Abstract
In G-protein signaling, an activated receptor catalyzes GDP/GTP exchange on the Gα subunit of a heterotrimeric G protein. In an initial step, receptor interaction with Gα acts to allosterically trigger GDP release from a binding site located between the nucleotide binding domain and a helical domain, but the molecular mechanism is unknown. In this study, site-directed spin labeling and double electron–electron resonance spectroscopy are employed to reveal a large-scale separation of the domains that provides a direct pathway for nucleotide escape. Cross-linking studies show that the domain separation is required for receptor enhancement of nucleotide exchange rates. The interdomain opening is coupled to receptor binding via the C-terminal helix of Gα, the extension of which is a high-affinity receptor binding element.
Keywords: signal transduction, structural polymorphism
The α-subunit (Gα) of heterotrimeric G proteins (Gαβγ) mediates signal transduction in a variety of cell signaling pathways (1). Multiple conformational states of Gα are involved in the signal transduction pathway shown in Fig. 1A. In the inactive state, the Gα subunit contains a bound GDP [Gα(GDP)] and has a high affinity for Gβγ. When activated by an appropriate signal, a membrane-bound G-protein coupled receptor (GPCR) binds the heterotrimer in a quaternary complex, leading to the dissociation of GDP and formation of an “empty complex” [Gα(0)βγ], which subsequently binds GTP. The affinity of Gα(GTP) for Gβγ is dramatically reduced relative to Gα(GDP), resulting in functional dissociation of active Gα(GTP) from the membrane-bound complex. The active Gα(GTP) subsequently binds downstream effector proteins to trigger a variety of regulatory events, depending on the particular system. Thus, the GPCR acts to catalyze GDP/GTP exchange via an empty complex. Crystallographic (2–7), biochemical (8), and biophysical (9–11) studies have elucidated details of the conformational states of Gα that correspond to the discrete steps indicated in Fig. 1A, but the mechanism by which receptor interaction leads to release of the bound GDP from Gα and the structure of the empty complex remain a major target of research in the field.
Fig. 1.
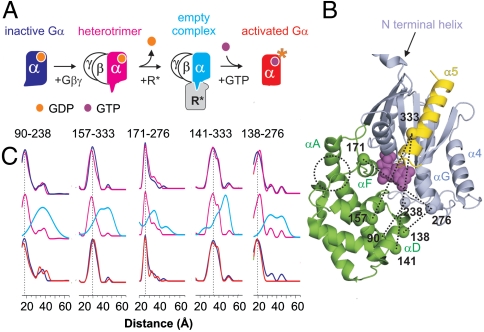
Receptor activation of G proteins leads to a separation between domains. (A) The pathway of Gα activation via activated rhodopsin (R*). The alpha subunit is color coded to denote the four different states investigated by SDSL/DEER spectroscopy. (B) Ribbon model of Gαi(GDP) (PDB ID code 1GP2). The helical and nucleotide binding domains are colored green and light blue, respectively, and GDP is shown as magenta spheres. Relevant secondary structural elements are noted for reference. The C-terminal helix α5 is colored yellow; six disordered residues at the C terminus are not displayed. The N-terminal helix is truncated for convenience. Sites from which R1 nitroxide side chains were selected pair wise for distance measurements are indicated by spheres; dotted traces indicated specific distances measured for each state in A. (C) Distance distributions for the indicated doubly spin-labeled mutants. (Top) Compares Gαi(GDP) and Gαi(GDP)βγ; (Middle) compares Gαi(GDP)βγ and R ∗ •Gαi(0)βγ; (Lower) compares Gαi(GDP) and Gαi(GTP); traces are color coded to match states in A.
The Gα subunit has two structural domains, namely a nucleotide binding domain and a helical domain that partially occludes the bound nucleotide (Fig. 1B). From the initial Gα crystal structure in 1993, Noel et al. (2) recognized that nucleotide release would probably require an opening between the two domains in the empty complex, but in the intervening 18 years there has been little compelling experimental support for this idea. Nevertheless, some constraints on the general topology of the complex are known. For example, numerous studies indicate that the C terminus of Gα is bound tightly to the receptor in the empty complex (9). In addition, the N-terminal helix of Gα is associated with Gβγ and with the membrane via N-terminal myristoylation (12, 13). Together, these constraints fix the position of the nucleotide domain with respect to the membrane. The helical domain is connected to the nucleotide domain through two flexible linkers, and linker 1 (switch I) undergoes conformational changes upon receptor binding (10). These observations provided the motivation to look for relative motion of the two Gα domains during formation of the empty complex.
