Abstract
The population structure of Toxoplasma gondii includes three highly prevalent clonal lineages referred to as types I, II, and III, which differ greatly in virulence in the mouse model. Previous studies have implicated a family of serine/threonine protein kinases found in rhoptries (ROPs) as important in mediating virulence differences between strain types. Here, we explored the genetic basis of differences in virulence between the highly virulent type I lineage and moderately virulent type II based on successful genetic cross between these lineages. Genome-wide association revealed that a single quantitative trait locus controls the dramatic difference in lethality between these strain types. Neither ROP16 nor ROP18, previously implicated in virulence of T. gondii, was found to contribute to differences between types I and II. Instead, the major virulence locus contained a tandem cluster of polymorphic alleles of ROP5, which showed similar protein expression between strains. ROP5 contains a conserved serine/threonine protein kinase domain that includes only part of the catalytic triad, and hence, all members are considered to be pseudokinases. Genetic disruption of the entire ROP5 locus in the type I lineage led to complete attenuation of acute virulence, and complementation with ROP5 restored lethality to WT levels. These findings reveal that a locus of polymorphic pseudokinases plays an important role in pathogenesis of toxoplasmosis in the mouse model.
Keywords: copy-number variation, parasite, quantitative trait locus mapping, genetic mapping
The protozoan Toxoplasma gondii is a widespread parasite of warm-blooded animals that causes zoonotic infection in humans (1). Although most human infections are mild, serious disease can develop in immunocompromised patients because of congenital infection (2) or in some localities, as a result of ocular infection in otherwise healthy adults (3). Toxoplasma is transmitted by water- or food-borne ingestion of oocysts that are shed by cats, the definitive host where sexual recombination occurs (1). Additionally, T. gondii is unique among tissue cyst-forming coccidians in that ingestion of tissue cysts (for example, in raw or undercooked meat) can lead to infection in intermediate hosts, including humans (1). In North America and Europe, the population structure of T. gondii is remarkably clonal, with the predominance of three lineages called types I, II, and III (4). These lineages are 98% similar at the DNA level, and polymorphisms comprise a biallelic pattern at virtually all loci, indicating that they arose through a few recombinations in the wild followed by clonal expansion within the last 10,000 y (5, 6).
Although strains of T. gondii are genetically quite similar, they show strong phenotypic differences in the laboratory mouse, which provides a model for acute and chronic infection. Type I strains are uniformly lethal at all doses (high virulence) in all strains of laboratory mice, whereas types II (intermediate virulence) and III (low virulence) show much lower levels of pathogenicity (7). Type I strains are also more highly motile in vitro and are proficient at transmigration across cellular barriers. Hence, they disseminate rapidly in vivo (8, 9), whereas type II strains rely on altering the trafficking of their host cells to assure dissemination (10). Type II strains also induce stronger proinflammatory responses, including very high levels of IL-12 in comparison with either types I or III (11). Defining genes that contribute to these phenotypic differences is complicated by the strong linkage disequilibrium that characterizes the parasite population.
Analysis of complex quantitative phenotypes such as virulence has been realized through a series of pair-wise genetic crosses of T. gondii performed in cats, the generation of genetic maps, and linkage analysis (12). Such studies have revealed that a single major quantitative trait locus (QTL) on chromosome VIIa controls virulence differences between highly virulent type I and avirulent type III (13, 14), whereas five separate loci contribute to the virulence differences between intermediate virulent type II and avirulent type III (15). Thus far, genes that have been identified as responsible for virulence encode members of a family of polymorphic serine/threonine (S/T) protein kinases that are found in rhoptries (ROP kinases) (16), which are apical secretory organelles that discharge their contents during invasion (17). Among these are polymorphic kinases called ROP16s (TGME49_062730) that directly phosphorylate signal transducer and activator of transcription 3 (STAT3) (18) and STAT6 (19), thus leading to altered cytokine profiles and repression of IL-12 signaling (20). In addition, a polymorphic kinase called ROP18 (TGME49_005250) contributes to phenotypic differences between the highly virulent type I or intermediate virulent II and avirulent type III strain parasites, a phenotype associated with underexpression in the type III background. Restoration of the expression of ROP18 alleles from either type I (14) or II (15) in the type III background greatly enhances virulence. Although South American lineages differ substantially from those in North America and Europe, most contain a type I-like allele at ROP18, and these variants are also capable of conferring high levels of virulence when expressed in the type III background (21).
Here, we report the successful cross between types I and II and map genes responsible for the dramatic differences in virulence between these two lineages. These studies reveal that the expected active ROP kinases identified previously play no significant role in the present genetic context, whereas virtually all of the virulence difference is attributable to a tandem cluster of polymorphic pseudokinases known as ROP5 (22).
Results
Generation of a I × II Genetic Cross and Phenotypic Analysis of Recombinant Progeny.
