8 [DOI] [PubMed] [Google Scholar]
- 11. Blenkiron C, Goldstein LD, Thorne NP, et al. MicroRNA expression profiling of human breast cancer identifies new markers of tumor subtype. Genome Biol. 2007;8:R214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Ma L, Reinhardt F, Pan E, et al. Therapeutic silencing of miR-10b inhibits metastasis in a mouse mammary tumor model. Nat Biotechnol. 2010;28:341-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Kota J, Chivukula RR, O’Donnell KA, et al. Therapeutic microRNA delivery suppresses tumorigenesis in a murine liver cancer model. Cell. 2009;137:1005-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Lu J, Getz G, Miska EA, et al. MicroRNA expression profiles classify human cancers. Nature. 2005;435:834-8 [DOI] [PubMed] [Google Scholar]
- 15. Volinia S, Calin GA, Liu CG, et al. A microRNA expression signature of human solid tumors defines cancer gene targets. Proc Natl Acad Sci U S A. 2006;103:2257-61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Calin GA, Croce CM. MicroRNA signatures in human cancers. Nat Rev Cancer. 2006;6:857-66 [DOI] [PubMed] [Google Scholar]
- 17. Gaur A, Jewell DA, Liang Y, et al. Characterization of microRNA expression levels and their biological correlates in human cancer cell lines. Cancer Res. 2007;67:2456-68 [DOI] [PubMed] [Google Scholar]
- 18. Kumar MS, Lu J, Mercer KL, et al. Impaired microRNA processing enhances cellular transformation and tumorigenesis. Nat Genet. 2007;39:673-7 [DOI] [PubMed] [Google Scholar]
- 19. Kumar MS, Pester RE, Chen CY, et al. Dicer1 functions as a haploinsufficient tumor suppressor. Genes Dev. 2009;23:2700-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Lambertz I, Nittner D, Mestdagh P, et al. Monoallelic but not biallelic loss of Dicer1 promotes tumorigenesis in vivo. Cell Death Differ. 2010;17:633-41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Merritt WM, Lin YG, Han LY, et al. Dicer, Drosha, and outcomes in patients with ovarian cancer. N Engl J Med. 2008;359:2641-50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Karube Y, Tanaka H, Osada H, et al. Reduced expression of Dicer associated with poor prognosis in lung cancer patients. Cancer Sci. 2005;96:111-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Zheng ZH, Sun XJ, Fu WN, et al. Decreased expression of DICER1 in gastric cancer. Chin Med J (Engl). 2007;120:2099-104 [PubMed] [Google Scholar]
- 24. Melo SA, Ropero S, Moutinho C, et al. A TARBP2 mutation in human cancer impairs microRNA processing and DICER1 function. Nat Genet. 2009;41:365-70 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 25. Martello G, Rosato A, Ferrari F, et al. A MicroRNA targeting dicer for metastasis control. Cell. 2010;141:1195-207 [DOI] [PubMed] [Google Scholar]
- 26. Blasi F. Proteolysis, cell adhesion, chemotaxis, and invasiveness are regulated by the u-PA-u-PAR-PAI-1 system. Thromb Haemost. 1999;82:298-304 [PubMed] [Google Scholar]
- 27. Blasi F, Sidenius N. The urokinase receptor: focused cell surface proteolysis, cell adhesion and signaling. FEBS Lett. 2010;584:1923-30 [DOI] [PubMed] [Google Scholar]
- 28. Foekens JA, Peters HA, Look MP, et al. The urokinase system of plasminogen activation and prognosis in 2780 breast cancer patients. Cancer Res. 2000;60:636-43 [PubMed] [Google Scholar]
- 29. Mazumdar A, Adam L, Boyd D, et al. Heregulin regulation of urokinase plasminogen activator and its receptor: human breast epithelial cell invasion. Cancer Res. 2001;61:400-5 [PubMed] [Google Scholar]
- 30. Sliva D, English D, Lyons D, et al. Protein kinase C induces motility of breast cancers by upregulating secretion of urokinase-type plasminogen activator through activation of AP-1 and NF-kappaB. Biochem Biophys Res Commun. 2002;290:552-7 [DOI] [PubMed] [Google Scholar]
- 31. Montero L, Nagamine Y. Regulation by p38 mitogen-activated protein kinase of adenylate- and uridylate-rich element-mediated urokinase-type plasminogen activator (uPA) messenger RNA stability and uPA-dependent in vitro cell invasion. Cancer Res. 1999;59:5286-93 [PubMed] [Google Scholar]
- 32. Huang S, New L, Pan Z, et al. Urokinase plasminogen activator/urokinase-specific surface receptor expression and matrix invasion by breast cancer cells requires constitutive p38alpha mitogen-activated protein kinase activity. J Biol Chem. 2000;275:12266-72 [DOI] [PubMed] [Google Scholar]
