Abstract
Extrinsic factors and the interactions of neurons with surrounding tissues are essential for almost every aspect of neuronal development. Here we describe a strategy of gene expression with an independent enhancer-driven cellular marker (GEEM) for studying roles of cell–cell interactions and extrinsic factors in the development of the Drosophila nervous system. Key to this strategy is robust expression of enhancer-driven transgenic markers in specific neurons. To this end, we have created vectors to achieve bright and even labeling of neuronal processes, easy cloning of enhancer elements, and efficient and flexible generation of transgenic animals. We provide examples of enhancer-driven membrane markers for specific neurons in both the peripheral and central nervous systems and their applications in the study of neuronal projections and connections in the Drosophila brain. We further applied GEEM to examine the wrapping of sensory neuron somas by glia during embryonic and larval stages, and neuron–glia interaction during dendrite pruning in live animals, leading to the discovery that glia play critical roles in the severing and degradation of proximal dendrites. The GEEM paradigm should be applicable to the studies of both cell-autonomous and nonautonomous regulations of any cell type.
Keywords: genetic tools, neuronal circuit, reporter
The formation of a mature nervous system relies on not only gene activities within neurons but also interactions with local environments, such as the intercellular signaling important for neuronal specification and differentiation (1, 2), and neuron–glia interactions crucial for axon pathfinding (3). In addition, the size and shape of dendritic trees are influenced by extracellular factors in the target fields (4). Moreover, synapse formation requires intimate interactions between pre- and postsynaptic neuronal partners (5).
Drosophila is a powerful model system for studying the biology and disease of the nervous system. However, tools for analyzing interactions of neurons with other tissues and the roles of extrinsic factors in neural development are limited, as it is difficult to simultaneously label different cell types and even more challenging to concurrently label neuronal structures and manipulate gene functions in adjacent tissues. In Drosophila, the Gal4/upstream activator sequence (UAS) (6) binary expression system provides a powerful way of transgene expression, with broad choices in Gal4 drivers and UAS-controlled transgenes. Whereas Gal4/UAS allows for labeling and manipulation of tissues that may affect neural development, Gal4-independent neuronal labeling is required for analyzing the resulting neuronal phenotypes. Two strategies for labeling neurons could be combined with Gal4/UAS to genetically manipulate other cell types. The first is to use binary systems, such as LexA/LexAop (7). Despite the success in certain cases (7, 8), this strategy has not been widely applied to study nonautonomous mechanisms in neural development, likely because of the less than optimal specificity of the LexA/LexAop system (9). The recent introduction of the Q system (9) may offer an alternative. The second strategy is to express a reporter in specific neurons under the control of a promoter or enhancer (10). However, effective application of this strategy is currently limited by the generally weak expression of enhancer-driven reporters.
Drosophila dendritic arborization (da) neurons are sensory neurons with dendritic arbors covering the body wall. Although peripheral glia are known to control axonal sprouting and dendritic branching (11), it is less clear how glia form associations with da neurons. Certain da neurons prune dendritic trees before growing new ones during metamorphosis (12, 13). The pruning of larval dendrites involves extrinsic factors (12–14). However, the sources of the extrinsic factors have not been identified.
To facilitate the study of nonautonomous mechanisms underlying neural development in Drosophila, we developed a strategy for generating enhancer-driven membrane markers with robust expression. This strategy combines a transgenic design for high expression, improved membrane markers for efficient labeling of neuronal processes, the Gateway system for convenient cloning of neuronal enhancers, and a dual transformation platform for comparing and establishing transgenic lines. By applying the method of GEEM (Gene Expression with an independent Enhancer-driven cellular Marker), we show the use of several CNS neuronal markers in studying neuronal projection and connection. In addition, we examined how glial membrane wraps the somas of sensory neurons, and analyzed neuron–glia interactions in dendrite pruning, revealing an unrecognized role of glia in the severing and degradation of proximal dendrites.
Results
Generation of High-Expression Membrane Markers.
