Abstract
Telomerases constitute a group of specialized ribonucleoprotein enzymes that remediate chromosomal shrinkage resulting from the “end-replication” problem. Defects in telomere length regulation are associated with several diseases as well as with aging and cancer. Despite significant progress in understanding the roles of telomerase, the complete structure of the human telomerase enzyme bound to telomeric DNA remains elusive, with the detailed molecular mechanism of telomere elongation still unknown. By application of computational methods for distant homology detection, comparative modeling, and molecular docking, guided by available experimental data, we have generated a three-dimensional structural model of a partial telomerase elongation complex composed of three essential protein domains bound to a single-stranded telomeric DNA sequence in the form of a heteroduplex with the template region of the human RNA subunit, TER. This model provides a structural mechanism for the processivity of telomerase and offers new insights into elongation. We conclude that the RNA∶DNA heteroduplex is constrained by the telomerase TEN domain through repeated extension cycles and that the TEN domain controls the process by moving the template ahead one base at a time by translation and rotation of the double helix. The RNA region directly following the template can bind complementarily to the newly synthesized telomeric DNA, while the template itself is reused in the telomerase active site during the next reaction cycle. This first structural model of the human telomerase enzyme provides many details of the molecular mechanism of telomerase and immediately provides an important target for rational drug design.
Keywords: polymerase, protein motions, structure prediction
Telomerases are essential for maintaining chromosome length and integrity (1–3). They complement the cellular DNA-dependent DNA polymerase replication machinery that is not capable of fully replicating chromosomal ends, leading to telomeric DNA sequence erosion. Loss of telomerase activity is tolerated to some extent in yeast, worm, plant, and mouse (4), but after a few generations, telomeres typically become too short to perform their essential functions. Excessive telomere shortening can result in chromosome degradation, illegitimate recombination and end-to-end fusion, the compromise of cell cycle regulation, and, ultimately, cell death (5, 6). Most eukaryotic organisms utilize telomerases for the successive synthesis and maintenance of telomeric DNA repeats at chromosome ends, replenishing the capability for further cell proliferation in stem cell lineages (7). Interestingly, the activity of telomerases in fully differentiated somatic cells is strongly down-regulated over time, in concert with aging, and a direct correlation between telomere shortening and aging has been demonstrated (8). In human cells, increased telomerase activity can increase renewal capacity in certain tissues, which has been interpreted as delaying the aging process. However, pathogenic overexpression of telomerases is also a hallmark of many human cancers (9), and several studies have shown that telomerase malfunction can lead to diseases in humans (4, 10), including dyskeratosis congenita (11). Thus, telomerase appears to be a key player critical in maintaining the balance between normal cellular differentiation (and aging) and the aberrant proliferation manifested in carcinogenic transformation (and immortality).
Influences of changes in telomerase activity have been observed in many biological processes not directly related to telomere maintenance (12, 13). For example, Gonzalez-Suarez et al. found that induced somatic expression of telomerase led to increased cellular proliferation and growth and, consequently, enhanced wound healing in mice (14). Studies on promyelocytic leukemia cells revealed that telomerase expression may also inhibit apoptosis (15). When overexpressed, telomerase is directed to the mitochondria and appears to help protect cells from H2O2-mediated damage (16). Recently, Blackburn and colleagues have shown that changes in telomerase activity are associated with human stress-related syndromes, including major depression (17, 18).
Telomerases function as specialized reverse transcriptases (19), RNA-dependent DNA polymerases capable of synthesizing multiple copies of the telomeric DNA repeat sequence by using an intrinsic RNA template to direct telomeric DNA synthesis (1, 13). Telomere repeats are often shown as 5′-(TTAGGG)n-3′, which is the DNA repeat unit. In this work we choose to focus on the RNA template, displaying its coding region as 5′-UAACCC-3′. For clarity, the alignment region 3′-AUC-5′ of hTR pairs to the DNA primer sequence 5′-TAG-3′ in order to synthesize the next DNA addition. The newly synthesized telomeric DNA repeats are added to the overhanging single-stranded 3′ end of the DNA at the chromosome termini. In other respects, the telomerase reverse transcriptase mechanism appears to be similar to that of well-studied retroviral reverse transcriptases (1, 12, 20).
The human telomerase enzyme contains a template-encoding RNA molecule, TER (TElomerase RNA or hTR) and a primary protein component, TERT (TElomerase Reverse Transcriptase) with several functional domains: TEN (Telomerase Essential N-terminal domain), TRBD (Telomerase RNA Binding Domain), RT (Reverse Transcriptase domain), and the C-Terminal Extension (CTE) (1, 12, 13). The RT domain can be further divided into two distinguishable subdomains: the “fingers” involved in nucleotide binding and processivity and the “palm” providing the polymerase catalytic residues and DNA primer grip. The C-terminal extension is responsible for interaction with DNA and has been proposed to correspond to the RT “thumb” domain (20). Because this CTE appears to be structurally equivalent to the C-terminal α-helical thumb of retroviral RT (21), it will be referred to as the RT thumb domain below. In describing the human TERT structure here, we thus refer to RT as fingers, palm, and thumb for clarity and simplicity. Both RT and TER are essential for telomerase activity, together forming the active site that catalyzes deoxynucleotide addition. The TRBD domain links the RT and TER components, and the TEN domain is proposed to facilitate the repetitive repeat addition mode of telomerases, which is one of the distinguishing features of telomerases, relative to classical reverse transcriptases (1, 12). Efficient repeat addition processivity is governed by multiple mechanisms involving both TER (22) and protein subdomains of TERT (23), as well as additional telomerase-associated processivity factors, notably TPP1-POT1 in humans, which enhances telomerase processivity by slowing primer dislocation and facilitating translocation (24). The TEN domain contains an “anchor” site (25) that is thought to help stabilize the bound single-stranded telomeric DNA substrate within the complex, while the intrinsic RNA template is realigned for the next, iterative reverse transcription cycle. Therefore, the complex is capable of processively synthesizing a long array of single-stranded telomeric DNA repeats by repeatedly copying the 6-nt long RNA template region within the TER component (1, 12). In addition to processivity factors, several species-specific accessory proteins are critical for telomerase assembly, subcellular localization, and function in vivo (12, 26). In human cell lines, for example, the catalytically active form of telomerase includes dyskerin (27), which together with NHP2 and NOP10, is required for stability and accumulation of the RNA component of human telomerase in vivo (28).
