Abstract
Respiratory syncytial virus (RSV), the main cause of infant bronchiolitis, remains a major unmet vaccine need despite more than 40 years of vaccine research. Vaccine candidates based on a chief RSV neutralization antigen, the fusion (F) glycoprotein, have foundered due to problems with stability, purity, reproducibility, and potency. Crystal structures of related parainfluenza F glycoproteins have revealed a large conformational change between the prefusion and postfusion states, suggesting that postfusion F antigens might not efficiently elicit neutralizing antibodies. We have generated a homogeneous, stable, and reproducible postfusion RSV F immunogen that elicits high titers of neutralizing antibodies in immunized animals. The 3.2-Å X-ray crystal structure of this substantially complete RSV F reveals important differences from homology-based structural models. Specifically, the RSV F crystal structure demonstrates the exposure of key neutralizing antibody binding sites on the surface of the postfusion RSV F trimer. This unanticipated structural feature explains the engineered RSV F antigen's efficiency as an immunogen. This work illustrates how structural-based antigen design can guide the rational optimization of candidate vaccine antigens.
Keywords: subunit, epitope
Respiratory syncytial virus (RSV) is the most common cause of acute lower respiratory infection among children worldwide and the leading cause of infant hospitalization for respiratory disease in developed countries (1, 2). There is currently no vaccine or specific therapeutic agent for RSV, although prophylaxis with a potently neutralizing monoclonal antibody, Palizivumab, is available for those infants at highest risk (3). Vaccine development has been hampered not only by a history of vaccine-mediated disease enhancement, but also by problems with the stability, purity, reproducibility, tolerability, and potency of vaccine candidates (4–6). The RSV fusion glycoprotein (F) is a conserved target of neutralizing antibodies (7), including Palivizumab and the closely related monoclonal antibody, Motavizumab (8). Therefore, F is a promising antigen for RSV candidate vaccines.
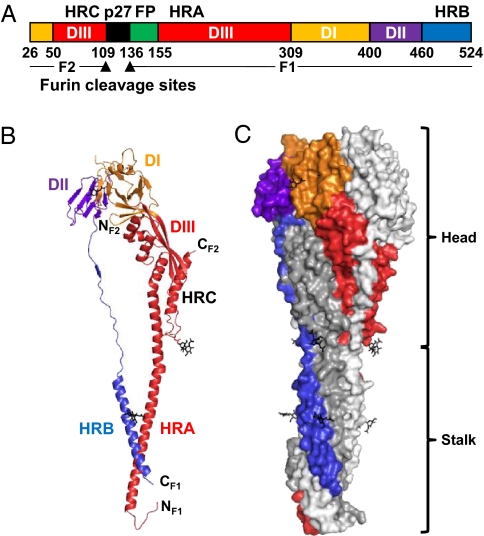
RSV F is a membrane anchored glycoprotein that mediates viral entry into host cells. The basic features of RSV F are shared with the fusion glycoproteins of other members of the Paramyxoviridae, such as parainfluenza virus 3 (PIV3), PIV5, and Newcastle disease virus (NDV). During cell entry, F glycoproteins undergo a conformational change that brings the viral and cellular membranes into proximity, ultimately leading to their fusion (9). Unlike parainfluenza F, which contain a single furin cleavage site, RSV F has two cleavage sites separated by a 27-amino-acid fragment (p27) (Fig. 1A). Activation of RSV F for membrane fusion requires cleavage by furin at the two sites, removing p27 and separating the protein into the disulfide-linked F1 and F2 (Fig. 1A and Fig. S1) (10). The resulting N terminus of F1 harbors a hydrophobic fusion peptide responsible for cellular membrane insertion, and the C terminus of F1 is anchored in the viral membrane by virtue of the transmembrane (TM) region.
Fig. 1.
RSV F ectodomain structure. (A) Linear diagram. Listed residue numbers correspond to the N terminus of each segment, the furin cleavage sites (arrowheads), and the C terminus. DI–III, domains I–III; p27, excised peptide; FP, fusion peptide; HRA, -B, and -C, heptad repeats A, B, and C. (B) Ribbon representation of one subunit. Domains colored as in A. Glycans are black. (C) Surface representation of the trimer. One subunit colored by domains as in A; the other two are white and gray.
The prefusion and postfusion forms of RSV F each have potential shortcomings as vaccine antigens. Large structural differences between the lollipop-shaped prefusion F trimer and the crutch-shaped postfusion F trimer are apparent even at the resolution of electron microscopy of negatively stained specimens, suggesting that prefusion and postfusion F may be antigenically distinct (11). To prevent viral entry, F-specific neutralizing antibodies presumably must bind the prefusion conformation of F on the virion, before the viral envelope fuses with a cellular membrane. Therefore, it might be expected that RSV F must be presented in the prefusion conformation to elicit neutralizing antibodies efficiently. However, prefusion F is a “metastable” structure that readily rearranges into the lower energy postfusion state, which aggregates due to exposure of a hydrophobic fusion peptide (12), and efforts to generate a soluble, stabilized prefusion F subunit antigen have not yet yielded candidates suitable for testing in humans.