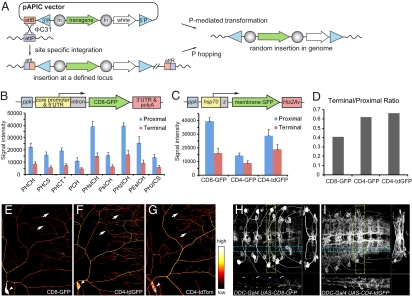
To create membrane markers expressed at high levels for in vivo studies of neural development in Drosophila, we generated a dual transforming vector, pAPIC (attB P-element insulated CaSpeR) (Fig. 1A) to compare different transgenic designs. The attB site is used for ϕC31-mediated integration (15) and is critical for comparing different transgenes at the same genomic locus. The P-element sites allow generation of additional insertions of selected transgenes at random loci by P-mediated transformation or mobilization (16). Two copies of the gypsy insulator flanking the transgene help to reduce line-to-line variations caused by position effects (17) and boost transgene expression (18).
Fig. 1.
Optimization of transgene components and fluorescent membrane markers. (A) Transgenesis based on the pAPIC vector. 5′P and 3′P, P-element sites; In, gypsy insulator. (B) Signal intensity of ppk-CD8-GFP reporters with various transgene components (Table 1). The reporters were integrated to attPVK19 and fluorescent signals from both proximal and terminal dendrites of ddaC neurons were compared. The asterisk indicates nonspecific expression in other classes of da neurons. (C) Signal intensity of CD8- and CD4-fusion reporters. The error bars represent SDs (B and C). (D) Ratios of terminal dendrite signals to proximal dendrite signals in CD8- and CD4-fusion reporter lines. (E–G) Distribution of CD8-GFP (E), CD4-tdGFP (F), and CD4-tdTom (G) in ddaC neurons. The arrowheads point to the somas and the arrows point to terminal dendrites. (H and I) Distribution of CD8-GFP (H) and CD4-tdGFP (I) in DDC neurons in the larval ventral nerve cord. In both images, the main panel shows the z-axis projection; the right panel shows the x-axis projection of the volume within the broken yellow lines; the bottom panel shows the y-axis projection of the volume within the broken blue lines.
For neuronal labeling, enhancer-driven membrane markers need to satisfy three criteria. First, the markers, even driven by a weak enhancer, should be robustly expressed at levels comparable to or exceeding those achieved using binary systems with traditional transgenic designs. Second, the markers should be bright and evenly distributed on the cell membrane so as to label all neuronal processes. Third, the markers should not cause cytotoxicity. To compare construct designs of membrane markers, we used class IV da (C4da) neurons of the Drosophila larva as a test case for several reasons. First, C4da neurons have highly complex dendritic trees comprised of more than six orders of dendritic branches (Fig. S1A) (19), which allows for assessment of the evenness of membrane markers at branches of different thickness. Second, the dendrites of C4da neurons cover the larval body wall and are amenable to live imaging in intact animals. Third, a C4da-specific enhancer has been identified from the promoter of ppk (10), which drives weak transgenic expression at most genomic loci examined, making it possible to evaluate construct designs with direct enhancer fusion.
To search for better designs for neuronal membrane markers, we first aimed to optimize the functional components of the transgene unit to achieve high expression, and then sought to improve the brightness and even distribution of the membrane markers. The expression level of a transgene is influenced by every component of the transgene unit, including the core promoter, the 5′UTR, the 3′UTR, and the mRNA cleavage and polyadenylation sequence (polyA). In Drosophila, most transgene designs use a small repertoire of functional components, such as the core promoter and 5′UTR from heat shock protein 70 (hsp70) and 3′UTR and polyA from SV40 early genes. A systematic comparison of the components used in various transgenic vectors on gene-expression levels in vivo, especially in neuronal tissues, has been lacking. We tested the effects of several core promoter 5′UTRs and 3′UTR-polyAs, as well as addition of an intron in the 5′UTR, on the expression level of a CD8-GFP reporter in C4da neurons (Table 1). We found that the combined use of hsp70 core promoter-5′UTR, an intron after 5′UTR, and His2Av 3′UTR-polyA (HIH cassette) gives the highest reporter expression (Fig. 1B), more than doubling that of the combination of hsp70 core promoter-5′UTR and SV40 early polyA used in most UAS transgenic lines (6).