Recently, X-ray structures of the full length T. castaneum telomerase (containing RT and TRBD domains) alone (PDB ID codes 3DU5 and 3DU6) (21) and in complex with an RNA∶DNA hairpin (PDB ID code 3KYL) (29) have been published. In addition, the crystal structures for separate TRBD (PDB ID code 2R4G) (30) and TEN (PDB ID code 2B2A) (31) domains from T. thermophila are now available. Despite much experimental effort, the detailed molecular mechanism of human telomerase enzymatic activity and the structural details of the interactions between the TEN domain and the other components of the complex (RT, TRBD, TER, and the telomeric DNA) are still not fully known. The absence of a high-resolution experimentally determined structure for the assembled telomerase core catalytic complex is a serious impediment to designing experiments that could further elucidate the molecular mechanism of telomerase action. Moreover, species-specific features of telomerase structure and function make obtaining a complete structure of the human telomerase enzyme particularly important. Thus we employ here a theoretical modeling approach to generate the entire 3D structure of the human TERT, TEN, and TRBD bound to a DNA substrate and its RNA template.
Automatic homology modeling using available web-based servers was not feasible because the amino acid sequence identities between the human telomerase domains and the corresponding RT and TRBD structures in the Protein Data Bank (PDB) are only 24% and 22%, respectively. This level of sequence similarity is near the limit for the reliable use of standard homology detection methods based on PSI-BLAST or RPS-BLAST. Furthermore, in our hands, such standard sequence comparison methods were unable to detect significant sequence similarity between the N-terminal domain of the human telomerase protein and the TEN domain of Tetrahymena, or any other known protein structure. To obviate this problem, we used an advanced meta-profile comparison method, Meta-BASIC (32), to map the human telomerase protein sequence onto sequences of the determined structures from Tribolium and Tetrahymena. The mappings obtained were confirmed by using a variety of fold recognition methods. Together with detailed manual inspection, these approaches allow us to generate highly accurate sequence-to-structure alignments between the human telomerase sequence and relevant structural templates. We then built three-dimensional models separately for TEN and the other components of the human telomerase complex including a hybrid RNA∶DNA double helix formed between the RNA template and the single-stranded telomeric DNA substrate and assembled them by using protein–protein docking, guided by relevant experimental data. Based on the resulting structural model of the telomerase enzyme, we propose a mechanism for human telomerase action in which interactions between TEN and the RT∶TRBD subcomplex play a critical role, and where the elongating telomeric DNA is stabilized by the TEN domain. We hypothesize that the helical structure of the heteroduplex formed between the RNA template and the telomeric DNA substrate is actively maintained during the individual repeated telomerase reactions producing a single copy of the template. Following this, the RNA template must be repositioned relative to the active site. We propose that its translocation proceeds along the extending helix due to constraints imposed by the TEN “anchoring” domain.
Results and Discussion
The present work was motivated by the lack of a complete structural model that could explain the detailed functional molecular mechanism of human telomerase. By using distant homology detection, comparative structural modeling, and computational docking, we developed a model of the human telomerase complex (Fig. 1). Then by using elastic network models, we investigated the intrinsic motions of the modeled structure. Our goals were twofold: (i) to understand how the individual telomerase protein domains and the intrinsic TER component interact in the assembled human telomerase RNP enzyme, and (ii) to generate a model illustrating how the telomerase RNP enzyme binds to and extends single-stranded telomeric DNA by reverse transcription of telomeric repeat sequences.
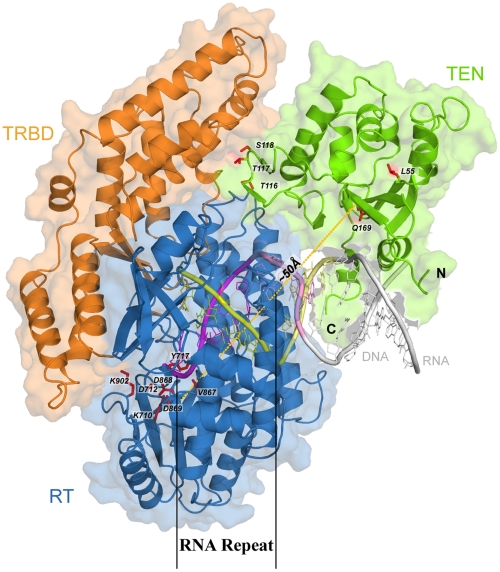
Fig. 1.
Partial model of human telomerase ribonucleoprotein complex. The model includes the TERT protein component composed of the catalytic reverse transcriptase domain (RT), the RNA binding domain (TRBD), and the N-terminal “anchor” domain (TEN), bound to a heteroduplex formed by oligomers corresponding to the template-encoding RNA (TER) and the single-stranded human telomeric DNA substrate. The template region (5′-UAACCC-3′) of the RNA is shown in yellow (labeled as RNA repeat) and the complementary region of DNA in magenta, while the partial template repeat (5′-UAAC-3′) in the RNA is shown in lighter yellow, and its complement in the DNA in pink. The N- and C-terminal α-helices of TEN interact with the major groove of the heteroduplex. Residues with experimentally determined influence on telomerase function or assembly are labeled and shown in red. The orange dotted line shows the approximately 50-Å distance between TEN (Q169) and the RT active site (D869).