Structures of the closely related PIV3 and NDV F proteins in their postfusion conformation and of the PIV5 F protein in its prefusion conformation have been determined previously (13–16). The structures confirm significant rearrangement of F between the prefusion and postfusion conformations. The largest difference between the two conformations is the packing of the heptad repeat A and B (HRA and HRB) regions (Fig. 1A). In the postfusion trimer, HRB helices and linkers pack against an HRA coiled coil, burying it in the center of a six-helix bundle. In the prefusion trimer, HRB forms a coiled-coil stalk, and the HRA residues pack into the globular head, making a significant contribution to the protein surface. Homology modeling of RSV F based on these PIV F structures and analysis of a Motavizumab–peptide complex suggested that the dominant neutralizing epitope recognized by Palivizumab and Motavizumab might be buried in trimeric RSV F, requiring at least a local dissociation for surface exposure (17).
Here we report the 3.2-Å resolution crystal structure of a stable, soluble, and well-behaved RSV F postfusion trimer. Contrary to expectations, immunization of mice or cotton rats with this antigen elicits high neutralizing antibody titers. The crystal structure reveals that, although RSV F shares the overall architecture of the PIV F glycoproteins, it also has important differences from structural models of RSV F that were based on homology modeling from PIV F (17). The unanticipated differences expose the key Palivizumab/Motavizumab neutralization site on the RSV F surface, even in the postfusion conformation, and explain this stable antigen's potency as a potential vaccine immunogen.
Results
RSV F Antigen Generation and Immunogenicity.
To produce a stable, nonaggregating RSV F candidate subunit antigen, we deleted the fusion peptide, transmembrane region, and cytoplasmic domain (Fig. 1A and Fig. S1) (18). This engineered F can be expressed efficiently and is readily purified. Because the construct retains the furin cleavage sites, the expressed glycoprotein is processed to F1 and F2 fragments. Electron microscopy of negatively stained specimens shows that it forms nonaggregated, homogeneous crutch-shaped molecules, consistent with postfusion F trimers (Fig. S2A). This engineered F trimer is very stable: Circular dichroism spectroscopy reveals no significant melting at temperatures up to 95 °C (Fig. S2 B and C).
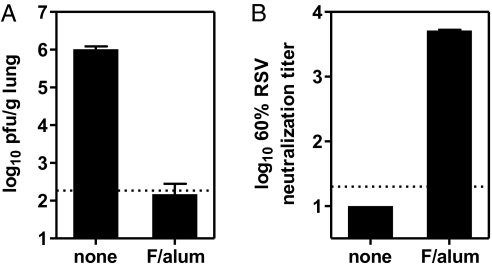
Two 5-μg doses of the alum-adsorbed RSV F protect cotton rats from intranasal RSV challenge (Fig. 2A), reducing lung RSV titers from ∼1 × 106 plaque forming units (pfu)/g in unimmunized animals to below the limit of detection (<200 pfu/g) in immunized animals. The immunized cotton rats had mean serum RSV neutralization titers of 1:5,150 (Fig. 2B), well above the 1:380 that correlates with protection in cotton rats (19) and similar passively acquired serum neutralizing titers that correlate with protection of human infants from severe RSV disease (20, 21).
Fig. 2.
Immunization of cotton rats with the postfusion RSV F trimer elicits neutralizing antibodies and protects from RSV challenge. Cotton rats were immunized intramuscularly with 5 μg of RSV F trimer adsorbed to alum on days 0 and 21 or were not immunized. Sera for neutralization assays were obtained on day 35. The cotton rats were challenged intranasally on day 49 with 1 × 105 pfu of RSV, strain Long. (A) Titers of RSV 5 d after challenge by plaque assay in the lungs of immunized (F/alum) or not immunized (none) cotton rats. Values shown are the means with SD of eight cotton rats per group. (B) Serum RSV neutralization titers. Values shown are the mean and range of two pools of four cotton rats per group.
Crystal Structure of the RSV F Postfusion Trimer.
To understand the molecular basis for the unexpectedly high immunogenicity of the postfusion F antigen, we crystallized the glycoprotein and determined its structure by molecular replacement and threefold noncrystallographic symmetry (NCS) averaging (Table S1, Fig. S3 and Materials and Methods). The structure does not include the p27 fragment (the peptide between the two furin sites that is lost upon cleavage), the fusion peptide, the transmembrane region, or the cytoplasmic domain (Fig. 1A and Fig. S1).
The overall architecture of postfusion RSV F is shared with postfusion parainfluenza virus F glycoproteins (Fig. 1). The glycoprotein is composed of three tightly intertwined subunits, forming a globular head and an elongated stalk. Each subunit contains three domains, designated I, II, and III (Fig. 1 A–C). Domains I and II are at the top of the trimeric head and form a triangular crown. Domain III forms the base of the head. A long helix, HRA, extends from domain III and forms the trimeric coiled coil in the center of the stalk. The HRB helix is tethered to domain II and reaches down to the head-distal end of the stalk, where it forms the outer coils of a six-helix bundle with the HRA interior coiled coil. In full-length F, the hydrophobic fusion peptide (N terminal to HRA) and the transmembrane region (C terminal to HRB) would be juxtaposed at the bottom of the stalk and inserted into the target cell membrane.