Table 1.
Components used in transgenic designs in Fig. 1B
| Construct | Core promoter-5′UTR | Intron | 3′UTR-PolyA |
| PHCH | hsp70 | — | his2Av |
| PHCS | hsp70 | — | SV40 early |
| PHCT | hsp70 | — | α-Tub |
| PCH | ppk | — | his2Av |
| PHsICH | hsp70 | Synthetic intron | his2Av |
| PsICH | ppk | Synthetic intron | his2Av |
| PHzICH | hsp70 | Zeste second intron | his2Av |
| PEsICH | eve | Synthetic intron | his2Av |
| PHzICS | hsp70 | Zeste second intron | SV40 early |
A limitation of CD8-GFP is the much weaker signals in thin terminal dendrites compared with those in the soma and thicker proximal dendrites, which is especially pronounced in neurons with extensive processes like C4da neurons (Fig. 1E). This weakness is possibly because of the presence of polar amino acid residues in the transmembrane domain (TM) of CD8, which may function as weak endoplasmic reticulum (ER) retention signals (20). We tried to solve this problem by improving both the plasma-membrane targeting of the marker and the brightness of the fluorescent tag. We chose the TM of human CD4 protein (21) as the membrane targeting sequence, as CD4 TM contains no polar residues and showed more efficient membrane delivery than CD8 (see below). To increase the brightness of the fluorescent marker, we made a tandem dimer GFP (tdGFP) and fused it with CD4 TM. To further reduce the accumulation in secretory compartments, we added an ER export signal from Kir2.1(22) to the C terminus of tdGFP. The final marker (referred to as CD4-tdGFP) is brighter than CD4-GFP in C4da neurons (Fig. 1C). More importantly, CD4-tagged markers, especially CD4-tdGFP, are distributed more evenly on dendrite membranes than CD8-GFP (Fig. 1D). As a result, the terminal dendrites are more readily detectable with CD4-tdGFP (Fig. 1F) than with CD8-GFP (Fig. 1E). The difference between CD8-GFP and CD4-tdGFP is even more obvious in the Drosophila CNS, where many neurons have very fine neuronal processes. For example, CD4-tdGFP labels significantly more neuronal processes than CD8-GFP in DOPA decarboxylase (DDC)-expressing neurons of the larval ventral nerve cord (Fig. 1 H and I) and tyrosine hydroxylase-expressing neurons of the larval brain (Fig. S2 A and B) (23). In addition, CD4-tdGFP is much more efficient than CD8-GFP in labeling membrane processes of two other cell types we examined, the basal filopodia of sensory-organ precursor cells of the notum (Fig. S2 C and D) (24) and apical microvilli, or cytonemes (25), of wing imaginal disk epithelia (Fig. S2 E and F). Based on the design of CD4-tdGFP, we also made a red membrane marker CD4-tdTomato (CD4-tdTom) (26), which also shows excellent brightness and even distribution on neuronal membranes (Fig. 1G). CD4-tdGFP and CD4-tdTom do not appear to be toxic to neurons, as high-level expression in C4da neurons does not alter the dendrite morphology.
High-Expression Membrane Markers for Labeling Neuronal Processes Driven by Tissue-Specific Enhancers.