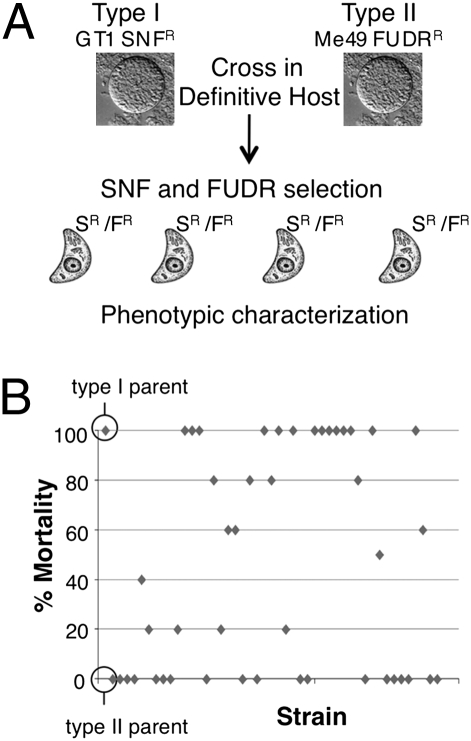
Previous studies have shown that a single type I parasite is lethal in outbred mice, whereas type II strain parasites are much less virulent (7). Because the available genetic crosses cannot be used to directly evaluate this phenotypic difference, we conducted a genetic cross between types I and II. A sinefungin-resistant line of the type I GT-1 strain (GT1 SNFR) was crossed with a 5-fluoro-2′-deoxyuridine–resistant line of the type II ME49 strain (ME49 FUDRR) by cofeeding of tissue cysts to four separate naïve cats (Fig. 1A). Progeny was cloned in the absence of drug and genotyped using 10 unlinked restriction fragment-length polymorphisms (RFLP) markers (Dataset S1), revealing that three of the cats shed recombinant progeny at the expected frequency (Fig. S1). Individual progeny clones were selected with drug selection or alternatively, without drug selection, and they were genotyped to identify 45 unique recombinant progeny Dataset S1 and Table S1). Consistent with previous studies, the lethal dose of the type I vs. type II parental lines differed by >3 logs in outbred mice based on i.p. challenge with tachyzoites (Fig. S2). To assess virulence of the progeny, mice were challenged with 100 parasites, a dose that readily distinguished between the parental lines (Fig. 1B). Most of the progeny clones matched one or the other parental lines, whereas others had intermediate values of survival (Fig. 1B).
Fig. 1.
Generation of a type I × II genetic cross and assessment of virulence. (A) Virulent type I GT-1 SNFR and avirulent type II ME49 FUDRR parental strains were crossed in the definitive host and recombinant progeny obtained using dual drug selection (SR/FR). Informative recombinants were identified using 10 RLFP markers located on 10 separate chromosomes. (B) The parental strains and 45 informative progeny were tested for virulence by inoculation of female CD-1 mice with 100 parasites each and monitoring for 30 d.
QTL Analysis Reveals a Single Virulence Association on Chromosome XII.
Previous genetic maps for T. gondii have relied on RFLP between parental lineages, and a composite genetic map has been assembled from previous crosses between types II and III and between types I and III (12). Here, we used a genotyping protocol (SI Results) based on hybridization of parental and recombinant progeny genomic DNA (gDNA) to T. gondii Affymetrix gene arrays (23). We identified 1,603 single-feature polymorphisms (SFP) based on probes that successfully discriminated perfect match (type II) from mismatch (type I) hybridization across all clones, thus providing reliable SFP markers for this cross. The resulting genetic map has 151 cross-over–defined genetic intervals and shows high-density marker coverage across the genome. Noninformative regions (∼8% of the total genome) consist of several small chromosomal regions that are nonpolymorphic in the parents, including chromosome Ia (which is monomorphic in the clonal lineages) (24) and three loci showing significant segregation distortion (Fig. S3). Despite this, genetic linkage analysis using this map is fully informative and sensitive for analyzing any virulence-related QTL that might occur in more than 85% of the genome, including loci previously implicated in virulence (i.e., ROP16 and ROP18).
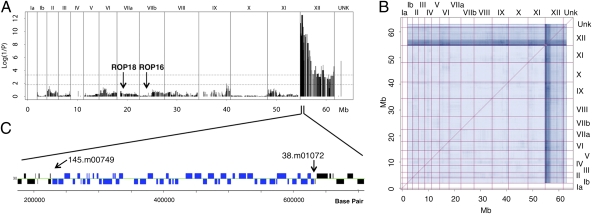
Genome-wide association analysis of the percent survival after challenge of naïve mice was used to define loci that contribute to virulence differences between types I and II. Remarkably, only a single high-probability QTL was identified on the left end of chromosome XII, accounting for ∼90% of the virulence phenotype variance (Fig. 2A). Notably, neither ROP16 nor ROP18, both of which have previously been implicated in acute virulence differences, showed any significant association in the current cross either in the whole-genome scan or when the QTL on chromosome XII was treated as a covariate (Fig. 2A). After statistical removal by regression of the chromosome XII locus variance contribution, the residual profile was not significantly different from background, reflecting a lack of detectable secondary associations. As well, no significant virulence-related epistasis was noted in whole-genome scans for nonadditive two-locus interactions (Fig. 2B). The QTL on chromosome XII comprises ∼400 kb containing 51 genes flanked by SFPs in the probes shown in Fig. 2C.
Fig. 2.
Genome-wide scan of the virulence phenotype reveals a single major QTL. (A) Progeny SFP marker genotypes were used to perform a genome-wide QTL association scan for virulence plotted on the y axis as log (1/P). A single major QTL on the left of chromosome XII explains 90.9% of the variance in the virulence phenotype. Markers are plotted on the x axis along the assembled genomic DNA sequence scaffolds of the 14 chromosomes (http://ToxoDB.org). Chromosomes are listed above the plot; UNK denotes markers on unassembled contigs. Dotted lines indicate marginal (lower line) and strongly significant (upper line) associations. (B) The matrix of two-locus associations shows a complete lack of nonadditive interactions or epistasis between pairs of loci. In A and B, Fisher's exact probability (P) was used for computing phenotype–genotype independence; results from other statistical strategies (t test statistic, Wilcoxon nonparametric, and mutual information) revealed the same QTL (SI Materials and Methods and SI Results). Chromosomes are listed at the top and right side. (C) The virulence QTL on XII is bound by SFP markers 145.m00749_at9 and 38.m01072_at10, which spans a region of ∼ 400 Kb containing 51 genes (blue).