Comparison of RSV F and Parainfluenza F Proteins.
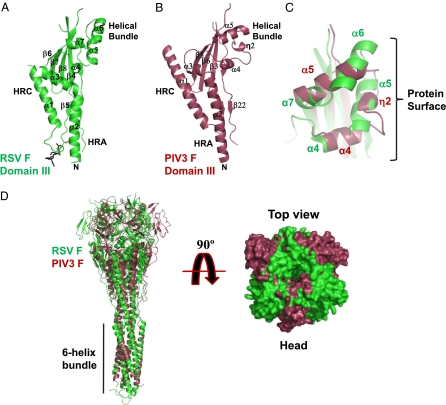
F domains I and II from RSV, PIV3 (Fig. S4 A and B), and PIV5 are structurally conserved. The only significant difference is in the orientation of the sole helix of domain I (η3 of RSV F and α6 of PIV3 or PIV5 F) relative to their common β sheets. In contrast, RSV domain III has features that were not predicted from PIV-based modeling (17) (Fig. 3). When the four-stranded β sheets of RSV domain III and PIV3 domain III are superimposed, key differences in the domains’ helical regions are apparent. Helix HRA kinks at a more N-terminal position in RSV F than in PIV3 F, causing an ∼60° difference in the rotation of the heads relative to the stalks (Fig. 3 A, B, and D). Influenza hemagglutinins also vary in the orientations of their heads relative to their stalks, with 30°–50° differences in rotation between subtypes (22).
Fig. 3.
RSV and PIV3 comparisons. (A) Ribbon diagram of RSV F domain III. (B) Ribbon diagram of the PIV3 domain III oriented as is the domain in A. (C) Detail of the RSV (green) and PIV3 (red) domain III helical bundles superimposed on the basis of domain III β sheets. Helix labels are colored by virus. (D) RSV and PIV3 F ectodomain trimers (colored as in A and B) superimposed on the basis of their six-helix bundles. (Left) Ribbon diagram viewed perpendicular to the threefold axis; (Right) surface representation viewed along the threefold axis from the top of the head.
Palivizumab and Motavizumab Binding Site Is Exposed in the Postfusion RSV F Trimer.
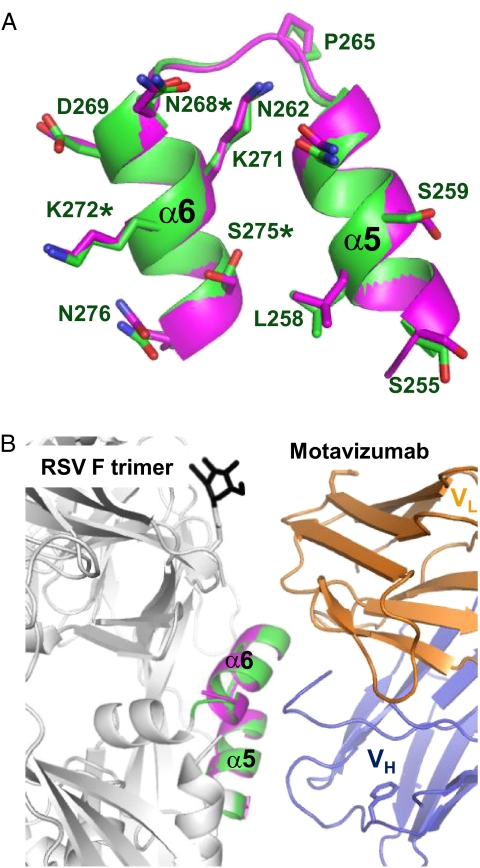
The RSV F domain III helical bundle region contains an extra helix (α6), changing the orientation of the bundled helices relative to those in parainfluenza F (Fig. 3 A–C and Fig. S1). RSV F helices α5 and α6 are almost parallel and are exposed on the trimer surface; the equivalent to RSV F α6 helix in the PIV3 helical bundle (α5, Fig. 3C) is buried in the intersubunit interface of the trimer. RSV F helices α5 and α6 form the epitope bound by neutralizing antibodies Palivizumab and Motavizumab. These antibodies bind cell surface-expressed, full-length, likely prefusion RSV F (23) and inhibit RSV cell entry and F-mediated cell–cell fusion (24), presumably by preventing conformational changes in F. The structure of a complex between the Motavizumab Fab and a 24-residue RSV F peptide that includes α5 and α6 has been reported (17). Comparison between this structure and the postfusion RSV F structure reveals a close match between the α5–α6 helices (rmsd for 23 α-carbon atoms = 0.52 Å; Fig. 4A). Superposition of the two structures based on these helices models a complex between Motavizumab and postfusion RSV F (Fig. 4B). This model reveals no steric hindrance that would prevent Motavizumab (or, presumably, Palivizumab) from binding to postfusion RSV F. We have confirmed binding of Palivizumab to the postfusion F ectodomain in solution using surface plasmon resonance (KD of 4.2 × 10−10 M) (Fig. S5). Notably, it has been reported that the affinity of Motavizumab for the isolated peptide is at least 6,000-fold lower than for the folded RSV F protein (8, 25), further suggesting that tight binding is dependent on the conformation of this epitope.