For this purpose, site-directed spin labeling (SDSL) and double electron–electron resonance (DEER) spectroscopy were employed to measure distances between pairs of spin labels, with one label in each domain. Distances were measured for each state of Gα along the activation pathway using activated rhodopsin (R*) as the GPCR. The results indicate that receptor-catalyzed nucleotide exchange in G proteins requires a large-scale reorientation of domains in the G protein α-subunit.
Results and Discussion
Using SDSL and DEER spectroscopy, distances were measured for each state of Gαi along the activation pathway using activated rhodopsin (R*) as the GPCR. In these experiments, the R1 nitroxide side chain (Fig. S1) was introduced via cysteine substitution mutagenesis into the background of Gαi with reactive cysteines removed, HexaI (Gαi HI) (14). Fig. 1B shows the set of sites from which pairs were selected and the five specific interdomain distances investigated.
All doubly spin-labeled proteins bind to R* to an extent similar to the Gαi HI parent protein as shown in direct endpoint binding assays (Fig. S2). In addition, they are all functional with respect to receptor-mediated nucleotide exchange, although mutants 138R1/276R1 and 157R1/333R1 have, respectively, about 40% and 55% of the receptor-catalyzed nucleotide exchange rate of the parent Gαi HI protein (Fig. S2). The reduced rates suggest that the residues involved are important in modulating receptor-mediated nucleotide exchange. In crystal structures of the inactive protein, residues Asn157 and Glu276 are involved in side chain H bonding and electrostatic interactions, respectively, and mutation of these may influence local conformation.
DEER spectroscopy relies on magnetic dipolar interactions between spin labels to measure interspin distances in the range of ≈17–60 Å (15, 16). Of particular importance is the ability to resolve multiple distances and the widths of the distributions. Fig. 1C compares the distance probability distributions for the five transdomain R1 pairs in each of the four states of Gαi, i.e., Gαi(GDP), Gαi(GDP)βγ, Gαi(0)βγ, and Gαi(GTP). For each pair, the measured most probable distances for Gαi(GDP) and Gαi(GDP)βγ agree well with expectations from the crystal structures (5–7) and models of the R1 side chain (17). In all cases there is little difference between Gαi(GDP) and Gαi(GDP)βγ.
Upon photoactivation of rhodopsin and formation of the R ∗ •Gαi(0)βγ complex, there is a remarkable increase in each interspin distance, with increases being as large as 20 Å (at 90/238) (for details, see SI Text and Figs. S3 and S4). Moreover, there is a dramatic increase in width of each distribution as well as multiple distances in most cases. It is of interest that distances present in the Gαi(0)βγ distributions correspond approximately to minor populations already present in Gαi(GDP) and Gαi(GDP)βγ, suggesting that activation may shift an existing equilibrium. Although the exact widths of the distributions in Gαi(0)βγ may not be well-determined in each case, they are clearly broader than possible from multiple rotamers of R1, suggesting spatial disorder of the Gα protein in the empty-pocket state of the activated complex (see SI Text). Finally, addition of GTPγS restores a state with a most probable distance and width of distribution similar to the GDP bound state. This is in agreement with expectations from GTPγS bound crystal structures (6).
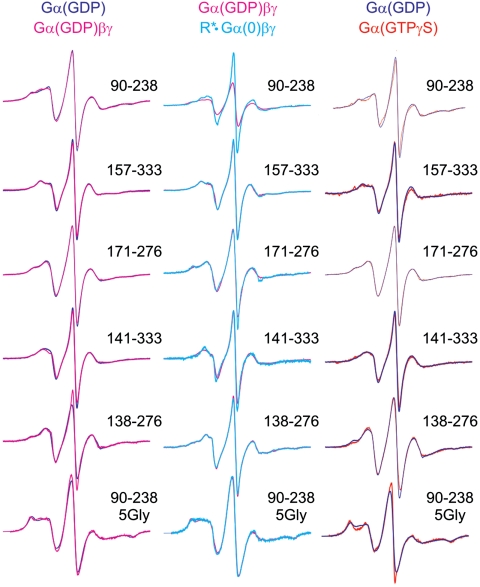
The EPR spectra of R1 residues at the sites shown in Fig. 1B have little or no changes upon receptor activation (Fig. 2). This result, taken together with the very large distance changes observed, ensure that the detected distance increases reflect global domain movement rather than simple R1 side chain rearrangements due to changes in local environment. Collectively, the data strongly support a model for a Gαi(0)βγ in which the helical domain is displaced relative to the nucleotide domain in the heterotrimer, and in which the structure is highly flexible with respect to the relative domain orientations.