Expression and Copy-Number Variation Within the Major Virulence QTL.
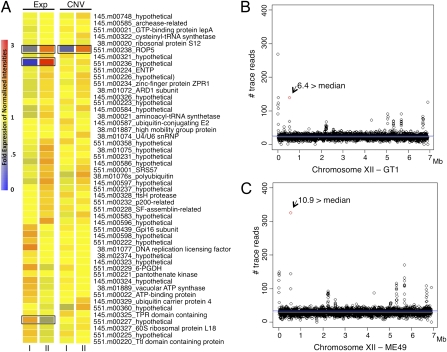
To identify genes within the major virulence QTL that might be responsible for virulence, we analyzed differences in expression between type I GT-1 and type II Me49 strains of T. gondii (http://toxodb.org/). Within the virulence QTL, 3 of 51 genes showed differences in expression between types I and II (Fig. 3A). In addition, we examined differences in relative copy-number variation (CNV) based on gDNA hybridizations used for genotyping. A single gene within the virulence QTL showed strong CNV, and this gene was annotated as ROP5 (TGGT1_042710 in the GT-1 genome) (Fig. 3A). Importantly, ROP5 also showed strong differences in expression level with it being higher in type II, which may be a reflection of the increased numbers of copies in this genetic background (Fig. 3A and Fig. S4). To further support the CNV data, we examined the trace reads from type I (GT-1) and type II (ME49) genomes for chromosome XII. ROP5 showed elevated signals in both strains, ∼6 times the median read density in type I and ∼11 times the median read density in type II (Fig. 3 B and C). The inheritance of ROP5 in the progeny closely followed this pattern, with the type I allele being associated with roughly one-half of the number of copies as the type II allele (Fig. S4). Collectively, these findings indicate that ROP5 is not a single gene as annotated in the genome but rather, a variable-copy tandem duplication, resulting in different mRNA expression levels between types I and II. Although CNV and mRNA data suggest that ROP5 is expressed at two times the level in type II strain parasites, protein expression levels seem to be roughly equal between the parental strains based on Western blot analysis using an antibody that recognizes both major alleles (Fig. S5).
Fig. 3.
Gene expression and CNV within the virulence QTL. (A) Gene expression (Exp) and CNV in type I and type II tachyzoites for the 51 genes within the XII locus. Heat map was created using the expression values, and colors represent fold expression of normalized intensities. Black boxes identify three genes with greater than or equal to twofold expression differences and one gene with a greater than or equal to twofold CNV difference between types I and II strains. (B and C) The number of GT-1 and ME49 trace reads (expect value = 0.0) per 2-kb segments across chromosome XII. The red dot denotes the fragment containing ROP5, and values are fold above the median number of trace reads across XII (gray line is the median and blue line is the mean).
Allelic Diversity of ROP5.
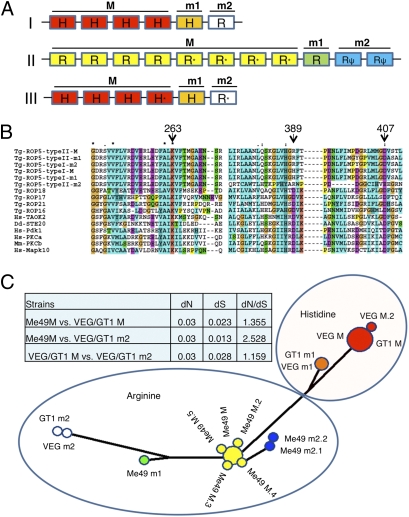
To examine the allelic diversity of ROP5, we compiled the different alleles from the trace reads by assembly in silico (Dataset S2 and Fig. S6). Analysis of the coding regions revealed significant polymorphism between the major allele shared by type I and III and the major allele found in type II (Fig. 4A and Fig. S7). Although types I and III were largely similar, each strain contains several minor alleles that contain a small number of polymorphisms; in the case of type II, several of these are pseudogenes caused by frame shifts or stop codons (Fig. 4A and Fig. S7). Differences between the major alleles are largely confined to the ATP-binding pocket and the substrate recognition regions of the kinase fold (Fig. S7). Within the ATP-binding pocket, ROP5 lacks an essential aspartate (D) found in the catalytic site within subdomain VIB, although it contains other conserved residues important for function including a critical lysine (K) in subdomain II and aspartate (D) in subdomain VII (Fig. 4B and Fig. S7). Based on the presence of a substitute histidine (H) or arginine (R) at this critical catalytic aspartate, we have designated the alleles as H or R and major and minor based on the relative copy number (Fig. 4 A and B). Network analysis of the genes encoding these polymorphic variants revealed that the major alleles in type I and III are most closely related, with all of the alleles found in type II forming a second separate node. Importantly, the minor alleles found in type I and III that encode R at the critical catalytic site (i.e., m2 alleles) were more closely related to type II than those typically found in type I and III (Fig. 4C). Analysis of synonymous and nonsynonymous polymorphisms in ROP5 reveals that most of the alleles have evolved under neutral selection [i.e., synonymous (dS)/nonsynonymous (dN) ∼ 1]. The exceptions are the m2 alleles found in type I and III and the major R alleles found in type II, which show that moderate selection pressure has influenced diversity between them (i.e., dN/dS > 2.5) (Fig. 4C).
Fig. 4.