Fig. 4.
Motavizumab epitope. (A) Superposition of the Motavizumab-binding helices, α5 and α6, from the RSV F postfusion trimer (green) and the peptide–Motavizumab complex (magenta, PDB code 3IXT) (17). RSV residues bound by Motavizumab are shown in stick representation. Asterisks denote Palivizumab escape mutations. (B) Ribbon representation modeling a Motavizumab–RSV F postfusion trimer complex. The VH and VL domains of the Fab are blue and orange, respectively; helices α5 and α6 from the RSV F structure and the peptide–Motavizumab structure colored green and magenta, respectively; a glycan on F is black; and the remainder of F is white.
Neutralization Sites on the RSV F Postfusion Structure and an RSV F Prefusion Model.
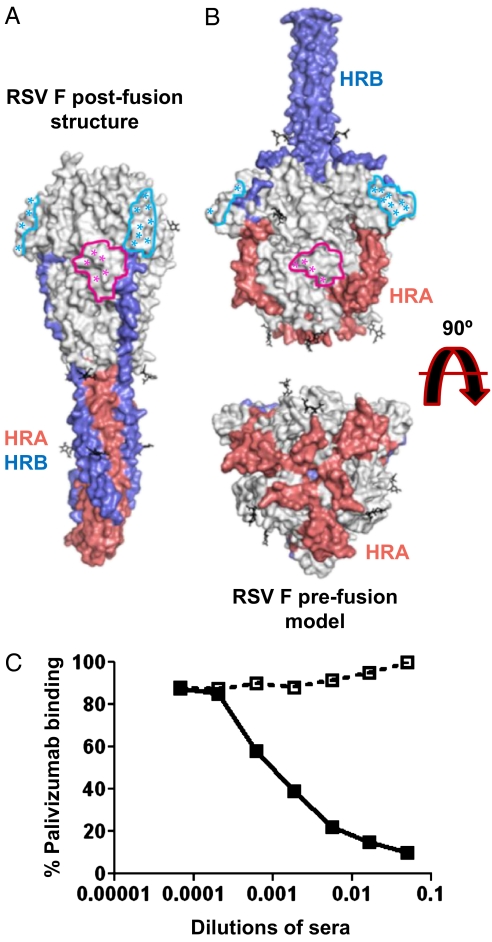
Prefusion and postfusion PIV F structures reveal en bloc shifts of domains and large rearrangements of HRA and HRB. In domain III of the prefusion PIV5 structure, HRA folds into three α helices and two β strands rather than the long postfusion HRA helix (15). However, when prefusion and postfusion conformations of individual PIV F domains are compared, the nonrearranging parts superimpose well. Superimposing postfusion RSV F domains on their prefusion PIV5 F counterparts does not result in major clashes and positions all of the pairs of cysteines that form interdomain disulfide bonds in proximity. The prefusion RSV F model obtained by thus combining information from the postfusion RSV F structure and the prefusion PIV5 F structure reveals a feature not apparent from homology modeling prefusion RSV F based solely on the PIV5 prefusion structure (17): The helices of the Palivizumab/Motivizumab epitope are exposed on the surface of the modeled prefusion RSV F trimer as they are on postfusion RSV F trimer structure (Fig. 5 A and B and Fig. S6). In our prefusion RSV F model, the loop connecting β4 and HRC (part of domain III) would hinder access of Palivizumab or Motavizumab to the epitope. However, it is likely that the loop has sufficient flexibility to adopt an alternative conformation that permits antibody binding (Fig. S6B).
Fig. 5.
RSV F conformational changes, antigenic structure, and Palivizumab binding. (A) Surface representation of the postfusion structure. Antigenic sites A and C are indicated by colored outlines (pink and cyan, respectively). Asterisks indicate residues selected in neutralization escape variants or forming contacts with an antibody in the determined structures of neutralizing antibody–peptide complexes. The HRA and HRB surfaces are red and blue, respectively. (B) Surface representation of a prefusion model, annotated as in A. (C) Inhibition of Palivizumab binding to postfusion RSV F by pooled sera from unimmunized mice (open boxes) or mice immunized with the RSV F antigen (filled boxes). Palivizumab binding (percentage of ELISA signal without competing sera) is plotted as a function of the dilution of competing pooled sera.
The RSV F construct does not include the fusion peptide. In the determined prefusion PIV5 structure, the fusion peptide packs into the globular F head, suggesting that fusion peptide deletion might destabilize the prefusion conformation (15). How then does fusion peptide-deleted RSV F achieve a stable postfusion conformation? Either fusion peptide-deleted RSV F folds directly into its postfusion state without passing through a prefusion intermediate or it folds into a transient prefusion conformation despite the lack of a fusion peptide. The presence of the p27 peptide immediately N terminal to the fusion peptide in uncleaved native RSV F but not PIV5 F suggests that the fusion peptides of RSV F and PIV5 F could pack differently in their prefusion conformations.