Several studies indicate that the TEN domain functions as an “anchor” to bind and stabilize the telomeric DNA substrate, contributing to the processivity of the repetitive reverse transcriptase activity (13, 23, 25). However, no structure-based explanation for how TEN contributes to the overall function of the telomerase enzyme or its processivity has been proposed previously. Here we suggest that TEN plays a critical role in controlling the processive step in which telomerase advances by one base on the strand being copied. Similarly, many experiments have determined the effects of specific amino acid substitutions or deletions on telomerase enzymatic activity and have provided us with useful information about key residues (see Table S1). It is important to reconcile these with a structural model and a structural mechanism. To date, however, it has not been possible to understand the effects of these changes due to the lack of a complete structural model for the telomerase RNP. The present structure will facilitate such investigations.
To derive a 3D model for human telomerase enzyme we used the available structures for telomerase components from T. castaneum and T. thermophila, including the recently released structure for RT and TRBD domains solved with RNA template and telomeric DNA (29). The modeled RNA∶DNA heteroduplex was extended as observed in the closely related HIV-1 reverse transcriptase structure (33) to construct interactions with the TEN domain (see Methods). The human TEN domain, which was modeled separately, seems to be designed to accommodate a nucleic acid double helix, as does the corresponding TEN structure from T. thermophila (31). It has a well-defined cleft to interact with the phosphate backbone and two helical segments that fit well into the major groove of the double helix. We used molecular docking to assemble the entire complex by fitting the TEN model into a position that ensured appropriate interaction with both the telomeric DNA and the other telomerase protein domains. We also considered available data suggesting plausible RNA binding sites, conservation of surface residues, and certain amino acid mutations that have been shown to impact the enzymatic activity (see references in Table S1). The final model shows that the central ring-shaped part of the human telomerase structure, formed by TRBD and RT domains, accommodates the RNA∶DNA heteroduplex and provides catalytic residues (Fig. S1). The ring is coupled with the TEN domain, which interacts with TRBD, the RT thumb, and the RNA∶DNA major groove and helps stabilize the substrate within the active site during repeated reaction cycles.
Reverse Transcriptase and Nucleic Acid Binding Domains.
The RT and TRBD domains form the central, ring-shaped core of the telomerase RNP complex (Fig. S1). They provide the catalytic activity of the enzyme by bringing together the necessary active site residues. Previous analyses of sequence conservation within these domains highlighted several motifs, shared by the majority of reverse transcriptases, including telomerases (1, 34). As shown in Fig. S2 starting from the N terminus, these motifs are: CP, T, 1, 2, 3, A, B′, C, D, and E. Motifs CP and T belong to the α-helical TRBD domain (Fig. S1), which mediates interactions between the template-carrying RNA component and the telomerase. The CP and T motifs have been shown to be directly involved in RNA binding (12, 35) and are required for the proper assembly of the complete telomerase ribonucleoprotein complex (13, 36). Motifs A and C in the RT palm contain the conserved active site signature sequence, KXD(X)nDD, in which three invariant aspartic acid residues (D712, D868, and D869) coordinate two Mg2+ ions, while the lysine (K710) provides the base for the deoxynucleotide condensation reaction (13, 21, 34) (see Fig. S3A).
The pocket surrounding the catalytic amino acids is lined with several residues that help position deoxynucleotide substrates (A, T, and G) with respect to the complementary RNA template-encoded ribonucleotides in the active site, as in the T. castaneum structure (21, 29). Three conserved uncharged residues—Y717, Q833, and V867 (from motifs A, B′, and C, respectively)—form a hydrophobic pocket adjacent to the catalytic aspartates and take part in nucleotide binding (29). Residue V867 has been shown to alter human telomerase substrate specificity (37), and residue Q833 corresponds to Q151 in HIV-1 reverse transcriptase, where mutations cause hypersensitivity to substrate analogs (38). This pocket appears to hold the incoming deoxynucleotide in close proximity to the active site, for coordination with one of the Mg2+ ions. In addition, Motif 2 residues K626 and R631 (from RT fingers), together with K902 from motif D (RT palm), may interact with both the sugar ring and phosphate groups, and provide stacking interactions with bases of the incoming deoxynucleotide. These interactions likely stabilize the telomeric DNA substrate during catalysis (21). The relatively conserved residues C931 and G932 from the RT palm define a “primer grip” (motif E) (29), which is essential for proper maintenance of telomeric DNA within telomerase active site (Fig. S3B). Additionally, R972 and K973 from the RT thumb, both located on an α-helix that packs into the minor groove of the RNA∶DNA heteroduplex, zinteract with the DNA backbone (Fig. S3C). These residues, present also in telomerases that lack the TEN domain, may contribute significantly to repetitive addition processivity.
The interactions between telomerase and telomeric DNA are mediated by a variety of critical residues grouped into motifs characteristic for this family of polymerases (1, 12, 13). The spatial arrangement of these motifs resembles the shape of a double-stranded nucleic acid helix (21). The recently described “motif 3” of the reverse transcriptase domain provides several residues that may interact directly with telomeric DNA (39). Notably, mutations of motif 3 residues (V658A and K659A from RT fingers, and R669A from RT palm) cause telomerase hyperactivity (39), apparently resulting from weaker interaction with the double-stranded heteroduplex, facilitating telomeric DNA release after reaction. In our model, the positively charged K659 and R669 side chains are directed toward both DNA and RNA backbones, compatible with their essential contribution to nucleic acid binding (Fig. S3A). Increased repeat addition rate reduces processivity, however, possibly because the telomeric DNA cannot be stabilized sufficiently while the template-carrying RNA is realigned and prepared for the next reaction cycle (39). In support of this, Xie et al. (39) were able to obtain hyperactive and hyperprocessive human telomerase mutants by combining the V658A mutation with the deletion of residues 643–649 in the RT fingers and the hTR-U57C substitution in the RNA. The loop containing amino acid residues 643–649 precedes the motif 3 α-helix and is an intriguing structural feature: It may freely interact with both the telomeric DNA and RNA and likely stabilizes the position of the DNA substrate in the telomerase complex (Fig. S3D). Deletion of the 643–649 loop weakens this interaction and likely allows for more rapid dissociation of the heteroduplex, making the template binding site available for the next substrate deoxynucleotide and the next round of synthesis. Additionally, the hTR-U57C substitution results in extension of the RNA∶DNA heteroduplex by an additional base, which could potentially form a classical Watson–Crick pair, possibly further stabilizing the telomerase–telomeric DNA association.