Composition of the ROP5 locus in type I and type II lineages. (A) Tandem array of 6 copies of ROP5 in types I and III vs. 11 copies in type II. Alignment of trace reads determined that types I and III have a four-copy major allele, M (red), and two single-copy minor alleles, m1 (orange) and m2 (white), and that type II ME49 has eight copies of a major allele, M (yellow), a single-copy minor allele, m1 (green), and two copies of a minor allele that encodes a pseudogene, m2 (blue). Alleles with a single nonsynonymous SNP have an asterisk. Pseudogenes are indicated by ψ. Copy numbers and sequences of individual alleles were reconstructed from the trace archive reads (Fig. S6), although the arrangement shown here is arbitrary. (B) ClustalX alignment of types I and II ROP5 alleles with active ROP kinases from T. gondii and host kinases (Fig. S7). Residues essential for catalysis (263K and 407D) are conserved in all ROP5 alleles. The catalytic aspartic residue (389D) is divergent in all ROP5 alleles, where instead, all type II ME49 alleles contain 389R and the majority of type I GT-1 alleles have 389H, except ROP5 type I m2, which has 389R (Fig. 4A). (C) Phylogenetic network of ROP5 sequences. The major allele in type I and III as well as m1 alleles are more closely related, whereas type II alleles form a second major group. The minor alleles m2 in types I and III show a closer ancestry to type II. Table of synonymous (dS) and nonsynonymous (dN) changes in ROP5 alleles. Alleles are as in A, with the suffix indicating copy number.
ROP5 Is Essential for Virulence in the Type I Lineage.
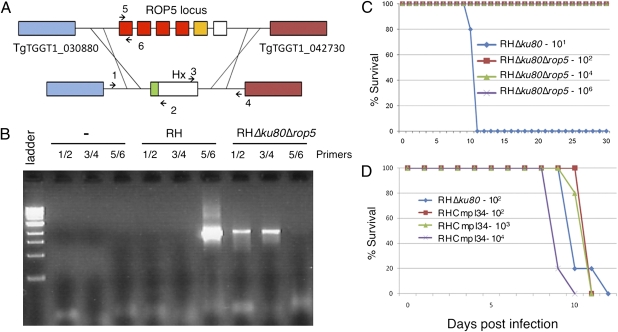
Based on differences in allelic composition and CNV, we reasoned that ROP5 was the most likely candidate within the virulence QTL to mediate the virulence differences between types I and II. To test this model, we generated a deletion of the entire ROP5 locus in the type I background using a recently described mutant of RH that lacks nonhomologous recombination because of deletion of ku80 (25) (Fig. 5A, Dataset S1, and Table S2). Deletion of the ROP5 locus and replacement with the selectable marker hypoxanthine xanthine guanosine phosphoribosyl transferase (HXGPRT) was confirmed by PCR using primers that flank the region (Fig. 5 A and B, Dataset S1, and Table S2) and by Western blotting using a rabbit anti-ROP5 antisera (Fig. S8). Absence of the ROP5 locus did not affect growth in vitro, because the deletion strain formed the same number and size of plaques as the absent WT parental strain (Fig. S9). Comparison of the virulence of the RHΔku80Δrop5 mutant to the parental RHΔku80 line revealed a >5-log increase in the lethal dose (Fig. 5C). Complementation of RHΔku80Δrop5 with ROP5 was achieved by transfection of a cosmid from the type 1 strain spanning the ROP5 locus (TOXOM52) and partially overlapping the adjacent gene TGGT1_042730. Western blot analysis revealed that a transformant called RHCmpl34 had restored levels of ROP5 expression (Fig. S8), and quantitative RT-PCR (qRT-PCR) data confirmed that adjacent gene TGGT1_042730 was not appreciably affected in either Δrop5 or the complemented clone (SI Results). Testing of the RHCmpl34 clone in outbred mice revealed that it fully restored the virulence phenotype seen in WT I parasites (Fig. 5D).
Fig. 5.
Deletion of ROP5 greatly reduces virulence. (A) Schematic of the approach used to create the RHΔku80Δrop5 parasite. Regions surrounding the ROP5 locus were used to replace the region with the selectable marker HXGPRT (Hx). (B) PCR analysis to confirm the knockout of ROP5 in RHΔku80Δrop5 by detecting of flanking regions (flanks) using the primer pairs depicted in A. Ladder is a 1-kb ladder (New England Biolabs). (C) Virulence of RHΔku80Δrop5 parasites in female CD-1 mice inoculated with 102, 104, or 106 parasites vs. 101 RHΔku80 strain parasites. (D) Infection of CD-1 mice with Δrop5 complement, RHCmpl34, restores virulence comparable with WT severity. Mice were inoculated with 102 RHΔku80 or 102, 103, or 104 RHCmpl34 parasites.
Discussion
We have extended the genetic analysis of complex traits in T. gondii by performing a genetic cross between types I and II, which differ substantially in several important biological traits. Genome-wide association studies revealed that a single locus controls differences in acute virulence between these strains. A polymorphic cluster of pseudokinases known as ROP5 was associated with significant differences in survival after acute challenge. Deletion of the entire locus in the type I strain confirms that it is essential for acute virulence, resulting in complete attenuation of the normally lethal type I RH strain. These findings suggest that the amplification of pseudokinases in the genome of T. gondii contributes to pathogenesis, although additional studies will be needed to explore the extent to which different alleles are important.