The antigenic structure of the RSV F trimer has been mapped by a variety of techniques (7, 17, 26–33) (Table S2 and Fig. S1). The best documented epitope clusters are designated A and C (34), and others have been proposed. The Motavizumab–peptide structure corroborated the location of site A, although it called into question the site's exposure on the RSV F trimer (17); a crystal structure of an RSV F peptide (residues 422–436) bound to the 101F neutralizing antibody corroborated the location of site C (33). The postfusion RSV F structure and the prefusion RSV F model suggest that sites A and C remain exposed and structurally similar in both conformations (Fig. 5 A and B and Fig. S6). Superposition of the 101F–peptide complex on the RSV F prefusion model and postfusion structure confirms that 101F would not clash with F in either conformation (Fig. S7). Although HRA and HRB do not contribute to antigenic sites A and C, some peptide binding data (7) suggest that these rearranging elements may contribute to less well-characterized neutralizing epitopes, which might only be presented by prefusion RSV F.
The preservation of sites A and C in both prefusion and postfusion RSV F plausibly explains the ability of a postfusion RSV F antigen to elicit high titer neutralizing antibodies in immunized animals. Consistent with this hypothesis, competition ELISA demonstrates that pooled sera of mice immunized with the alum-adsorbed postfusion RSV F antigen (but not sera of unimmunized mice) inhibit Palivizumab binding (Fig. 5C).
Discussion
We have determined the crystal structure of an RSV F postfusion antigen that was structurally engineered for easy purification, homogeneity, and stability. On the basis of PIV F homology models, it was thought that neutralizing epitopes would be disrupted on the postfusion RSV F conformer and that the key neutralizing epitope bound by Palivizumab and Motavizumab might not be exposed, even on the surface of the prefusion RSV F trimer (17). However, we found that immunization of cotton rats with the fusion peptide-deleted, postfusion RSV F ectodomain elicits high neutralizing antibody titers and protects the animals from RSV challenge. The crystal structure of the RSV F postfusion trimer revealed that the Palivizumab/Motavizumab epitope, site A, is exposed and accessible on the postfusion trimer structure. Indeed, tight binding of the RSV F postfusion trimer by Palivizumab was confirmed by surface plasmon resonance. Sera of mice immunized with the RSV F postfusion trimer, but not sera from nonimmunized animals, competed with Palivizumab for binding to F. Site C, an additional important neutralizing epitope recognized by neutralizing antibodies such as 101F is also well exposed on the postfusion RSV F trimer. Therefore, the crystal structure of the postfusion RSV F trimer demonstrates that key neutralizing binding sites are present and exposed on this form of F, providing the basis for eliciting high titer neutralizing antibodies. Notably, an improved model of the prefusion F structure based on combining information from the postfusion RSV F structure and the prefusion PIV5 F structure shows that the Palivizumab/Motavizumab and 101F binding sites would likely be available on the prefusion protein surface as well.
In summary, our results demonstrate that it is possible to engineer an optimized RSV F antigen for a candidate subunit vaccine. The postfusion conformation confers high thermal stability and homogeneity. The deletion of the fusion peptide and transmembrane domain confers solubility and lack of aggregation (18). The presentation of neutralizing epitopes on the surface of postfusion F trimers enables this biochemically tractable antigen to elicit the high titer neutralizing antibodies that are associated with protection from disease (19–21). The structural approach to antigen design for a candidate RSV vaccine provides a model for optimizing and evaluating new vaccine antigens to address unmet medical needs.
Materials and Methods
RSV F Protein Expression and Purification.
A DNA construct encoding RSV F residues 1–136 and 146–524 (corresponding to the F ectodomain without the fusion peptide) with a C-terminal histidine tag was codon optimized and synthesized (Geneart). Recombinant F was expressed using a baculovirus expression vector in HiFive cells (Invitrogen) and purified by nickel affinity and size exclusion chromatography. Although the construct lacks the fusion peptide, HRA and HRC are still separated by a 48-amino-acid-long tether (Fig. 1A and Fig. S1). These residues could span the 130-Å distance that separates HRA and HRC in the postfusion form of F (Fig. 1B), potentially allowing folding of the protein into the postfusion conformation before furin cleavage.
Crystallization.
Crystals were grown at 15 °C with the hanging-drop vapor-diffusion method by mixing equal volumes of protein solution (∼10 mg/mL) and precipitant solution (4.2 M sodium formate, 100 mM sodium acetate, pH 5.1). Crystals were flash frozen directly from the hanging drop in liquid nitrogen. All crystals belong to the orthorhombic space group P212121 (Table S1).
Structure Determination.