In contrast to T. castaneum telomerase (PDB ID code 3KYL), the human protein contains two additional α-helices (residues 415–456) within the TRBD domain (similar to T. thermophila; PDB ID code 2R4G, residues 333–371), which, according to our model, together with the final α-helix in TRBD, form a three-helix bundle that packs tightly against the RT fingers (Fig. S4). This structural feature additionally stabilizes the interaction of RT and TRBD.
Essential N-Terminal Domain.
The TEN domain, composed of a central β-sheet flanked by α-helices on both sides, is the most divergent domain within the telomerases (1, 12, 13). Nevertheless, it appears to be essential for proper telomere maintenance, because it “anchors” telomeric DNA (25). A recent study used a combination of comparative modeling and machine learning to identify several residues in TEN that are likely to play a role in nucleic acid binding (40). The sequence diversity among TEN domains of different species may be related to differences in telomeric DNA repeat sequences. TEN recognizes telomeric DNA in a sequence-specific manner, and several experiments have revealed differences in DNA binding affinity to different telomeric repeat sequences (41, 42). The TEN domain is separated from TRBD by a linker region, predicted to be largely unstructured, ranging in length from 20 to more than 500 amino acids, depending on the species (1). The TEN domain is believed to contribute to the processivity of the enzyme, because several studies have identified mutations or deletions of TEN residues that lead to a reduced ability of the telomerase to synthesize more than one telomeric repeat (23, 43). The crystal structure of the TEN domain from T. thermophila (PDB ID code 2B2A) (31) revealed certain features that adapt it for interaction with telomeric DNA. In particular, a deep cleft in the TEN domain surface is closely complementary to the shape of a double helix. Mutation of Q169, which is located in the central part of the cleft, compromises human telomerase processivity by hindering proper protein-DNA interaction (42, 44). In our modeled human TEN, Q169 forms a hydrogen bond with the backbone carbonyl group of P174 and the backbone amine group of L175, stabilizing the intervening loop, which establishes hydrogen bonds with another element of the TEN structure (Fig. S5). These interactions thus bridge adjacent structural elements and stabilize the overall shape of this region. The cleft is flanked by α-helical extensions on both sides and engages the RNA∶DNA double helix, fitting into its major groove (Fig. 1). Jurczyluk et. al. (45) recently performed mutational analyses on TEN. Two mutations of particular interest involve residues 8–13 and 170–175 of TEN. The former exhibits wild type processivity but a decreased Km, and the latter has significantly decreased processivity and an increased Km. In our model, residues 8–13 make close contact with the heteroduplex, supporting this observation of decreased affinity. Residues 170–175 do have heteroduplex interactions in the model but are also partly buried in the TEN domain.
Interaction with the RNA Template.
According to our model the template-carrying RNA component (TER) interacts with the TRBD, RT, and TEN domains of telomerase. Human TER is a structurally complex RNA molecule of 451 nucleotides, containing several conserved sequence and structural motifs (46). A characteristic pseudoknot domain, located in close proximity to the template repeat sequence and to an RNA loop domain designated CR4-CR5, is essential for telomerase catalytic activity (47). A 3'-terminal “H/ACA box” in TER contributes to the assembly and maturation of the ribonucleoprotein complex (48). The RNA pseudoknot region was shown to interact with a C-terminal region (residues 150–159) of TEN, providing further insight into the localization of the template region within the human telomerase complex (49). Our modeled structure of TRBD exposes a wide cleft opening toward the C terminus of TEN, which might bind the pseudoknot domain. Furthermore, the surface of the cleft presents several lysines (K492, K493, K511) that could interact with RNA. The TER CR4-CR5 domain has been shown to bind to the CP and T motifs of the TRBD domain (1), which are located at a considerable distance (approximately 23 Å) from the putative pseudoknot-binding site in our model. Recent work by Egan and Collins provides insight into hTERT-hTR interactions and will be useful for future studies that attempt to model the full length hTR (50).
RT∶TRBD-TEN Interaction.
The shape of the TEN domain restricts its possible orientation with respect to the other domains and the RNA∶DNA heteroduplex. Mutual positioning of the telomeric DNA substrate and RT, to ensure proper interactions within the reverse transcriptase active site, determines the distance between the surface of TRBD and the major groove of the bound double helix. Therefore, the possible TEN orientations are dramatically limited. Together, these constraints aided in the assembly of our model of the human telomerase RNP complex. The surfaces of RT, TRBD, and TEN expose poorly conserved residues, hindering the modeling of interactions between TEN and the other two domains. However, detailed analysis of surface residues in the model reveals increased conservation of uncharged amino acids at the domain interfaces (e.g., G100, F101 in TEN and G967, V1025 in the RT thumb; T117, S118 in TEN and S504, L505, A542 in TRBD; see Fig. S6). Despite poor surface conservation in TEN, the proposed interface represents an optimal structural fit between TEN and RT∶TRBD and the RNA∶DNA heteroduplex. Additional support for our proposed assembly is provided by Sealey et al. (42), who reported that T116A, T117A, and S118A mutations in TEN compromise repetitive addition processivity but do not alter DNA binding affinity and thus probably affect interaction with the remaining protein domains of the telomerase. In the model, these three residues are located at the binding surface between TEN and the TRBD and RT thumb domains. This interface is far from the nucleotide binding region, with the closest nucleotide atom approximately 16 Å away. It is, however, involved in the slower motions, presumed to be functionally relevant (see below), and this may explain the role of these residues in the processivity.