The current genetic cross between types I and II strains of T. gondii completes the set of pair-wise genetic crosses between these three lineages and expands our ability to compare them genetically and phenotypically. A comprehensive analysis of the genetic parameters among the various pair-wise crosses is planned for a future report, although such details are not essential for robust mapping of complex phenotypes, such as the virulence examined here. Comparison of the virulence of type I and II strains in laboratory mice indicates that they differ by more than 3 logs in LD100 in outbred mice in the present study (Fig. S2); however, this difference is likely to be greater, because a single type I parasite is normally sufficient to cause lethal infection (7). Remarkably, this difference can be attributed to a single locus containing a repetitive and polymorphic cluster of pseudokinases, previously named ROP5 (22). Although this difference was mapped in the type I strains GT-1, here, we have disrupted the entire locus in the related type I strain RH and observed a strong phenotype consistent with the expected mapping data. Despite small differences previously identified among type I strains (26), these findings argue that ROP5 likely plays a similar important role in the virulence of all type I strains. The ROP5 locus mapped here also corresponds to that previously identified in the cross between types II and III, denoted VIR1 (15), and also, an accompanying report (27) confirms that ROP5 also contributes to virulence differences between types II and III.
The absence of a role for either ROP18 or ROP16 was somewhat surprising given that these active kinases have been found to be highly significant in other genetic crosses (14, 15). ROP16 phosphorylates STAT3 and STAT6, leading to differences in gene expression and cytokine induction between types II and III (20). Although ROP16 contributes to the more subtle virulence differences between II and III in inbred mice (15), it was not a major factor in the survival of outbred mice challenges with types I vs. II strain parasites as shown here. More surprisingly, we did not observe a contribution for allelic variants of ROP18 that are encoded by type I vs. II strains (14, 15). Both type I and II alleles have previously been found to confer enhanced virulence when expressed in the type III background (14, 15). The diversification of ROP18 between types I and II, which has occurred under strong selective pressure (21), suggests these two alleles may play an important role in other hosts of T. gondii (for example, in the wild). Regardless, under the condition examined here, neither ROP16 nor ROP18 seems to play any role in differences between types I and II.
Although previous studies have highlighted the importance of ROP kinases in mediating pathogenesis of T. gondii, this is an example of a pseudokinase participating in pathogen virulence. The ROP5 alleles contain a predicted kinase domain containing the typical subdomains of S/T protein kinases (22). However, ROP5 lacks the key catalytic aspartate residue involved in transfer of the γ-PO4 from ATP to its targets; this residue is conserved among all active protein kinases (28). Hence, it is extremely unlikely that ROP5 is an active protein kinase. ROP5 has previously been predicted to fold with the topology of a kinase (22), and the crystal structures of two related ROP pseudokinases, ROP2 and ROP8 (29, 30), confirm this prediction. Similar to other members of the ROP2 family, ROP5 contains an N-terminal extension that contains several low complexity regions. Arginine-rich amphipathic regions in the N terminus have been implicated in targeting of ROP family members to the parasitophorous vacuole membrane (29, 31), although the role of ROP5 at this interface remains uncertain. Pseudokinases have recently gained attention for their role as allosteric regulators of active kinases (32). Although potential partners of ROP5 remain undefined, an appealing possibility is that it regulates other parasite kinases that are responsible for modulating virulence, such as ROP18. In this regard, it is interesting to note that ROP5 is epistatic on ROP18 in a previous cross between types II and III (15, 27) and that transfer of either ROP18 allele into type III strain, which contains the virulent ROP5 cluster, restores virulence to this lineage (14, 15). Defining the relative contributions of the various alleles of ROP5 found in respective strains and their interactions with ROP18 and possibly other partners will require further biochemical and molecular studies. Collectively, it seems likely that the expansion and diversification of pseudokinases, which are a prominent feature of the ROP family in T. gondii (16), are not simply a relict caused by genome decay but rather, play an important role in the biology of infection.
Materials and Methods
Propagation of Parasite Strains.
Strains of T. gondii used in this study include type I GT-1, type II ME49, and type I RHΔku80ΔHx (referred to as RHΔku80) (25). Parasites were grown in human foreskin fibroblast (HFF) cell monolayers sustained in DMEM supplemented with 10% FBS at 37 °C under 5% CO2 as described previously (13). Parasites were harvested from host cells after natural egress as described previously (13).
Genetic Cross.
Drug-resistant parental lines were generated by treating parasites with N-ethyl-N-nitrosourea (200 μg/mL for 2 h at 37 °C) (Sigma-Aldrich) as described (33). Selection with SNF (3 × 10−7 M) for GT-1 or FUDR (1 × 10−5) for ME49 (parental clone B7) was used to obtain the drug-resistant lines GT1 SNFR and ME49 FUDRR, respectively. Tissue cysts containing bradyzoites of each strain were obtained from chronically infected CD-1 mice maintained in the case of GT1 SNFR parasites by treatment with 0.5 mg/mL sulfadiazine in the drinking water. Strains were crossed by cofeeding tissue cysts to four naïve cats, and oocysts were purified, hatched, and used to infect monolayers of HFF cells as described previously (34). To determine the frequency of recombination, randomly selected clones were genotyped using 10 unlinked genetic markers (Dataset S1 and Table S1) as previously characterized (12). Additional recombinant progeny was obtained by growth in the presence of both drugs and cloning by limiting dilution (Dataset S1 and Table S1). A total of 45 clones exhibiting unique recombinant genotypes was cryo-preserved and retained for additional analysis.
gDNA Hybridizations.
Freshly egressed parasites grown in HFF cells were harvested as described above, and gDNA was purified using the DNAeasy kit (Qiagen) with the addition of RNase Cocktail (Ambion). gDNA was sheared by nebulization (Invitrogen) for 3 min at ∼ 40 psi, precipitated, washed in 70% ethanol, and resuspended in water. The BioPrime Labeling System (Invitrogen) was used to label 600 ng gDNA per reaction for 2 h at 37 °C. A total of 5 μg labeled DNA was hybridized to Affymetrix T. gondii GeneChips (http://ancillary.toxodb.org/docs/Array-Tutorial.html) performed at the Functional Genomics Core Facility at Montana State University (http://cores.montana.edu/index.php?page=functional-genomics-core-facility). Hybridization intensities were processed and normalized with the software package R (http://www.r-project.org/) using the following R library functions: bg.correct = rma and normalize = quantiles. Replicate hybridizations were performed for the parental clones to establish SFP markers, and single hybridizations were performed for the progeny.