X-ray diffraction data were collected at the 17-ID beamline (Industrial Macromolecular Crystallography Association - Collaborative Access Team, Advanced Photon Source, Argonne National Laboratory, Argonne, IL) on a Pilatus detector. The data were integrated with XDS (35) and scaled with SCALA (36, 37). Details of the structure determination and refinement are reported in SI Materials and Methods. Briefly, initial phases were obtained by molecular replacement with PHASER (36, 38) using as a search model a modified postfusion PIV3 F protein [Protein Data Bank (PDB) code 1ZTM] in which the six-helix bundle had been replaced with the corresponding region from RSV F (PDB code 3KPE).
Model building was performed with “O” (39) and Refmac (36, 40) was used for refinement. The final model has Rwork and Rfree of 23.1 and 26.6%, respectively (Table S1).
Mouse Immunization.
BALB/c mice were immunized intramuscularly with 5 μg of RSV F trimers adsorbed to aluminum hydroxide. Sera were collected 2 wk after the second immunization (given 4 wk after the first).
Cotton Rat Immunization and RSV Challenge.
Cotton rats were immunized intramuscularly on days 0 and 21 with 5 μg of RSV F trimers adsorbed to aluminum hydroxide. Sera were collected on day 35. Cotton rats were challenged intranasally with 1 × 105 pfu of RSV strain Long on day 49. Cotton rat lungs were harvested on day 54. Harvested lungs were homogenized and clarified. Virus in the lung samples was titered by plaque assay on Hep-2 cells by infecting for 2 h, removing the inoculum, and overlaying with 1.25% SeaPlaque agarose (Lonza) in Eagle's minimum essential medium. After 3–4 d, cells were stained with neutral red. Plaques were counted 1 d later. Samples with titers less than the limit of detection (∼200 pfu/g of lung tissue) were assigned a titer of 100 pfu/g.
Neutralization Assay.
The RSV microneutralization assay was performed in 96-well microplates using Hep-2 cells and the RSV Long strain. Details of the assay are described in the SI Materials and Methods.
Electron Microscopy.
RSV F protein sample (50 μg/mL in 25 mM Tris, 300 mM NaCl) was absorbed onto a 400-mesh carbon-coated grid (Electron Microscopy Sciences) and stained with 0.75% uranyl formate. A Jeol 1200EX microscope, operated at 80 kV, was used to analyze the samples. Micrographs were taken at 65,000× magnification.
Circular Dichroism Spectroscopy.
The CD spectrum was collected using a 1-mm cuvettete on a Jasco J-815 CD spectrometer from 320 to 190 nm at 20 °C. The RSV F trimer sample (500 μg/mL in 25 mM Tris pH 7.5 buffer and 50 mM sodium chloride) was heated from 20 °C to 95 °C in 1 °C/min steps, and a CD rotation at 210 nm was recorded at 5 °C intervals. When the sample reached 95 °C, the CD spectrum was collected again from 320 to 190 nm for comparison.
Binding Studies by SPR.
The affinity of the monoclonal antibody Palivizumab for the RSV F trimer was measured by SPR with a Biacore T100 instrument. Palivizumab was directly immobilized on a CM5 sensor chip using amine coupling at very low levels (50 response units) and RSV F was injected at a high flow rate (100 μL/min) to avoid avidity effects and higher than 1:1 binding interaction. The data were processed using Biacore T100 evaluation software and double referenced by subtraction of the blank surface and buffer-only injection before global fitting of the data to a 1:1 binding model.
Competition ELISA.
A total of 100 ng of purified RSV F in PBS was coated onto each well of Nunc MaxiSorp plates by overnight incubation at 4 °C. The uncoated surfaces were blocked with PBS containing 1% BSA. Then, varying concentrations (threefold dilutions from a 1:25-starting dilution) of mouse sera (immunized or naive) were added and incubated at room temperature for 1 h, followed by addition of 100 ng/well of Palivizumab. Plates were again incubated for 1 h at room temperature. Bound Palivizumab in presence of varying concentrations of either RSV F-immunized mice sera or naive sera was detected using antihuman HRP-conjugated antibody (Jackson ImmunoResearch) and the TMB Peroxidase Substrate system (KPL) measured at 450 nm (Optimax microplate reader). 100% Palivizumab binding is defined as the binding in presence of the lowest dilution (1:25) of naive sera.
Supplementary Material
Acknowledgments
We thank the RSV Project Team at Novartis Vaccines and Diagnostics and the staff at beamline 17-ID at Industrial Macromolecular Crystallography Association - Collaborative Access Team, Advanced Photon Source, Argonne National Laboratory, Argonne, IL. In addition, we thank the Protein Structure Unit of the Novartis Institutes for Biomedical Research and, in particular, Rajiv Chopra, for help with data collection and processing. We thank Kara Balabanis for cloning the RSV F construct and Claire Metrick for protein expression and purification. Electron microscopy was performed at the Harvard Medical School Electron Microscopy Facility, Boston, MA.
Footnotes
Conflict of interest statement: The authors are Novartis shareholders and employees of Novartis Vaccines and Diagnostics.
Database deposition: Coordinates of the RSV F structure have been deposited in the Research Collaboratory for Structural Bioinformatics Protein Database with the accession code 3rki.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1106536108/-/DCSupplemental.