Fig. 2.
CW EPR spectra of the spin-labeled double mutants in Gαi at the indicated states along the activation pathway. (Left) Compares EPR spectra of the doubly labeled Gαi(GDP) and Gαi(GDP)βγ mutants; (Middle) compares Gαi(GDP)βγ and R ∗ •Gαi(0)βγ; (Right) compares Gαi(GDP) and Gαi(GTP).
To visualize the domain opening, a model of the empty complex on the receptor was constructed that is consistent with the DEER and other available experimental data (see SI Text). To generate the model, the heterotrimeric Gi was docked with the photoreceptor using crystal structures of Gαi(GDP)βγ (4, 7) and opsin in complex with the high-affinity Gαt C-terminal peptide (18). The Gαi C-terminal helix was fused with the high-affinity Gα C-terminal peptide bound to opsin (for details, see SI Text and Figs. S5 and S6), which provided a convenient starting point for the model (19). The myristoylated N-terminal amphipathic helix was placed parallel to the membrane surface and the heterotrimer oriented such that both the myristoyl group and the nearby farnesylated C terminus of the Gγ-subunit can be inserted into the membrane; together these hydrophobic interactions cooperatively drive membrane binding of the intact heterotrimer (20). The procedure required chain breaks within the linker regions of the α-subunit (between residues 59–60 and 184–185) and resulted in clashes in loop regions within the heterotrimer that were then resolved through loop reconstruction and model relaxation in Rosetta (21, 22). A rigid body docking protocol was executed to find placements of the helical domain consistent with the DEER distance restraints (SI Text, Fig. S7, and Table S1). An ensemble of models was found to be in agreement with the experimental distances from DEER data, consistent with the increase in width of the distance distributions (Fig. S8). The model that agrees best with the most probable distances from DEER data (Fig. 3B) fulfills all distance restraints within the error of the experiment and involves an approximately 8-Å motion of the helical domain away from the nucleotide domain as well as an approximately 29° rotation relative to its starting position (Fig. S9).
Fig. 3.
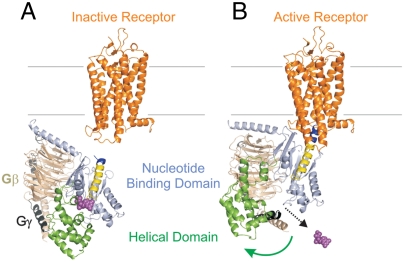
A model showing the opening of the interdomain cleft in formation of the empty complex. (A) The inactive receptor (1U19.pdb) and inactive G protein (see SI Text), with color coding as in Fig. 1. (B) Model of the complex with active receptor (3DQB.pdb) showing the reorientation of the helical domain (Movie S1).
The model shown in Fig. 3 incorporates a constraint gleaned from an interesting feature of the Gαi structure. In the structure, helix αA has a pronounced kink (dotted circle, Fig. 1B) that is not due to proline or glycine residues in the sequence. Rather, the strained kink may be stabilized by a three-element network of packing interactions between the α5/β6 turn, the αF helix, and the helix αA. Previous results showed that receptor interaction with Gαi moves the α5/β6 turn, a change that could weaken the three-element interaction and trigger kink relaxation, thus moving the body of the helical domain relative to the nucleotide domain. Coupling between α5 and αF was suggested by several Gαi proteins that act as functional mimetics of the receptor-bound state (23). Kink relaxation is incorporated into the preliminary model of Fig. 3, but the actual relative movement of the helical domain shown in the figure does not depend on this mechanism, which will be examined in future studies. An animation showing the features of this model and the avenue for nucleotide escape can be found in SI Text.