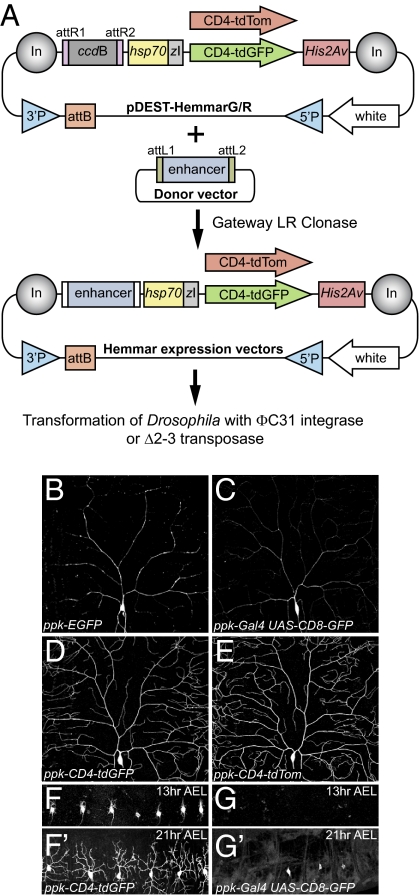
To build on these improved markers, we created two versatile cloning vectors, which we named pDEST-HemmarG/R (destination vectors with high expression membrane marker, green or red), for expressing CD4-tdGFP and CD4-tdTom in specific cell populations. By combining the attB/P-element dual-transformation platform, gypsy insulators, the HIH cassette, CD4 membrane markers, as well as the Gateway system, these vectors offer great flexibility in cloning enhancer elements and generating transgenic lines (Fig. 2A).
Fig. 2.
The Hemmar vectors for making enhancer-driven membrane markers. (A) Diagram of enhancer cloning and transgenesis with the Hemmar vectors. (B–E) Comparison of existing C4da reporters and new ones generated with Hemmar vectors. Neurons in B–D were imaged at identical settings. (F–G′) Comparison of expression timing of ppk-CD4-tdGFP (F and F′) and ppk-Gal4 UAS-CD8-GFP (G and G′) in embryos. DdaC neurons in A1-A7 (F and G) and A1-6 segments (F′ and G′) are shown.
The Hemmar vectors are suited for testing candidate enhancers and for generating additional insertions of selected reporters. To illustrate the versatility of these vectors, we performed P-mediated mobilization of ppk-CD4-tdGFP and ppk-CD4-tdTom and selected new insertions that offer stronger expression, which label the entire dendritic trees much more efficiently than ppk-EGFP (10) and ppk-Gal4 UAS-CD8-GFP (13) (Fig. 2 B–E). In contrast to binary systems, the direct enhancer-fusion strategy does not require expression of intermediate transcription factors, and therefore allows for much quicker expression of reporters. For example, ppk-CD4-tdGFP brightly labels newly extended dendrites of C4da neurons at 13 h after egg laying (AEL) (Fig. 2F), while the earliest detectable expression of ppk-Gal4 UAS-CD8-GFP is at 21 h AEL in some segments (Fig. 2G′).
Enhancer-Driven Membrane Markers for Studying Neuronal Projections and Connections of CNS Neurons.
Enhancer-driven membrane markers may be used for analyzing interactions of neurons with neighboring cells. In the presence of a cellular marker exclusively expressed in specific neurons, the influence of surrounding tissues and the roles of extrinsic factors can be studied when coupled with binary systems, such as Gal4/UAS, to manipulate tissues that may impact neural development. In such a setup, the cells interacting with the labeled neurons can be labeled by cellular marker expression, or manipulated by gene overexpression and misexpression, and gene silencing with dominant negative proteins or RNAi constructs. We call this type of analysis GEEM.