Mouse Infectivity Studies.
To determine the virulence of progeny clones compared with the parental lines, parasites (n = 100) were injected i.p. into 8- to 10-wk-old female CD-1 mice (5–15 mice per isolate) (Dataset S1 and Table S1), and survival was monitored for 30 d as described previously (13). To examine the virulence of the knockout, RHΔku80Δrop5 parasites were injected i.p. into 8- to 10-wk-old female CD-1 mice at doses of 102, 104, and 106 (five mice per dose). In comparison, 101 RHΔku80 was injected into five female CD-1 mice. Similarly, to assess the virulence of the complement, RHCmpl34 parasites were injected i.p. into 8- to 10-wk-old female CD-1 mice at doses of 102, 103, and 104 (five mice per dose). As a control, five CD-1 mice were injected with 102 RHΔku80. Mice were monitored until there were no remaining survivors or for 30 d.
Genotyping and Genome-Wide Scan of Virulence.
A total of 1,603 reliable SFP genetic markers was identified by genotyping of parental and progeny clones based on gDNA hybridization to the Affymetrix array (SI Materials and Methods). Array data are available at the National Center for Biotechnology Information (NCBI; Gene Expression Omnibus accession number GSE24905). On average, there are 10.6 markers per 151 genetic intervals defined by cross-overs. Genotypic and phenotypic information for 45 informative progeny was used to perform genome-wide scans of virulence using the cumulative survival data from mouse challenges. For QTL association mapping, the virulence trait showed the same association whether treated as a continuous trait or (Fig. 2A) partitioned into four categories (high = 100%, intermediate to high = 50–80%, intermediate to low = 20–40%, and low = 0%). Segregation distortion was mapped by statistical association of the observed genome-wide genotypes to two virtual pseudophenotype vectors in which all of the progeny were assigned only one of the parental values.
Gene Expression Analysis.
Microarray data for type I GT-1 and type II ME49 strains available at ToxoDB (http://toxodb.org/toxo/) were downloaded from GEO (GSE16037 - http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE16037) and used to examine tachyzoite gene expression differences for the 51 genes within the major virulence QTL. Gene expression differences were compared, and a heat map was created from the expression values using software package GeneSpring 7.2 (Agilent Technologies) as described previously (35).
CNV.
Genomic hybridization data for type I GT-1 SNFR and type II ME49 FUDRR strains were analyzed using the software package GeneSpring 7.2 using robust multiarray averaging (RMA) and normalized (per chip per gene) to the median. Genes with greater than or equal to twofold differences between types I and II were identified. Trace Archive reads from T. gondii genomes type I GT-1, type II ME49, and type III VEG were obtained from NCBI (http://www.ncbi.nlm.nih.gov/Traces/trace.cgi) and compared with the ME49 chromosome XII sequence available at ToxoDB (http://toxodb.org/common/downloads/) (36). Briefly, chromosome XII was segmented into 2-kb fragments that were compared with individual trace reads using BLASTN (expect value = 0.0). The number of trace reads matching each chromosome XII fragment was determined and graphed with the software package R. Reads that matched ROP5 were compared with the median number of trace reads across chromosome XII.
Characterization of ROP5 Alleles.
Trace archive reads matching the ROP5 coding sequence (expect value = 0.0) were assembled using Vector NTI ContigExpress (Invitrogen) for each of the parental strains using the following variables: overlap cutoff = 35, overlap percent identity = 0.9, and overlap similarity = 500. Manual inspection of the trace read assemblies was conducted to define separate alleles of ROP5 (Fig. S6 and Dataset S2). Alignment of ROP5 and host kinases was performed using Clustal X (37) (Fig. S7).
Phylogenetic Analysis of ROP5.
A phylogenetic network of ROP5 gene sequences was derived using the median-joining algorithm (38) (with ε = 0) as implemented in NETWORK 4.1 (http://fluxus-engineering.com/). ROP5 sequences were clustered using DNA Alignment v1.1.2.1 (http://fluxus-engineering.com/) and used to construct a network using Kruskal's algorithm for minimum spanning trees and Farris's maximum parsimony heuristic algorithm. dS and dN substitution rates in ROP5 sequences and positive selection were estimated using PAML.4.4 (39).
RHΔku80Δrop5 Generation and Complementation.
To disrupt the ROP5 locus (i.e., TGGT1_042710), sequences from the type I RH strain corresponding to regions downstream of TGGT1_042730 (ChrXII: 600,519–601,909 bp) and upstream of TGGT1_030880 (ChrXII: 614,958–616,405 bp) were used to construct a knockout plasmid for homologous integration into the parasite line RHΔku80Δhx as described previously (25). A plasmid containing these flanking sequences on either side of the HXGPRT gene was created using the MultiSite three-fragment gateway system (Invitrogen) (Dataset S1 and Table S2). The 5′ flanking sequence was cloned into pDONR-p4p1r, the HXGPRT gene was cloned into pDONR-p1p2, and the 3′ flanking region was cloned into pDONR-p2rp3 using standard BP reactions. All three plasmids were combined with pDest-p4p3 into a MultiSite LR reaction to create the knockout plasmid pDest-p4p3-Δrop5. RHΔku80 parasites were electroporated with pDest-p4p3-Δrop5, inoculated onto monolayers of HFF cells, and allowed to recover for ∼12 h before selection with 25 μg/mL mycophenolic acid (Sigma-Aldrich) supplemented 50 μg/mL xanthine (Sigma-Aldrich). Resistant parasites were cloned by limiting dilution in complete medium with mycophenolic acid and xanthine. Knockout clones were screened by PCR using primers specific for homologous recombination (Fig. 5 A and B, Dataset S1 and Table S2), leading to replacement of the HXGPRT gene at the ROP5 locus; this is designated as RHΔku80Δrop5-Hx+ (referred to as RHΔku80Δrop5). Complementation of the RHΔku80Δrop5 parasite was achieved by transfection with the TOXOM52 cosmid that spans the ROP5 locus in the type I genome and hence, likely contains approximately six copies of the ROP5 gene (SI Materials and Methods). A representative transformant referred to as RHCmpl34, which reexpresssed ROP5 at WT levels (Fig. S8), was chosen for analysis.