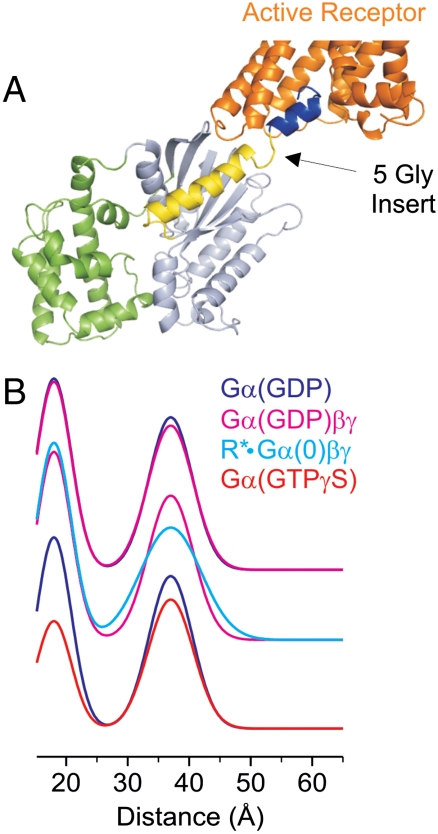
The C terminus of Gα is a critical interaction site between the G protein and the receptor (9, 24–26) as illustrated in the model of Fig. 3. Previous studies demonstrate that the C terminus undergoes a disorder-to-order transition upon binding to activated receptors, inducing structural changes that are important for efficient GDP release (27–29). Gαi with a flexible 5-glycine linker inserted at the base of the α5 helix (at residue 343, Fig. 4A) binds to R* but eliminates a receptor-mediated movement of this helix, increases basal exchange, and uncouples nucleotide exchange from binding (9, 30). We have introduced the same 5-glycine insertion into the interdomain pair, R90R1/E238R1. Fig. 4 shows the distance distribution for the various states of Gαi, to be compared with those of the parent protein shown in Fig. 1C. Remarkably, the 5-Gly insertion results in a bimodal distance distribution in all states, the components of which correspond approximately to the open and closed positions of the helical domain. However, the distribution for the population at longer distances (approximately 40 Å) is substantially sharper than that in Fig. 1C. Apparently, the perturbation of α5 by the insertion uncouples movement of the helical domain from receptor interaction. Although additional studies would be required to characterize the states of the insertion mutant, the result suggests a critical role of the C terminus in allosteric communication from the receptor to helical domain opening and the nucleotide binding pocket.
Fig. 4.
A 5-Gly insertion in α5 of Gαi uncouples domain opening from receptor binding. (A) Ribbon model of Gαi(GDP) showing the location of the 5-Gly insertion between residues 343–344; additional residues (345–354, blue ribbon) from the opsin/peptide crystal structure (3DQB.pdb) were added after the insert to suggest the subunit bound to activated rhodopsin. (B) Distance distributions of 90R1/238R1 compared for Gαi(GDP) and Gαi(GDP)βγ (Top), Gαi(GDP)βγ and R ∗ •Gαi(0)βγ (Middle), and Gαi(GDP) and Gαi(GTP) (Lower). The 5-Gly insert bearing the 90R1/238R1 double mutation binds to R* in native disc membranes to approximately the same extent as the GαiHI parent.