- 33. Salvi A, Sabelli C, Moncini S, et al. MicroRNA-23b mediates urokinase and c-met downmodulation and a decreased migration of human hepatocellular carcinoma cells. FEBS J. 2009;276:2966-82 [DOI] [PubMed] [Google Scholar]
- 34. Li XF, Yan PJ, Shao ZM. Downregulation of miR-193b contributes to enhance urokinase-type plasminogen activator (uPA) expression and tumor progression and invasion in human breast cancer. Oncogene. 2009;28:3937-48 [DOI] [PubMed] [Google Scholar]
- 35. Lewis BP, Shih IH, Jones-Rhoades MW, et al. Prediction of mammalian microRNA targets. Cell. 2003;115:787-98 [DOI] [PubMed] [Google Scholar]
- 36. Friedman RC, Farh KK, Burge CB, et al. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res. 2009;19:92-105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Mahanivong C, Yu J, Huang S. Elevated urokinase-specific surface receptor expression is maintained through its interaction with urokinase plasminogen activator. Mol Carcinog. 2007;46:165-75 [DOI] [PubMed] [Google Scholar]
- 38. Dass K, Ahmad A, Azmi AS, et al. Evolving role of uPA/uPAR system in human cancers. Cancer Treat Rev. 2008;34:122-36 [DOI] [PubMed] [Google Scholar]
- 39. Johnson SM, Grosshans H, Shingara J, et al. RAS is regulated by the let-7 microRNA family. Cell. 2005;120:635-47 [DOI] [PubMed] [Google Scholar]
- 40. Mayr C, Hemann MT, Bartel DP. Disrupting the pairing between let-7 and Hmga2 enhances oncogenic transformation. Science. 2007;315:1576-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Tavazoie SF, Alarcon C, Oskarsson T, et al. Endogenous human microRNAs that suppress breast cancer metastasis. Nature. 2008;451:147-52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Yu Z, Wang C, Wang M, et al. A cyclin D1/microRNA 17/20 regulatory feedback loop in control of breast cancer cell proliferation. J Cell Biol. 2008;182:509-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Lujambio A, Calin GA, Villanueva A, et al. A microRNA DNA methylation signature for human cancer metastasis. Proc Natl Acad Sci U S A. 2008;105:13556-61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Kozaki K, Imoto I, Mogi S, et al. Exploration of tumor-suppressive microRNAs silenced by DNA hypermethylation in oral cancer. Cancer Res. 2008;68:2094-105 [DOI] [PubMed] [Google Scholar]
- 45. Saito Y, Jones PA. Epigenetic activation of tumor suppressor microRNAs in human cancer cells. Cell Cycle. 2006;5:2220-2 [DOI] [PubMed] [Google Scholar]
- 46. Lujambio A, Ropero S, Ballestar E, et al. Genetic unmasking of an epigenetically silenced microRNA in human cancer cells. Cancer Res. 2007;67:1424-9 [DOI] [PubMed] [Google Scholar]
- 47. Dedes KJ, Natrajan R, Lambros MB, et al. Down-regulation of the miRNA master regulators Drosha and Dicer is associated with specific subgroups of breast cancer. Eur J Cancer. 2011;47:138-50 [DOI] [PubMed] [Google Scholar]
- 48. Irvin-Wilson CV, Chaudhuri G. Alternative initiation and splicing in dicer gene expression in human breast cells. Breast Cancer Res. 2005;7:R563-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Degryse B, Orlando S, Resnati M, et al. Urokinase/urokinase receptor and vitronectin/alpha(v)beta(3) integrin induce chemotaxis and cytoskeleton reorganization through different signaling pathways. Oncogene. 2001;20:2032-43 [DOI] [PubMed] [Google Scholar]
- 50. Sidenius N, Blasi F. The urokinase plasminogen activator system in cancer: recent advances and implication for prognosis and therapy. Cancer Metastasis Rev. 2003;22:205-22 [DOI] [PubMed] [Google Scholar]
- 51. Shapiro RL, Duquette JG, Roses DF, et al. Induction of primary cutaneous melanocytic neoplasms in urokinase-type plasminogen activator (uPA)-deficient and wild-type mice: cellular blue nevi invade but do not progress to malignant melanoma in uPA-deficient animals. Cancer Res. 1996;56:3597-604 [PubMed] [Google Scholar]
- 52. Subramanian R, Gondi CS, Lakka SS, et al. siRNA-mediated simultaneous downregulation of uPA and its receptor inhibits angiogenesis and invasiveness triggering apoptosis in breast cancer cells. Int J Oncol. 2006;28:831-9 [PMC free article] [PubMed] [Google Scholar]
- 53. Chen H, Zhu G, Li Y, et al. Extracellular signal-regulated kinase signaling pathway regulates breast cancer cell migration by maintaining slug expression. Cancer Res. 2009;69:9228-35 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