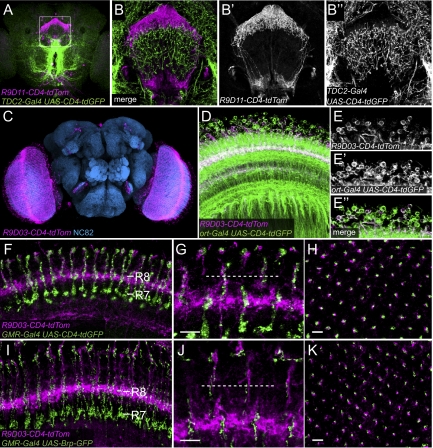
For example, one can simultaneously label two groups of neurons and determine whether their processes contact one another. For this purpose, we generated several CNS neuronal markers by using enhancers from a large collection established at the Janelia Farm Research Campus (27). These CNS enhancers are in Gateway donor vectors that are compatible with pDEST-HemmarG/R. These resulting CNS-CD4-tdTom reporters label discrete neuronal populations in larval and adult brains (Fig. 3 A–B′′ and Fig. S3), yielding readily detectable processes. In the same animal, the neurites from two groups of neurons can be labeled in different colors and the projections of these neurons can be simultaneously analyzed (Fig. 3 A–B′′ and Fig. S3 I–L′′). We further tried to identify the neuronal types and presynaptic partners of the neurons labeled by one marker, R9D03-CD4-tdTom, which labels many neurons in the medulla of the adult brain (Fig. 3C). We found that R9D03-CD4-tdTom overlaps with ort-Gal4 UAS-CD4-tdGFP in a group of medulla neurons (Fig. 3 D–E′′). ort-Gal4 labels histamine receptor neurons in medulla that directly receive visual information from color sensitive photoreceptors R7 and R8 (28). To test if R9D03 neurons form connections with R7/R8, we labeled the axon terminals and presynaptic sites of R7/R8 with GMR-Gal4 UAS-CD4-tdGFP and GMR-Gal4 UAS-Brp-GFP, respectively. R9D03 neurons show ramifications at the R8 termination layer (Fig. 3F) and their processes are closely associated with R8 axon terminals (Fig. 3 G and H). R8 synaptic compartments labeled by Brp-GFP are also in close proximity to R9D03 neuronal processes (Fig. 3 I–K), suggesting that R9D03 neurons likely receive inputs from photoreceptor R8. These data suggest that the GEEM method can be used to identify neuronal types and potential synaptic partners of neurons labeled by Hemmar reporters.
Fig. 3.
Analysis of neuronal projection and connection with enhancer-driven CNS neuronal markers. (A-B′′) R9D11-CD4-tdTom and TDC2-Gal4 UAS-CD4-tdGFP in the central brain (A) and fan-shaped body (B–B′′) (boxed in A) of the adult. (C) R9D03-CD4-tdTom (magenta) in an adult brain counter stained with NC82 (blue). (D) R9D03-CD4-tdTom (magenta) and ort-Gal4 UAS-CD4-tdGFP (green) in the medulla. (E–E′′) Overlap of R9D03 neurons and ort neurons in the medulla. (F) Projection of horizontal sections showing R9D03-CD4-tdTom (magenta) ramifications and axon terminals of R7/R8 labeled by GMR-Gal4 UAS-CD4-tdGFP (green) in outer medulla layers. The R7 and R8 termination layers are indicated. (G and H) High-magnification views showing R7/R8 axon terminals (green) and R9D03 neurites (magenta) in sections parallel (G) and perpendicular (H) to medulla columns. The broken line in G indicates the proximate position of the section shown in H. (I–K) Colabeling of R9D03 neurites (magenta) and R7/R8 synaptic sites labeled by GMR-Gal4 UAS-Brp-GFP (green) in outer medulla layers. I, J, and K are annotated similar to F, G, and H, respectively. (Scale bars, 5 μm.)
Analysis of the Wrapping of C4da Neurons by Glia During the Embryonic and Larval Development Using Enhancer-Driven Membrane Markers.