Supplementary Material
Acknowledgments
We are grateful to John Boothroyd, Jon Boyle, and Michael Reese for discussions of unpublished data; David Roos, David Kulp, and Amit Bahl for their design of the Toxoplasma gene chip; Julie Nawas and Jennifer Barks for expert technical assistance; Mae Huynh and Vern Carruthers for the Δku80-deficient parasite line; Paul H. Davis for guidance with gDNA preparation; Kate McInnerney for hybridization of the ToxoGeneChip microarrays; and the public databases provided at http://ToxoDB.org and http://www.ncbi.nlm.nih.gov/Traces/trace.cgi. J.C.W is supported by the Intramural Program of National Center for Biotechnology Information, National Library of Medicine, National Institutes of Health. J.P.D. was partially supported by US Department of Agriculture, Agriculture Research Service, Current Research Information System (CRIS) 1265-32000-076. This work was supported by National Institutes of Health Grants AI082423 and AI036629 (to L.D.S.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, www.ncbi.nlm.nih.gov/geo (accession no. GSE24905). The sequence files for ROP5 alleles have been deposited in GenBank.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1015338108/-/DCSupplemental.
References
- 1.Dubey JP. Toxoplasmosis of Animals and Humans. Boca Raton, FL: CRC; 2010. [Google Scholar]
- 2.Joynson DH, Wreghitt TJ. Toxoplasmosis: A Comprehensive Clinical Guide. Cambridge, UK: Cambridge University Press; 2001. [Google Scholar]
- 3.Jones LA, Alexander J, Roberts CW. Ocular toxoplasmosis: In the storm of the eye. Parasite Immunol. 2006;28:635–642. doi: 10.1111/j.1365-3024.2006.00874.x. [DOI] [PubMed] [Google Scholar]
- 4.Sibley LD, Ajioka JW. Population structure of Toxoplasma gondii: Clonal expansion driven by infrequent recombination and selective sweeps. Annu Rev Microbiol. 2008;62:329–351. doi: 10.1146/annurev.micro.62.081307.162925. [DOI] [PubMed] [Google Scholar]
- 5.Boyle JP, et al. Just one cross appears capable of dramatically altering the population biology of a eukaryotic pathogen like Toxoplasma gondii. Proc Natl Acad Sci USA. 2006;103:10514–10519. doi: 10.1073/pnas.0510319103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Su C, et al. Recent expansion of Toxoplasma through enhanced oral transmission. Science. 2003;299:414–416. doi: 10.1126/science.1078035. [DOI] [PubMed] [Google Scholar]
- 7.Sibley LD, Boothroyd JC. Virulent strains of Toxoplasma gondii comprise a single clonal lineage. Nature. 1992;359:82–85. doi: 10.1038/359082a0. [DOI] [PubMed] [Google Scholar]
- 8.Barragan A, Sibley LD. Transepithelial migration of Toxoplasma gondii is linked to parasite motility and virulence. J Exp Med. 2002;195:1625–1633. doi: 10.1084/jem.20020258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Barragan A, Sibley LD. Migration of Toxoplasma gondii across biological barriers. Trends Microbiol. 2003;11:426–430. doi: 10.1016/s0966-842x(03)00205-1. [DOI] [PubMed] [Google Scholar]
- 10.Lambert H, Barragan A. Modelling parasite dissemination: Host cell subversion and immune evasion by Toxoplasma gondii. Cell Microbiol. 2010;12:292–300. doi: 10.1111/j.1462-5822.2009.01417.x. [DOI] [PubMed] [Google Scholar]
- 11.Robben PM, et al. Production of IL-12 by macrophages infected with Toxoplasma gondii depends on the parasite genotype. J Immunol. 2004;172:3686–3694. doi: 10.4049/jimmunol.172.6.3686. [DOI] [PubMed] [Google Scholar]
- 12.Khan A, et al. Composite genome map and recombination parameters derived from three archetypal lineages of Toxoplasma gondii. Nucleic Acids Res. 2005;33:2980–2992. doi: 10.1093/nar/gki604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Su C, Howe DK, Dubey JP, Ajioka JW, Sibley LD. Identification of quantitative trait loci controlling acute virulence in Toxoplasma gondii. Proc Natl Acad Sci USA. 2002;99:10753–10758. doi: 10.1073/pnas.172117099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Taylor S, et al. A secreted serine-threonine kinase determines virulence in the eukaryotic pathogen Toxoplasma gondii. Science. 2006;314:1776–1780. doi: 10.1126/science.1133643. [DOI] [PubMed] [Google Scholar]