References
- 1.Nair H, et al. Global burden of acute lower respiratory infections due to respiratory syncytial virus in young children: A systematic review and meta-analysis. Lancet. 2010;375:1545–1555. doi: 10.1016/S0140-6736(10)60206-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hall CB. Respiratory syncytial virus. In: Mandell GL, Bennett JE, Dolin R, editors. Mandell, Douglas, and Bennett's Principles and Practice of Infectious Diseases. 7th Ed. New York: Churchill Livingstone; 2010. pp. 2207–2221. [Google Scholar]
- 3.Johnson S, et al. Development of a humanized monoclonal antibody (MEDI-493) with potent in vitro and in vivo activity against respiratory syncytial virus. J Infect Dis. 1997;176:1215–1224. doi: 10.1086/514115. [DOI] [PubMed] [Google Scholar]
- 4.Kim HW, et al. Respiratory syncytial virus disease in infants despite prior administration of antigenic inactivated vaccine. Am J Epidemiol. 1969;89:422–434. doi: 10.1093/oxfordjournals.aje.a120955. [DOI] [PubMed] [Google Scholar]
- 5.Munoz FM, Piedra PA, Glezen WP. Safety and immunogenicity of respiratory syncytial virus purified fusion protein-2 vaccine in pregnant women. Vaccine. 2003;21:3465–3467. doi: 10.1016/s0264-410x(03)00352-9. [DOI] [PubMed] [Google Scholar]
- 6.Wright PF, et al. The absence of enhanced disease with wild type respiratory syncytial virus infection occurring after receipt of live, attenuated, respiratory syncytial virus vaccines. Vaccine. 2007;25:7372–7378. doi: 10.1016/j.vaccine.2007.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Anderson R, Huang Y, Langley JM. Prospects for defined epitope vaccines for respiratory syncytial virus. Future Microbiol. 2010;5:585–602. doi: 10.2217/fmb.10.22. [DOI] [PubMed] [Google Scholar]
- 8.Wu H, et al. Development of motavizumab, an ultra-potent antibody for the prevention of respiratory syncytial virus infection in the upper and lower respiratory tract. J Mol Biol. 2007;368:652–665. doi: 10.1016/j.jmb.2007.02.024. [DOI] [PubMed] [Google Scholar]
- 9.Lamb RA, Jardetzky TS. Structural basis of viral invasion: Lessons from paramyxovirus F. Curr Opin Struct Biol. 2007;17:427–436. doi: 10.1016/j.sbi.2007.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.González-Reyes L, et al. Cleavage of the human respiratory syncytial virus fusion protein at two distinct sites is required for activation of membrane fusion. Proc Natl Acad Sci USA. 2001;98:9859–9864. doi: 10.1073/pnas.151098198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Calder LJ, et al. Electron microscopy of the human respiratory syncytial virus fusion protein and complexes that it forms with monoclonal antibodies. Virology. 2000;271:122–131. doi: 10.1006/viro.2000.0279. [DOI] [PubMed] [Google Scholar]
- 12.Begoña Ruiz-Argüello M, et al. Effect of proteolytic processing at two distinct sites on shape and aggregation of an anchorless fusion protein of human respiratory syncytial virus and fate of the intervening segment. Virology. 2002;298:317–326. doi: 10.1006/viro.2002.1497. [DOI] [PubMed] [Google Scholar]
- 13.Chen L, et al. The structure of the fusion glycoprotein of Newcastle disease virus suggests a novel paradigm for the molecular mechanism of membrane fusion. Structure. 2001;9:255–266. doi: 10.1016/s0969-2126(01)00581-0. [DOI] [PubMed] [Google Scholar]
- 14.Yin HS, Paterson RG, Wen X, Lamb RA, Jardetzky TS. Structure of the uncleaved ectodomain of the paramyxovirus (hPIV3) fusion protein. Proc Natl Acad Sci USA. 2005;102:9288–9293. doi: 10.1073/pnas.0503989102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yin HS, Wen XL, Paterson RG, Lamb RA, Jardetzky TS. Structure of the parainfluenza virus 5 F protein in its metastable, prefusion conformation. Nature. 2006;439:38–44. doi: 10.1038/nature04322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Swanson K, et al. Structure of the Newcastle disease virus F protein in the post-fusion conformation. Virology. 2010;402:372–379. doi: 10.1016/j.virol.2010.03.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.McLellan JS, et al. Structural basis of respiratory syncytial virus neutralization by motavizumab. Nat Struct Mol Biol. 2010;17:248–250. doi: 10.1038/nsmb.1723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Martín D, Calder LJ, García-Barreno B, Skehel JJ, Melero JA. Sequence elements of the fusion peptide of human respiratory syncytial virus fusion protein required for activity. J Gen Virol. 2006;87:1649–1658. doi: 10.1099/vir.0.81715-0. [DOI] [PubMed] [Google Scholar]
- 19.Prince GA, Horswood RL, Chanock RM. Quantitative aspects of passive immunity to respiratory syncytial virus infection in infant cotton rats. J Virol. 1985;55:517–520. doi: 10.1128/jvi.55.3.517-520.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Groothuis JR, et al. The Respiratory Syncytial Virus Immune Globulin Study Group. Prophylactic administration of respiratory syncytial virus immune globulin to high-risk infants and young children. N Engl J Med. 1993;329:1524–1530. doi: 10.1056/NEJM199311183292102. [DOI] [PubMed] [Google Scholar]