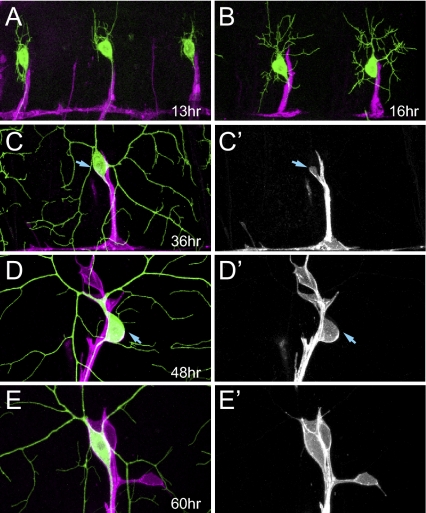
To demonstrate the usefulness of enhancer-driven membrane markers in analyzing the morphogenesis of the peripheral nervous system, we examined the association of glia with dorsal C4da neuron ddaC during development in live animals by using repo-Gal4 UAS-CD4-tdTom to label the glial membrane and ppk-CD4-tdGFP to label C4da neurons. By 13 h AEL, glia migrating dorsally have reached the proximal segment of ddaC axon (Fig. 4A). The dorsal tip of glia continues to extend until 16 h AEL (Fig. 4B). After this period of dynamic growth, the relative positions of the glial membrane and the ddaC neuron remain constant for the rest of embryonic development and during the early phase of first instar lava. Between 30 and 36 h AEL, glia begin to wrap the ddaC soma (Fig. 4 C and C′). By 55 to 60 h AEL, glia have completed wrapping the soma and begin to extend thin processes along proximal dendrites of ddaC (Fig. 4 E and E′). It thus appears that ddaC neurons can function without glial wrapping in the early larva.
Fig. 4.
(A–E′) The wrapping of ddaC somas by glia during embryonic and larval development. Glia are labeled with repo-Gal4 UAS-CD4-tdTom (magenta in merged images) and ddaC neurons are labeled with ppk-CD4-tdGFP (green in merged images). Animals are oriented anterior left, dorsal up. All times indicated are after egg laying. Arrows in C and C′ point to the glial membrane extended from the root of the ddaC axon. Arrows in D and D′ point to the boundary of the incomplete glial wrapping of the soma.
Analysis of Glia–Neuron Interactions During the Pruning of C4da Dendrites Using Enhancer-Driven Membrane Markers.
During metamorphosis, the Drosophila nervous system undergoes extensive remodeling; some neurons cull their processes before regrowing new ones and others die and are replaced by new neurons (29). The C4da neurons ddaC and v'ada survive during metamorphosis and remodel their entire dendritic arbors (13). To examine the role of glia in dendrite pruning of C4da neurons, we first analyzed the time course of glia-dendrite interaction during early phases of the metamorphosis. In early prepupae, glia not only wrap the soma of ddaC but also portions of proximal dendrites (Fig. 5 A–C′). By 12 h after puparium formation (APF), all proximal dendrites are severed from the soma and the dendrite segments previously wrapped by glia are degraded (Fig. S4 C and C′). By16 h APF, the glial wrap has retracted and begins to disintegrate, and new neurites start to extend from the soma of ddaC (Fig. 6 C and C′). By 18 h APF, the soma is completely exposed and more extensive dendritic arbors have formed (Fig. S4 D and D′).
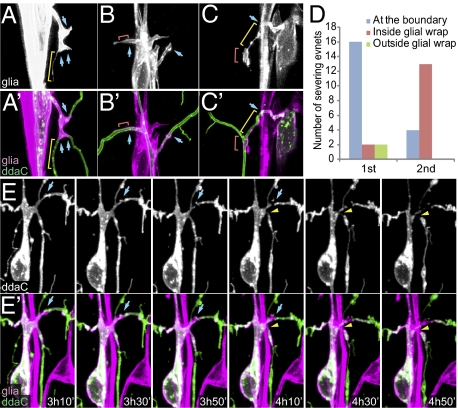
Fig. 5.