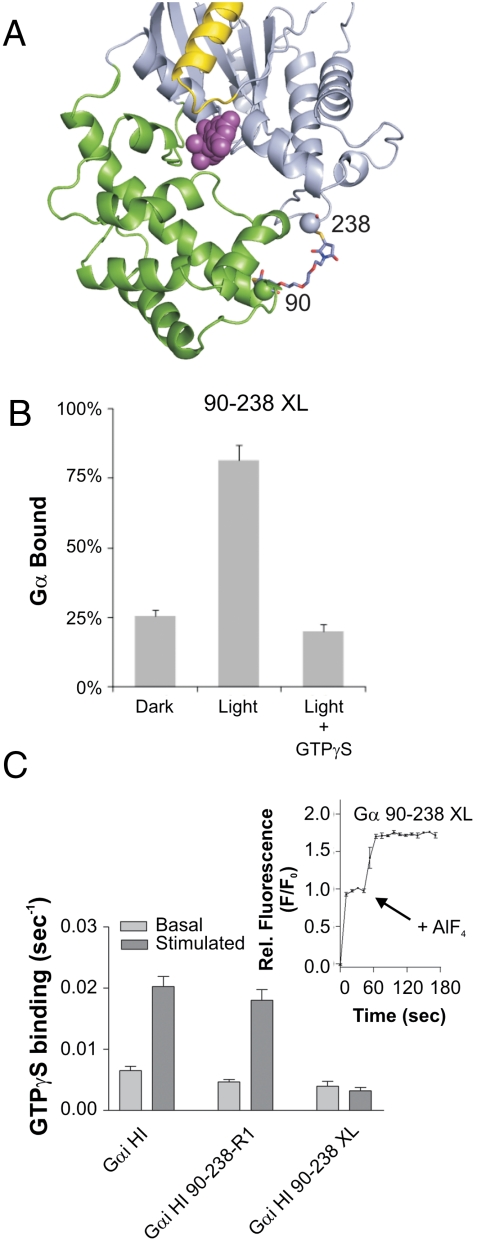
Is the domain rearrangement required for GDP release? To address this question, the two domains were cross-linked, disallowing the domain opening. For this purpose, a bifunctional, thiol-directed bis-maleimide was selected to cross-link cysteine residues in the R90C-E238C protein, based on the predicted proximity between these thiols in the Gαi(GDP) protein (Fig. 5A). Cross-linking resulted in a Gαi(GDP)βγ-protein competent to bind activated receptors to approximately the same extent as the parent protein (Fig. 5B). Moreover, the cross-linked protein undergoes aluminum fluoride-dependent conformational changes (Fig. 5C, Inset) consistent with an active, properly folded protein. On the other hand, this protein exhibited severely impaired rates of receptor-mediated nucleotide exchange as compared to either the parent or uncross-linked protein (Fig. 5C), demonstrating the essential nature of the domain separation in receptor-mediated G-protein activation. The basal nucleotide exchange rate was only slightly reduced (Fig. 5C), suggesting an effect specific to receptor-mediated nucleotide release, the slow step in G-protein activation.
Fig. 5.
Cross-linking of the helical and nucleotide domains of a R90C-E238C Gαi double mutant. (A) Model of the bis-maleimide interdomain cross-linker; the color code is as in Fig. 1. (B) Binding of the cross-linked mutant to rhodopsin in disc membranes. (C) Basal and receptor-stimulated nucleotide exchange rates for the bis-maleimido cross-linked (XL) Gαi. For comparison, the GαiHI and R90R1/E238R1 nucleotide exchange rates are shown. (Inset) Tryptophan fluorescence changes of the XL Gαi subunit upon aluminum fluoride addition.
Conclusions
This study demonstrates that the result of G-protein interaction with an activated receptor is propagated allosterically to reorient the distant helical domain of Gαi, opening the domain interface in formation of a flexible ternary receptor–G-protein complex. Preventing the large interdomain movement through cross-linking markedly reduces the rate of catalyzed nucleotide exchange, demonstrating the crucial role of the interdomain opening in receptor-mediated G-protein activation. Although the detailed mechanism is currently under further investigation, this domain opening would be predicted to reduce the GDP binding energy as interactions are lost upon opening of the domain interface. Together these changes help broaden our understanding of the conformational changes in the G protein that lead to GDP release, the slow step in G-protein activation.
Methods
Membrane Binding Assays.
The ability of wild-type and Gαi proteins containing the side chain R1 (Fig. S1) to bind rhodopsin was tested as described previously (31). For additional details, see SI Methods.
Cross-Linking.
The bifunctional cross-linking reagent 1,11-bis(maleimido)triethylene glycol (Pierce Biotechnology) was incubated in a 2∶1 molar ratio with Gαi Hexa I-R90C-E238C at 4 °C for 2 h in 50 mM Tris, 130 mM NaCl, 10 mM MgCl2, 5 mM EDTA, 100 μM GDP, at pH 7.0. After 2 h, reaction was quenched with chromatography buffer (50 mM Tris, 130 mM NaCl, 2 mM MgCl2, 10 uM GDP, 1 uM EDTA, 1 mM DTT, pH 7.5) and concentrated in this buffer. The concentrated, cross-linked monomeric protein was then purified by gel filtration FPLC on a calibrated SW2000 column (Sigma). Calibration was performed under the same conditions as purification, using a broad range of molecular weight standards (Biorad).