- 15.Saeij JPJ, et al. Polymorphic secreted kinases are key virulence factors in toxoplasmosis. Science. 2006;314:1780–1783. doi: 10.1126/science.1133690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Peixoto L, et al. Integrative genomic approaches highlight a family of parasite-specific kinases that regulate host responses. Cell Host Microbe. 2010;8:208–218. doi: 10.1016/j.chom.2010.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Håkansson S, Charron AJ, Sibley LD. Toxoplasma evacuoles: A two-step process of secretion and fusion forms the parasitophorous vacuole. EMBO J. 2001;20:3132–3144. doi: 10.1093/emboj/20.12.3132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yamamoto M, et al. A single polymorphic amino acid on Toxoplasma gondii kinase ROP16 determines the direct and strain-specific activation of Stat3. J Exp Med. 2009;206:2747–2760. doi: 10.1084/jem.20091703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ong YC, Reese ML, Boothroyd JC. Toxoplasma rhoptry protein 16 (ROP16) subverts host function by direct tyrosine phosphorylation of STAT6. J Biol Chem. 2010;285:28731–28740. doi: 10.1074/jbc.M110.112359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Saeij JPJ, et al. Toxoplasma co-opts host gene expression by injection of a polymorphic kinase homologue. Nature. 2007;445:324–327. doi: 10.1038/nature05395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Khan A, Taylor S, Ajioka JW, Rosenthal BM, Sibley LD. Selection at a single locus leads to widespread expansion of Toxoplasma gondii lineages that are virulent in mice. PLoS Genet. 2009;5:e1000404. doi: 10.1371/journal.pgen.1000404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.El Hajj H, Lebrun M, Fourmaux MN, Vial H, Dubremetz JF. Inverted topology of the Toxoplasma gondii ROP5 rhoptry protein provides new insights into the association with the parasitophorous vacuole membrane. Cell Microbiol. 2007;9:54–64. doi: 10.1111/j.1462-5822.2006.00767.x. [DOI] [PubMed] [Google Scholar]
- 23.Bahl A, et al. A novel multifunctional oligonucleotide microarray for Toxoplasma gondii. BMC Genomics. 2010;11:603. doi: 10.1186/1471-2164-11-603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Khan A, et al. Common inheritance of chromosome Ia associated with clonal expansion of Toxoplasma gondii. Genome Res. 2006;16:1119–1125. doi: 10.1101/gr.5318106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Huynh MH, Carruthers VB. Tagging of endogenous genes in a Toxoplasma gondii strain lacking Ku80. Eukaryot Cell. 2009;8:530–539. doi: 10.1128/EC.00358-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Khan A, Behnke MS, Dunay IR, White MW, Sibley LD. Phenotypic and gene expression changes among clonal type I strains of Toxoplasma gondii. Eukaryot Cell. 2009;8:1828–1836. doi: 10.1128/EC.00150-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Reese ML, Zeiner GM, Saeij JPJ, Boothroyd JC, Boyle JP. Polymorphic family of injected pseudokinases is paramount in Toxoplasma virulence. Proc Natl Acad Sci USA. 2011;108:9625–9630. doi: 10.1073/pnas.1015980108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hanks SK, Hunter T. Protein kinases 6. The eukaryotic protein kinase superfamily: Kinase (catalytic) domain structure and classification. FASEB J. 1995;9:576–596. [PubMed] [Google Scholar]
- 29.Labesse G, et al. ROP2 from Toxoplasma gondii: A virulence factor with a protein-kinase fold and no enzymatic activity. Structure. 2009;17:139–146. doi: 10.1016/j.str.2008.11.005. [DOI] [PubMed] [Google Scholar]
- 30.Qiu W, et al. Novel structural and regulatory features of rhoptry secretory kinases in Toxoplasma gondii. EMBO J. 2009;28:969–979. doi: 10.1038/emboj.2009.24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Reese ML, Boothroyd JC. A helical membrane-binding domain targets the Toxoplasma ROP2 family to the parasitophorous vacuole. Traffic. 2009;10:1458–1470. doi: 10.1111/j.1600-0854.2009.00958.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Boudeau J, Miranda-Saavedra D, Barton GJ, Alessi DR. Emerging roles of pseudokinases. Trends Cell Biol. 2006;16:443–452. doi: 10.1016/j.tcb.2006.07.003. [DOI] [PubMed] [Google Scholar]
- 33.Pfefferkorn ER, Pfefferkorn LC. Quantitative studies of the mutagenesis of Toxoplasma gondii. J Parasitol. 1979;65:364–370. [PubMed] [Google Scholar]
- 34.Dubey JP. Oocyst shedding by cats fed isolated bradyzoites and comparison of infectivity of bradyzoites of the VEG strain Toxoplasma gondii to cats and mice. J Parasitol. 2001;87:215–219. doi: 10.1645/0022-3395(2001)087[0215:OSBCFI]2.0.CO;2. [DOI] [PubMed] [Google Scholar]
- 35.Behnke MS, Radke JB, Smith AT, Sullivan WJ, Jr, White MW. The transcription of bradyzoite genes in Toxoplasma gondii is controlled by autonomous promoter elements. Mol Microbiol. 2008;68:1502–1518. doi: 10.1111/j.1365-2958.2008.06249.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gajria B, et al. ToxoDB: An integrated Toxoplasma gondii database resource. Nucleic Acids Res. 2008;36:D553–D556. doi: 10.1093/nar/gkm981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Larkin MA, et al. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- 38.Bandelt HJ, Forster P, Röhl A. Median-joining networks for inferring intraspecific phylogenies. Mol Biol Evol. 1999;16:37–48. doi: 10.1093/oxfordjournals.molbev.a026036. [DOI] [PubMed] [Google Scholar]
- 39.Yang Z. PAML 4: Phylogenetic analysis by maximum likelihood. Mol Biol Evol. 2007;24:1586–1591. doi: 10.1093/molbev/msm088. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.