Correlation of glial wrapping and initial severing of proximal dendrites during dendrite pruning of ddaC. (A–C′) Glial wrapping of proximal dendrites of ddaC in white pupae. Blue arrows indicate the boundaries of complete glial wrapping. Yellow brackets indicate long, thin glial processes. Red brackets indicate glial membrane patches that partially wrap dendrites. (D) Locations of observed first and second dendritic severing points in relation to glial wrapping. (E and E′) Time-lapse images of a ddaC neuron showing the first and second severing events. Blue arrows indicate the first severing at a glial wrapping boundary and the yellow arrowheads indicate the second severing within the glial wrapping. In all panels, the glial membrane is labeled by repo-Gal4 UAS-CD4-tdTom and ddaC neurons are labeled by ppk-CD4-tdGFP.
Fig. 6.
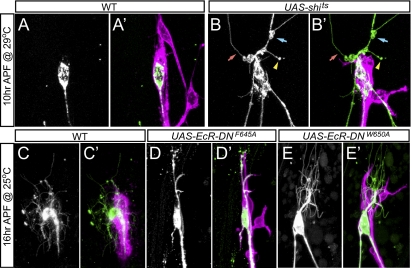
Glia are involved in determining the location of initial dendrite severing and degrading the wrapped dendrite segments. (A–B′) Dendrite pruning at 10 h APF in repo-Gal4 (A and A′) and repo-Gal4 UAS-shits (B andB′) after incubation at 29 °C from 3 to 10 h APF. Blue and red arrows indicate the swelling and thinning of proximal dendrites, respectively. The yellow arrowhead indicated a breaking point at an unwrapped dendrite fragment. (C–E′) Dendrite pruning at 16 h APF in repo-Gal4 (C and C′) and repo-Gal4 UAS-EcR-DN (D–E′) at 25 °C. In all panels, the glial membrane is labeled by repo-Gal4 UAS-CD4-tdTom and ddaC neurons are labeled by ppk-CD4-tdGFP.
A key step in dendrite pruning is the severing of proximal dendrites from the soma beginning at 4 to 6 h APF (12, 14, 30, 31). The observation that glia wrap proximal dendrites of C4da neurons in early prepupae prompted us to ask whether glial wrapping and dendrite severing are correlated. To address this question, we conducted time-lapse analyses to monitor the membrane dynamics of glia and ddaC during dendrite pruning. Consistent with a previous study (12), we found that the proximal dendrites are highly dynamic before severing and display various forms of constrictions and swellings (Fig. S4 A and A′ and Movie S1). In addition, we found that the proximal dendrites are first severed at the boundary of complete glial wrapping in most cases (80%, n = 20) (Fig. 5 D–E′ and Movie S1). Following the first severing event, the proximal dendrites were quickly degraded within the glial wrap (Fig. S4 B and B′ and Movie S1), typically resulting in the second severing within the glial wrap near the first severing point (76%, n = 17). To test whether glia play a role in determining the location of the initial severing and subsequent degradation of wrapped dendrite segments, we analyzed the effects of perturbing glial activities using two strategies. First, we expressed UAS-Shits (32), a temperature-sensitive allele of Drosophila Dynamin, in glia to block endocytosis at the restrictive temperature of 29 °C. In control animals, most ddaC neurons have proximal dendrites completely severed (80%, n = 50) and the wrapped dendrite segments degraded by 10 h APF at 29 °C (Fig. 6 A and A′). In contrast, in animals with Shits expression, most ddaC neurons showed no apparent dendrite severing at the glial wrapping boundary or within the wrap at 10 h APF (94%, n = 70) (Fig. 6 B and B′), despite the appearance of thinning and swelling of dendrites. Instead, proximal dendrites were often severed at unwrapped segments (43%, n = 70). Despite this delay of early steps of dendrite pruning, pruning is completed by 14 h APF in these animals (Fig. S4 F and F′), as in the wild-type (Fig. S4 E and E′). These data suggest that glial endocytosis is important for determining the location of the initial dendrite severing, possibly by mediating glia-dendrite communications.