Nucleotide Exchange Assays.
G proteins and rod outer segment (ROS) membranes were prepared essentially as previously described (10). The rates of basal and receptor-mediated nucleotide exchange of the spin-labeled Gα proteins were measured at excitation/emission wavelengths of 290/340 nm in buffer containing 50 mM Tris, 130 mM NaCl, 2 mM MgCl2, 1 mM DTT, 1 μM EDTA, pH 7.5, for 40 min at 18 °C after addition of 10 μM GTPγS. For receptor-stimulated exchange, proteins were reconstituted with an equimolar amount of Gβ1γ1 (200 nM each) prior to measurement of exchange; the experiments were performed in the presence of light activated rhodopsin (100 nM) obtained from urea washed ROS membranes. Basal exchange was carried out in the absence of rhodopsin and Gβγ. The data were normalized to the baseline and the fluorescence maximum, and rate of exchange was determined by fitting the data to an exponential association curve. Rates shown in Fig. S2B are from a minimum of four independent experiments (± SEM).
Spin Labeling and Electron Paramagnetic Resonance Measurements.
Spin labeling was carried out in buffer containing 20 mM 3-(N-morpholino)propanesulfonic acid (pH 6.8), 100 mM NaCl, 2 mM MgCl2, 50 μM GDP, and 10% glycerol (vol/vol). The Gαi double mutants were incubated with the sulfhydryl spin-label S-(1-oxy-2,2,5,5,-tetramethylpyrroline-3-methyl)-methanethiosulfonate in a 2∶1 molar ratio at room temperature for 5 min. Noncovalently bound nitroxide was removed by extensive washing with labeling buffer using a 30-kDa molecular weight concentrator.
A series of EPR spectra were recorded for each spin-labeled mutant. Continuous wave (CW) EPR spectra were recorded at room temperature on a Bruker E580 spectrometer using a high-sensitivity resonator (HS0118) at X-band microwave frequencies. Each spectrum was collected using a 100-G field scan at a microwave power of 19.92 mW. Optimal field-modulation amplitudes were selected to give maximal signal intensity without line-shape distortion. The data were typically averages of approximately 20 scans.
Four-Pulse DEER Measurements.
The spin-labeled proteins were flash frozen within quartz capillaries in a liquid nitrogen bath. After freezing, they were loaded into a 2-mm split-ring resonator, and DEER measurements were performed at 80 K on a Bruker Elexsys 580 spectrometer. Four-pulse DEER was carried out as previously described (32), with the π-pump pulse (16 ns) was positioned at the absorption maximum of the field swept nitroxide center line and the observer π (16 ns) and π/2 (8 ns) pulses at the absorption maximum of the low-field line.
The buffer used for DEER measurements was similar to the CW EPR experiments. Four different states of each double-labeled mutant were measured to determine conformational changes along the G-protein activation pathway. All DEER data were analyzed with the DEER Analysis 2011 software package freely available at the Web site http://www.epr.ethz.ch/, and with a Labview software package provided by Christian Altenbach (Jules Stein Eye Institute, Los Angeles, CA). Details for utilization of the DEER Analysis 2011 software package were previously described (33). Background correction of the primary dipolar evolution data was performed as described (33). For distance distributions below 20 Å, excitation bandwidth corrections were applied (34). These corrections had very little effect on the computed distributions. Tikhonov regularization techniques were used for fitting the data using L-curve methods for determining the regularization parameter (35). In some instances, Gaussian fitting was also employed where distribution widths of the Gaussian fits were guided by Tikhonov results. Figs. S3 and S4 show the background-corrected dipolar evolution data, the dipolar spectra, and the normalized integral representations of the distance distributions. For the distances between the nucleotide and helical domains in the receptor-bound empty complex, the width of the distribution may not be well determined due to the limited collection time of the dipolar evolution. Nevertheless, the fact that the distribution is indeed broad is revealed by the lack of well-defined oscillations in the dipolar evolution.
Modeling of the Complex Based on Available Information, Including DEER Distances.
For details about the modeling of the complex, see SI Methods.
Supplementary Material
Acknowledgments.