- 21.Piedra PA, Jewell AM, Cron SG, Atmar RL, Glezen WP. Correlates of immunity to respiratory syncytial virus (RSV) associated-hospitalization: Establishment of minimum protective threshold levels of serum neutralizing antibodies. Vaccine. 2003;21:3479–3482. doi: 10.1016/s0264-410x(03)00355-4. [DOI] [PubMed] [Google Scholar]
- 22.Ha Y, Stevens DJ, Skehel JJ, Wiley DC. H5 avian and H9 swine influenza virus haemagglutinin structures: Possible origin of influenza subtypes. EMBO J. 2002;21:865–875. doi: 10.1093/emboj/21.5.865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wu H, et al. Ultra-potent antibodies against respiratory syncytial virus: Effects of binding kinetics and binding valence on viral neutralization. J Mol Biol. 2005;350:126–144. doi: 10.1016/j.jmb.2005.04.049. [DOI] [PubMed] [Google Scholar]
- 24.Huang K, Incognito L, Cheng X, Ulbrandt ND, Wu H. Respiratory syncytial virus-neutralizing monoclonal antibodies motavizumab and palivizumab inhibit fusion. J Virol. 2010;84:8132–8140. doi: 10.1128/JVI.02699-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tous G, Schenerman M, Casas-Finet J, Wei Z, Pfarr DS. 2010 Antibodies against and methods for producing vaccines for respiratory syncytial virus. United States Patent: 7,700,720. [Google Scholar]
- 26.López JA, Peñas C, García-Barreno B, Melero JA, Portela A. Location of a highly conserved neutralizing epitope in the F glycoprotein of human respiratory syncytial virus. J Virol. 1990;64:927–930. doi: 10.1128/jvi.64.2.927-930.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Arbiza J, et al. Characterization of two antigenic sites recognized by neutralizing monoclonal antibodies directed against the fusion glycoprotein of human respiratory syncytial virus. J Gen Virol. 1992;73:2225–2234. doi: 10.1099/0022-1317-73-9-2225. [DOI] [PubMed] [Google Scholar]
- 28.Crowe JE, et al. Monoclonal antibody-resistant mutants selected with a respiratory syncytial virus-neutralizing human antibody fab fragment (Fab 19) define a unique epitope on the fusion (F) glycoprotein. Virology. 1998;252:373–375. doi: 10.1006/viro.1998.9462. [DOI] [PubMed] [Google Scholar]
- 29.Zhao X, Chen FP, Sullender WM. Respiratory syncytial virus escape mutant derived in vitro resists palivizumab prophylaxis in cotton rats. Virology. 2004;318:608–612. doi: 10.1016/j.virol.2003.10.018. [DOI] [PubMed] [Google Scholar]
- 30.Liu C, et al. Relationship between the loss of neutralizing antibody binding and fusion activity of the F protein of human respiratory syncytial virus. Virol J. 2007;4:71. doi: 10.1186/1743-422X-4-71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wu SJ, et al. Characterization of the epitope for anti-human respiratory syncytial virus F protein monoclonal antibody 101F using synthetic peptides and genetic approaches. J Gen Virol. 2007;88:2719–2723. doi: 10.1099/vir.0.82753-0. [DOI] [PubMed] [Google Scholar]
- 32.López JA, et al. Antigenic structure of human respiratory syncytial virus fusion glycoprotein. J Virol. 1998;72:6922–6928. doi: 10.1128/jvi.72.8.6922-6928.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.McLellan JS, et al. Structure of a major antigenic site on the respiratory syncytial virus fusion glycoprotein in complex with neutralizing antibody 101F. J Virol. 2010;84:12236–12244. doi: 10.1128/JVI.01579-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Beeler JA, van Wyke Coelingh K. Neutralization epitopes of the F glycoprotein of respiratory syncytial virus: Effect of mutation upon fusion function. J Virol. 1989;63:2941–2950. doi: 10.1128/jvi.63.7.2941-2950.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kabsch W. XDS. Acta Crystallogr D Biol Crystallogr. 2010;66:125–132. doi: 10.1107/S0907444909047337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Collaborative Computational Project, Number 4. The CCP4 suite: Programs for protein crystallography. Acta Crystallogr D Biol Crystallogr. 1994;50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 37.Evans P. Scaling and assessment of data quality. Acta Crystallogr D Biol Crystallogr. 2006;62:72–82. doi: 10.1107/S0907444905036693. [DOI] [PubMed] [Google Scholar]
- 38.McCoy AJ, et al. Phaser crystallographic software. J Appl Cryst. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jones TA, Zou JY, Cowan SW, Kjeldgaard M. Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr A. 1991;47:110–119. doi: 10.1107/s0108767390010224. [DOI] [PubMed] [Google Scholar]
- 40.Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr D Biol Crystallogr. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.