Mechanism of Telomerase Action.
To investigate the processive mechanism of telomerase, we used elastic network models to generate the mechanistic step shown in close-up in Fig. 2. We utilized our Anisotropic Network Model (ANM) (51), with one node for each Cα, P and O4′ atom, and with identical springs placed between pairs of these atoms within 13 Å. To generate the processive conformations shown, we used the global mode of motion to move the original structure. This reveals in detail how the telomerase structure effects the motion of the template. Views of the whole structure are shown in further detail in Fig. S7 and in Movies S1 and S2.
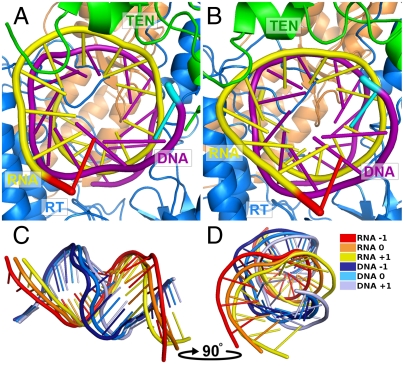
Fig. 2.
Structural model for the processive motion of human telomerase. The procession of the DNA∶RNA heteroduplex is a critical aspect of telomerase function. Rotation and translation of the heteroduplex is evident in the global mode of the elastic network. The effects of following the global mode in the (A) negative (-1) and (B) positive (+1) directions are shown. Termini closest to the viewer are highlighted: the 3′ end of RNA in red and the 5′ end of DNA in cyan. See Fig. S7 for corresponding views of the entire structure and Movies S1 and S2 for two dynamic views of this motion. C offers a side view and D a face view of the heteroduplex for three states of the negative global mode (-1), the original state (0), and the positive mode deformation (+1).
Our 3D model assumes that the intrinsic template-carrying RNA molecule forms a heteroduplex with the single-stranded telomeric DNA, while the architecture of both the RT active site and the TEN anchor domain are adapted for double-stranded nucleic acid binding. The template-encoding region of TER contains one complete 6-nt repeat complementary to the human telomeric (TTAGGG)n repeat sequence, which can initiate RNA∶DNA heteroduplex formation. Multiple sequence alignment of telomerase RNAs from several phyla (extracted from the Telomerase Database, http://telomerase.asu.edu) (52) reveals that the telomere repeat template sequence is partially repeated, extending the potential length of the helical heteroduplex region, as was previously proposed (53). In the human enzyme, such an extended RNA∶DNA heteroduplex would contain at least 10 base pairs, exactly filling the space between the RT active site and the experimentally supported nucleic acid binding region of TEN (Fig. 1). Thus, our model provides a strong structural basis for the mechanism shown in Fig. 2, in which, after completion of a single telomere repeat synthesis cycle, the template RNA is moved ahead at the polymerase site to the next base. The TER template region must then shift relative to the substrate DNA, while maintaining the RNA∶DNA heteroduplex (Fig. S8). The second, partial repeat (5′-UAAC-3′) adjacent to the TER template region complements the newly synthesized telomeric DNA repeat, while the TER template region itself (5′-UAACCC-3′) forms a 5′ overhang ready for the next reaction cycle. Our model strongly supports an essential role of TEN in stabilizing the assembling heteroduplex in an orientation that promotes proper interaction with the RT active site. Further, our dynamics simulations suggest the important role of the structure in controlling the shift along the template to the next base to be copied. The current model does not provide insight into the mechanism by which the nucleic acid translocates after the complete synthesis of each complete template sequence; however, there is the possibility that the protein can extend together with the RNA through the repeated synthesis steps for one template cycle before recoiling to activate and reposition the template for the next cycle.
Notably, the template-encoding region of TER is followed by a sequence rich in uracil (U) residues that are capable of forming wobble base pairs with guanine (G). Therefore, the proposed RNA∶DNA heteroduplex formed during reverse transcription may actually be longer, to help stabilize the helical structure during the RNA template realignment step between reaction cycles. Such a possible extended helix could contribute significantly to the extension mechanism.
Several telomerases (e.g., those from C. elegans and T. castaneum) lack a distinguishable TEN domain, which is essential for activity and processivity of the human and Tetrahymena enzymes (1, 12, 13). Despite this, the T. castaneum enzyme appears to be active in vitro (29) and genetic evidence suggests that the C. elegans telomerase is capable of synthesizing multiple telomeric repeats in vivo (54). Meier and colleagues found that TRBD domain in C. elegans is preceded by a domain that might be distantly homologous to TEN (54); however, our methods failed to detect significant similarity of this region to any known protein domains. The RT thumb and IFD motif (RT fingers) have been shown to play roles in repetitive addition processivity (20, 25) and, in the absence of TEN, could provide substrate stabilization during subsequent reaction cycles. Species-specific accessory factors that influence processivity could also compensate for the lack of a TEN domain (12, 13). Interestingly, we found that telomerases lacking the TEN domain (e.g., PDB ID code 3KYL) also lack the α-helical insertion within the TRBD (Fig. S4), which would allow for a more elastic RT:TRBD interface (discussed above). Telomerases possessing TEN domains possibly do not require such elasticity because TEN aids the nucleic acid binding and regulates the processivity.
Conclusions
The availability of a structural model of the assembled human telomerase complex presented here provides information necessary for investigating its mechanism further, as well as for locating its interactions within the complex cellular signaling networks in which it is known to participate (8, 17, 18). A complete structural model of telomerase may also accelerate the development of new anticancer therapies that aim to abolish telomerase activity in proliferating tumor cells, or to augment enzymatic activity in cases of telomerase insufficiency diseases.
Methods
We present a brief overview of our methods here with further details given in SI Methods.