The authors gratefully acknowledge helpful discussions with C. Altenbach and thank C. Hubbell, C. J. López, V. V. Gurevich, and C. R. Sanders for carefully reading the manuscript. Research reported here was supported by National Institutes of Health Grants GM080403 (to J.M.), EY006062 (to H.E.H.), EY005216 (to W.L.H.) and the Jules Stein Professorship endowment (to W.L.H.). N.A. was supported by National Research Service Award MH086222.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1105810108/-/DCSupplemental.
References
- 1.Tesmer JJ. The quest to understand heterotrimeric G protein signaling. Nat Struct Mol Biol. 2010;17:650–652. doi: 10.1038/nsmb0610-650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Noel JP, et al. The 2.2 Å crystal structure of transducin-alpha complexed with GTP gamma S. Nature. 1993;366:654–663. doi: 10.1038/366654a0. [DOI] [PubMed] [Google Scholar]
- 3.Lambright DG, et al. Structural determinants for activation of the alpha-subunit of a heterotrimeric G protein. Nature. 1994;369:621–628. doi: 10.1038/369621a0. [DOI] [PubMed] [Google Scholar]
- 4.Lambright DG, et al. The 2.0 Å crystal structure of a heterotrimeric G protein. Nature. 1996;379:311–319. doi: 10.1038/379311a0. [DOI] [PubMed] [Google Scholar]
- 5.Coleman DE, et al. Crystal structures of the G protein Gi alpha 1 complexed with GDP and Mg2+: A crystallographic titration experiment. Biochemistry. 1998;37:14376–14385. doi: 10.1021/bi9810306. [DOI] [PubMed] [Google Scholar]
- 6.Coleman DE, et al. Structures of active conformations of Gi alpha 1 and the mechanism of GTP hydrolysis. Science. 1994;265:1405–1412. doi: 10.1126/science.8073283. [DOI] [PubMed] [Google Scholar]
- 7.Wall MA, et al. The structure of the G protein heterotrimer Gi alpha 1 beta 1 gamma 2. Cell. 1995;83:1047–1058. doi: 10.1016/0092-8674(95)90220-1. [DOI] [PubMed] [Google Scholar]
- 8.Higashijima T, et al. Effects of Mg2+ and the beta gamma-subunit complex on the interactions of guanine nucleotides with G proteins. J Biol Chem. 1987;262:762–766. [PubMed] [Google Scholar]
- 9.Oldham WM, et al. Mechanism of the receptor-catalyzed activation of heterotrimeric G proteins. Nat Struct Mol Biol. 2006;13:772–777. doi: 10.1038/nsmb1129. [DOI] [PubMed] [Google Scholar]
- 10.Oldham WM, et al. Mapping allosteric connections from the receptor to the nucleotide-binding pocket of heterotrimeric G proteins. Proc Natl Acad Sci USA. 2007;104:7927–7932. doi: 10.1073/pnas.0702623104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Van Eps N, et al. Structural and dynamical changes in an alpha-subunit of a heterotrimeric G protein along the activation pathway. Proc Natl Acad Sci USA. 2006;103:16194–16199. doi: 10.1073/pnas.0607972103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Linder ME, et al. Lipid modifications of G protein subunits. Myristoylation of Go alpha increases its affinity for beta gamma. J Biol Chem. 1991;266:4654–4659. [PubMed] [Google Scholar]
- 13.Resh MD, et al. Fatty acylation of proteins: New insights into membrane targeting of myristoylated and palmitoylated proteins. Biochim Biophys Acta. 1999;1451:1–16. doi: 10.1016/s0167-4889(99)00075-0. [DOI] [PubMed] [Google Scholar]
- 14.Medkova M, et al. Conformational changes in the amino-terminal helix of the G protein alpha(i1) following dissociation from Gbetagamma subunit and activation. Biochemistry. 2002;41:9962–9972. doi: 10.1021/bi0255726. [DOI] [PubMed] [Google Scholar]
- 15.Pannier M, et al. Dead-time free measurement of dipole-dipole interactions between electron spins. J Magn Reson. 2000;142:331–340. doi: 10.1006/jmre.1999.1944. [DOI] [PubMed] [Google Scholar]
- 16.Jeschke G. Distance measurements in the nanometer range by pulse EPR. Chemphyschem. 2002;3:927–32. doi: 10.1002/1439-7641(20021115)3:11<927::AID-CPHC927>3.0.CO;2-Q. [DOI] [PubMed] [Google Scholar]