We have utilized the human telomerase protein sequence (GenBank accession no. NM_198253.2) and PSI-BLAST (55) to study the sequence conservation within the telomerase protein family. Multiple sequence alignments of collected sequences were prepared with PCMA (56), while PSI-PRED (57) was used for secondary structure prediction. Templates for comparative modeling of human telomerase domains were identified from the full-length sequence and individual domains using the Gene Relational Database (GRDB) system, which stores pre-calculated Meta-BASIC mappings (32) between Pfam families, conserved domains, and PDB structures. The results were validated by 3D-Jury (58) and manual inspection followed by 3D assessment (59).
Three-dimensional models of human telomerase protein domains were generated with Modeller (60) based on manually curated, high confidence sequence-to-structure alignments. These models were built separately for (i) the TEN domain, using PDB ID code 2B2A (31); (ii) the RT∶TRBD subcomplex, using PDB ID code 3KYL (29) and the superimposed TRBD domain from PDB ID code 2R4G (30) as templates. The resulting 3D models were then assembled manually after careful consideration of the CABS (61) results for protein domain docking, published experimental data (Table S1), and conservation of surface residues from ConSurf (62) (Fig. S6). Assembly of the modeled domains proceeded by first rigidly docking using an exhaustive global search in a six-dimensional space of “ligand” rotations and translations against the frozen structure of the “receptor” using FTDOCK (63). The resulting 10,000 FTDOCK top-scoring structures were rescored with the CABS force field and grouped using hierarchical clustering. From each cluster, a representative with the lowest energy was selected, leaving 30 models.
Positions of the intrinsic RNA template and single-stranded telomeric DNA substrate in the human telomerase model were copied from the T. castaneum telomerase structure (PDB ID code 3KYL) after superposition of their RT and TRBD domains. The 3D partial model comprising all three protein domains and the RNA∶DNA heteroduplex was energy-minimized with Tripos SYBYL using an AMBER force field (64), followed by a short molecular dynamics run to relieve steric clashes and improve internal packing.
The final model was used to investigate the motions of the structure using coarse-grained elastic network models in conjunction with normal mode analysis. This approach has been widely used to investigate important functional motions of biomolecular structures (65). Notably the computed motions are quite insensitive to the details of the structure, which means the computed motions reported here are robust and unlikely to be changed by any minor errors in the model.
Supplementary Material
Acknowledgments.
We thank the reviewers for their insightful suggestions and Peter Zaback for critical reading of the manuscript. This work was supported by EMBO Installation, National Science Foundation Grant MCB-1021785, National Institutes of Health grants R01GM072014 and R01GM081680, Foundation for Polish Science (Focus, Team), Ministry of Science and Higher Education (N N301 159435) and European Social Fund (UDA-POKL.04.01.01-00-072/09-00), and Center for Integrated Animal Genomics (Iowa State University) grants.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1015399108/-/DCSupplemental.
References
- 1.Autexier C, Lue NF. The structure and function of telomerase reverse transcriptase. Annu Rev Biochem. 2006;75:493–517. doi: 10.1146/annurev.biochem.75.103004.142412. [DOI] [PubMed] [Google Scholar]
- 2.Blackburn EH, Collins K. Telomerase: An RNP enzyme synthesizes DNA. Cold Spring Harb Perspect Biol. 2010 doi: 10.1101/cshperspect.a003558. 10.1101/csshperspect.a003558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.McEachern MJ, Krauskopf A, Blackburn EH. Telomeres and their control. Annu Rev Genet. 2000;34:331–358. doi: 10.1146/annurev.genet.34.1.331. [DOI] [PubMed] [Google Scholar]
- 4.Lansdorp PM. Telomeres and disease. EMBO J. 2009;28:2532–2540. doi: 10.1038/emboj.2009.172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Armanios M. Syndromes of telomere shortening. Annu Rev Genomics Hum Genet. 2009;10:45–61. doi: 10.1146/annurev-genom-082908-150046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Murnane JP. Telomeres and chromosome instability. DNA Repair (Amst) 2006;5:1082–1092. doi: 10.1016/j.dnarep.2006.05.030. [DOI] [PubMed] [Google Scholar]
- 7.Flores I, Blasco MA. The role of telomeres and telomerase in stem cell aging. FEBS Lett. 2010;584:3826–3830. doi: 10.1016/j.febslet.2010.07.042. [DOI] [PubMed] [Google Scholar]
- 8.Aubert G, Lansdorp PM. Telomeres and aging. Physiol Rev. 2008;88:557–579. doi: 10.1152/physrev.00026.2007. [DOI] [PubMed] [Google Scholar]
- 9.Artandi SE, DePinho RA. Telomeres and telomerase in cancer. Carcinogenesis. 2010;31:9–18. doi: 10.1093/carcin/bgp268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Calado RT, Young NS. Telomere diseases. N Engl J Med. 2009;361:2353–2365. doi: 10.1056/NEJMra0903373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mitchell JR, Wood E, Collins K. A telomerase component is defective in the human disease dyskeratosis congenita. Nature. 1999;402:551–555. doi: 10.1038/990141. [DOI] [PubMed] [Google Scholar]
- 12.Wyatt HD, West SC, Beattie TL. InTERTpreting telomerase structure and function. Nucleic Acids Res. 2010;38:5609–5622. doi: 10.1093/nar/gkq370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sekaran VG, Soares J, Jarstfer MB. Structures of telomerase subunits provide functional insights. Biochim Biophys Acta. 2010;1804:1190–1201. doi: 10.1016/j.bbapap.2009.07.019. [DOI] [PubMed] [Google Scholar]
- 14.Gonzalez-Suarez E, et al. Increased epidermal tumors and increased skin wound healing in transgenic mice overexpressing the catalytic subunit of telomerase, mTERT, in basal keratinocytes. EMBO J. 2001;20:2619–2630. doi: 10.1093/emboj/20.11.2619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dudognon C, et al. Death receptor signaling regulatory function for telomerase: hTERT abolishes TRAIL-induced apoptosis, independently of telomere maintenance. Oncogene. 2004;23:7469–7474. doi: 10.1038/sj.onc.1208029. [DOI] [PubMed] [Google Scholar]
- 16.Santos JH, Meyer JN, Van Houten B. Mitochondrial localization of telomerase as a determinant for hydrogen peroxide-induced mitochondrial DNA damage and apoptosis. Hum Mol Genet. 2006;15:1757–1768. doi: 10.1093/hmg/ddl098. [DOI] [PubMed] [Google Scholar]
- 17.Epel ES, et al. Dynamics of telomerase activity in response to acute psychological stress. Brain Behav Immun. 2010;24:531–539. doi: 10.1016/j.bbi.2009.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wolkowitz OM, et al. Resting leukocyte telomerase activity is elevated in major depression and predicts treatment response. Mol Psychiatry. 2011 doi: 10.1038/mp.2010.133. 10.1038/mp.2010.133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Greider CW, et al. Identification of a specific telomere terminal transferase activity in Tetrahymena extracts. Cell. 1985;43:405–413. doi: 10.1016/0092-8674(85)90170-9. [DOI] [PubMed] [Google Scholar]
- 20.Hossain S, Singh S, Lue NF. Functional analysis of the C-terminal extension of telomerase reverse transcriptase. A putative “thumb” domain. J Biol Chem. 2002;277:36174–36180. doi: 10.1074/jbc.M201976200. [DOI] [PubMed] [Google Scholar]
- 21.Gillis AJ, Schuller AP, Skordalakes E. Structure of the Tribolium castaneum telomerase catalytic subunit TERT. Nature. 2008;455:633–637. doi: 10.1038/nature07283. [DOI] [PubMed] [Google Scholar]
- 22.Chen JL, Greider CW. Determinants in mammalian telomerase RNA that mediate enzyme processivity and cross-species incompatibility. EMBO J. 2003;22:304–314. doi: 10.1093/emboj/cdg024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lue NF. Adding to the ends: What makes telomerase processive and how important is it? Bioessays. 2004;26:955–962. doi: 10.1002/bies.20093. [DOI] [PubMed] [Google Scholar]
- 24.Latrick CM, Cech TR. POT1-TPP1 enhances telomerase processivity by slowing primer dissociation and aiding translocation. EMBO J. 2010;29:924–933. doi: 10.1038/emboj.2009.409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lue NF. A physical and functional constituent of telomerase anchor site. J Biol Chem. 2005;280:26586–26591. doi: 10.1074/jbc.M503028200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lingner J, Price C. Conservation of telomere protein complexes: Shuffling through evolution. Crit Rev Biochem Mol Biol. 2009;44:434–446. doi: 10.3109/10409230903307329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cohen SB, et al. Protein composition of catalytically active human telomerase from immortal cells. Science. 2007;315:1850–1853. doi: 10.1126/science.1138596. [DOI] [PubMed] [Google Scholar]
- 28.Fu D, Collins K. Purification of human telomerase complexes identifies factors involved in telomerase biogenesis and telomere length regulation. Mol Cell. 2007;28:773–785. doi: 10.1016/j.molcel.2007.09.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mitchell M, Gillis A, Futahashi M, Fujiwara H, Skordalakes E. Structural basis for telomerase catalytic subunit TERT binding to RNA template and telomeric DNA. Nat Struct Mol Biol. 2010;17:513–518. doi: 10.1038/nsmb.1777. [DOI] [PubMed] [Google Scholar]
- 30.Rouda S, Skordalakes E. Structure of the RNA-binding domain of telomerase: Implications for RNA recognition and binding. Structure. 2007;15:1403–1412. doi: 10.1016/j.str.2007.09.007. [DOI] [PubMed] [Google Scholar]
- 31.Jacobs SA, Podell ER, Cech TR. Crystal structure of the essential N-terminal domain of telomerase reverse transcriptase. Nat Struct Mol Biol. 2006;13:218–225. doi: 10.1038/nsmb1054. [DOI] [PubMed] [Google Scholar]
- 32.Ginalski K, von Grotthuss M, Grishin NV, Rychlewski L. Detecting distant homology with Meta-BASIC. Nucleic Acids Res. 2004;32:W576–581. doi: 10.1093/nar/gkh370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sarafianos SG, et al. Crystal structure of HIV-1 reverse transcriptase in complex with a polypurine tract RNA∶DNA. EMBO J. 2001;20:1449–1461. doi: 10.1093/emboj/20.6.1449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lingner J, et al. Reverse transcripase motifs in the catalytic subunit of telomerase. Science. 1997;276:561–567. doi: 10.1126/science.276.5312.561. [DOI] [PubMed] [Google Scholar]
- 35.Weinrich SL, et al. Reconstitution of human telomerase with the template RNA component hTR and the catalytic protein subunit hTRT. Nat Genet. 1997;17:498–502. doi: 10.1038/ng1297-498. [DOI] [PubMed] [Google Scholar]
- 36.Bosoy D, Peng Y, Mian IS, Lue NF. Conserved N-terminal motifs of telomerase reverse transcriptase required for ribonucleoprotein assembly in vivo. J Biol Chem. 2003;278:3882–3890. doi: 10.1074/jbc.M210645200. [DOI] [PubMed] [Google Scholar]