- 17.Fleissner MR, et al. Structural origin of weakly ordered nitroxide motion in spin-labeled proteins. Protein Sci. 2009;18:893–908. doi: 10.1002/pro.96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Scheerer P, et al. Crystal structure of opsin in its G-protein-interacting conformation. Nature. 2008;455:497–502. doi: 10.1038/nature07330. [DOI] [PubMed] [Google Scholar]
- 19.Scheerer P, et al. Structural and kinetic modeling of an activating helix switch in the rhodopsin-transducin interface. Proc Natl Acad Ssi USA. 2009;106:10660–10665. doi: 10.1073/pnas.0900072106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hermann R, et al. Signal transfer from GPCRs to G proteins: Role of the G alpha N-terminal region in rhodopsin-transducin coupling. J Biol Chem. 2006;281:30234–30241. doi: 10.1074/jbc.M600797200. [DOI] [PubMed] [Google Scholar]
- 21.Kaufmann KW, et al. Practically useful: What the Rosetta protein modeling suite can do for you. Biochemistry. 2010;49:2987–2998. doi: 10.1021/bi902153g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hirst SJ, et al. RosettaEPR: An integrated tool for protein structure determination from sparse EPR data. J Struct Biol. 2011;173:506–514. doi: 10.1016/j.jsb.2010.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Preininger AM, et al. Helix dipole movement and conformational variability contribute to allosteric GDP release in Gαi subunits. Biochemistry. 2009;48:2630–2642. doi: 10.1021/bi801853a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hamm HE, et al. Site of G protein binding to rhodopsin mapped with synthetic peptides from the alpha subunit. Science. 1988;241:832–835. doi: 10.1126/science.3136547. [DOI] [PubMed] [Google Scholar]
- 25.Marin EP, et al. Disruption of the alpha5 helix of transducin impairs rhodopsin-catalyzed nucleotide exchange. Biochemistry. 2002;41:6988–6994. doi: 10.1021/bi025514k. [DOI] [PubMed] [Google Scholar]
- 26.Martin EL, et al. Potent peptide analogues of a G protein receptor-binding region obtained with a combinatorial library. J Biol Chem. 1996;271:361–366. doi: 10.1074/jbc.271.1.361. [DOI] [PubMed] [Google Scholar]
- 27.Dratz EA, et al. NMR structure of a receptor-bound G-protein peptide. Nature. 1993;363:276–281. doi: 10.1038/363276a0. [DOI] [PubMed] [Google Scholar]
- 28.Kisselev OG, et al. Light-activated rhodopsin induces structural binding motif in G protein alpha subunit. Proc Natl Acad Sci USA. 1998;95:4270–4275. doi: 10.1073/pnas.95.8.4270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Van Eps N, et al. Electron paramagnetic resonance studies of functionally active, nitroxide spin-labeled peptide analogs of the C-terminus of a G-protein alpha subunit. Biochemistry. 2010;49:6877–6886. doi: 10.1021/bi100846c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Natochin M1, et al. Probing the mechanism of rhodopsin-catalyzed transducin activation. J Neurochem. 2001;77:202–210. doi: 10.1046/j.1471-4159.2001.t01-1-00221.x. [DOI] [PubMed] [Google Scholar]
- 31.Preininger AM, et al. Receptor-mediated changes at the myristoylated amino terminus of Gαil proteins. Biochemistry. 2008;47:10281–10293. doi: 10.1021/bi800741r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hanson SM, et al. Structure and function of the visual arrestin oligomer. EMBO J. 2007;26:1726–1736. doi: 10.1038/sj.emboj.7601614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jeschke G, et al. DeerAnalysis2006: A comprehensive software package for analyzing pulsed ELDOR. Appl Magn Reson. 2006;30:473–498. [Google Scholar]
- 34.Banham JE, et al. Distance measurements in the borderline region of applicability of CW EPR and DEER: A model study on a homologous series of spin-labeled peptides. J Magn Reson. 2008;191:202–218. doi: 10.1016/j.jmr.2007.11.023. [DOI] [PubMed] [Google Scholar]
- 35.Chiang YW, et al. The determination of pair distance distributions by pulsed ESR using Tikhonov regularization. J Magn Reson. 2005;172:279–295. doi: 10.1016/j.jmr.2004.10.012. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.