- 37.Drosopoulos WC, Prasad VR. The active site residue Valine 867 in human telomerase reverse transcriptase influences nucleotide incorporation and fidelity. Nucleic Acids Res. 2007;35:1155–1168. doi: 10.1093/nar/gkm002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Smith RA, Anderson DJ, Preston BD. Hypersusceptibility to substrate analogs conferred by mutations in human immunodeficiency virus type 1 reverse transcriptase. J Virol. 2006;80:7169–7178. doi: 10.1128/JVI.00322-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Xie M, Podlevsky JD, Qi X, Bley CJ, Chen JJ. A novel motif in telomerase reverse transcriptase regulates telomere repeat addition rate and processivity. Nucleic Acids Res. 2009;38:1982–1996. doi: 10.1093/nar/gkp1198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lee JH, et al. Striking similarities in diverse telomerase proteins revealed by combining structure prediction and machine learning approaches. Pac Symp Biocomput. 2008;13:501–512. [PMC free article] [PubMed] [Google Scholar]
- 41.Wyatt HD, Lobb DA, Beattie TL. Characterization of physical and functional anchor site interactions in human telomerase. Mol Cell Biol. 2007;27:3226–3240. doi: 10.1128/MCB.02368-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sealey DC, et al. The N-terminus of hTERT contains a DNA-binding domain and is required for telomerase activity and cellular immortalization. Nucleic Acids Res. 2010;38:2019–2035. doi: 10.1093/nar/gkp1160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zaug AJ, Podell ER, Cech TR. Mutation in TERT separates processivity from anchor-site function. Nat Struct Mol Biol. 2008;15:870–872. doi: 10.1038/nsmb.1462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wyatt HD, Tsang AR, Lobb DA, Beattie TL. Human telomerase reverse transcriptase (hTERT) Q169 is essential for telomerase function in vitro and in vivo. PLoS One. 2009;4:e7176. doi: 10.1371/journal.pone.0007176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Jurczyluk J, et al. Direct involvement of the TEN domain at the active site of human telomerase. Nucleic Acids Res. 2011;39:1774–1788. doi: 10.1093/nar/gkq1083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Feng J, et al. The RNA component of human telomerase. Science. 1995;269:1236–1241. doi: 10.1126/science.7544491. [DOI] [PubMed] [Google Scholar]
- 47.Theimer CA, Feigon J. Structure and function of telomerase RNA. Curr Opin Struct Biol. 2006;16:307–318. doi: 10.1016/j.sbi.2006.05.005. [DOI] [PubMed] [Google Scholar]
- 48.Collins K. Physiological assembly and activity of human telomerase complexes. Mech Ageing Dev. 2008;129:91–98. doi: 10.1016/j.mad.2007.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Moriarty TJ, Marie-Egyptienne DT, Autexier C. Regulation of 5′ template usage and incorporation of noncognate nucleotides by human telomerase. RNA. 2005;11:1448–1460. doi: 10.1261/rna.2910105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Egan ED, Collins K. Specificity and stoichiometry of subunit interactions in the human telomerase holoenzyme assembled in vivo. Mol Cell Biol. 2010;30:2775–2786. doi: 10.1128/MCB.00151-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Atilgan AR, et al. Anisotropy of fluctuation dynamics of proteins with an elastic network model. Biophys J. 2001;80:505–515. doi: 10.1016/S0006-3495(01)76033-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Podlevsky JD, Bley CJ, Omana RV, Qi X, Chen JJ. The telomerase database. Nucleic Acids Res. 2008;36:D339–343. doi: 10.1093/nar/gkm700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Gilley D, Lee MS, Blackburn EH. Altering specific telomerase RNA template residues affects active site function. Genes Dev. 1995;9:2214–2226. doi: 10.1101/gad.9.18.2214. [DOI] [PubMed] [Google Scholar]
- 54.Meier B, et al. trt-1 is the Caenorhabditis elegans catalytic subunit of telomerase. PLoS Genet. 2006;2:e18. doi: 10.1371/journal.pgen.0020018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Altschul SF, et al. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Pei J, Sadreyev R, Grishin NV. PCMA: Fast and accurate multiple sequence alignment based on profile consistency. Bioinformatics. 2003;19:427–428. doi: 10.1093/bioinformatics/btg008. [DOI] [PubMed] [Google Scholar]
- 57.Jones DT. Protein secondary structure prediction based on position-specific scoring matrices. J Mol Biol. 1999;292:195–202. doi: 10.1006/jmbi.1999.3091. [DOI] [PubMed] [Google Scholar]
- 58.Ginalski K, Elofsson A, Fischer D, Rychlewski L. 3D-Jury: A simple approach to improve protein structure predictions. Bioinformatics. 2003;19:1015–1018. doi: 10.1093/bioinformatics/btg124. [DOI] [PubMed] [Google Scholar]
- 59.Ginalski K, Rychlewski L. Protein structure prediction of CASP5 comparative modeling and fold recognition targets using consensus alignment approach and 3D assessment. Proteins. 2003;53(Suppl 6):410–417. doi: 10.1002/prot.10548. [DOI] [PubMed] [Google Scholar]
- 60.Eswar N, et al. Curr Protoc Bioinformatics. NY: John Wiley & Sons, Inc.; 2006. Comparative protein structure modeling using Modeller. Chapter 5:Unit 5.6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kolinski A. Protein modeling and structure prediction with a reduced representation. Acta Biochim Pol. 2004;51:349–371. [PubMed] [Google Scholar]
- 62.Landau M, et al. ConSurf 2005: The projection of evolutionary conservation scores of residues on protein structures. Nucleic Acids Res. 2005;33:W299–302. doi: 10.1093/nar/gki370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Jackson RM, Gabb HA, Sternberg MJ. Rapid refinement of protein interfaces incorporating solvation: Application to the docking problem. J Mol Biol. 1998;276:265–285. doi: 10.1006/jmbi.1997.1519. [DOI] [PubMed] [Google Scholar]
- 64.Cornell WD, et al. A second generation force field for the simulation of proteins, nucleic acids, and organic molecules. J Am Chem Soc. 1995;117:5179–5197. [Google Scholar]
- 65.Bahar I, Lezon TR, Yang LW, Eyal E. Global dynamics of proteins: Bridging between structure and function. Annu Rev Biophys. 2010;39:23–42. doi: 10.1146/annurev.biophys.093008.131258. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.