Second, we blocked the ecdysone signaling in glia by expressing dominant-negative ecdysone receptors (EcR-DN) (33). In these animals, dendrite severing occurred, albeit with some delay. However, the glial membrane that wrapped the proximal dendrites showed very little sign of retraction or disintegration by 16 h APF, and all ddaC neurons examined had the wrapped dendrite segments intact (n = 54) (Fig. 6 D–E′). This defect persisted until the death of these EcR-DN–expressing animals at various stages of metamorphosis. These data suggest that ecdysone signaling in glia play a crucial role in promoting the degradation of the wrapped dendrites in pruning of C4da neuron dendrites.
Our time-lapse analyses further revealed that, throughout pruning, C4da dendrites shed membrane vesicles through bulging and extensions of dendrite surfaces (Fig. S5A). The shedding of these “shedosomes” starts shortly after pupariation and lasts until the complete fragmentation of all dendrites. At proximal dendrites wrapped by glia, the attached glial wrap that accompanies shedosomes was degraded first (Fig S5 B–B′′). We interpret the shedding of dendritic membranes as a means to reduce membrane contents.
Together, our GEEM analyses revealed the dynamic interactions between glia and C4da neurons during dendrite pruning and the critical roles of glia in determining the location of initial dendrite severing and degrading the wrapped dendrite segments.
Discussion
Despite the abundant genetic tools available for studying gene functions within neurons, we have limited means for dissecting the in vivo roles of extrinsic factors in neural development. GEEM analysis provides a simple yet powerful solution by allowing for gain-of-function and loss-of-function analysis in any tissue with concurrent examination of the effects on relevant neurons. The numerous Gal4 drivers and UAS transgenes, and the recent establishment of several comprehensive transgenic RNAi libraries (National Institute of Genetics, the Vienna Drosophila RNAi Center, and Transgenic RNAi Project) make this strategy especially suited for large scale gain-of-function and loss-of-function screens for candidate genes involved in the morphogenesis of specific neurons.
Gene manipulation can also be combined with independent neuronal labeling by using dual binary systems. There are several advantages of the GEEM method based on enhancer-driven neuronal markers. First, with the versatility of the single-component system, neuronal markers based on Hemmar vectors offer robust expression at levels similar to or higher than those using Gal4/UAS with traditional designs. Second, enhancer-driven markers avoid the lag time in transgene expression associated with binary systems, which could be problematic for neuronal labeling in early development. Third, binary systems show inherent limitations under some circumstances. For example, QF may be toxic when highly expressed (9). The LexA/LexAop system has higher rate of nonspecific transgene induction (9). The enhancer-fusion strategy circumvents these potential problems. During the past decade, mosaic-based strategies, such as dual-expression control MARCM (mosaic analysis by repressible cell marker) (7), twin-spot MARCM (34), twin-spot generator (35), and coupled MARCM (9), have been devised for simultaneous labeling of two different populations of cells. Although offering great value for lineage analysis and neuronal study at single-cell resolution, these systems can only label cell populations derived from the same progenitors, limiting their use as a general approach for studying nonautonomous regulation of neural development.
In this study, we provide three examples of GEEM analysis. First, we simultaneously labeled two groups of neurons using CD4 membrane markers, which allowed us to determine the identity of the neurons labeled by an enhancer-driven marker and to identify their potential synaptic partners. Second, we examined the wrapping of ddaC neurons by glia during embryonic and larval development. Third, we explored the role of glia–neuron interactions in dendrite pruning of da neurons. In addition, the improved membrane markers have allowed us to identify shedomsomes as a mechanism of dendrite destruction. Shedosomes bear a resemblance to the “axosomes” produced by retracting axons during synapse elimination (36). Although axosomes are formed by engulfment of axon tips by Schwann cells, it is yet to be determined whether nonneural tissues participate in the formation of shedosomes.
Key to the GEEM analysis is the high transgene expression conferred by Hemmar vectors, which can be modified to express other cellular markers and functional reporters for studying nonautonomous mechanisms, including markers localized in specific cellular compartments, GFP reconstitution across synaptic partners (21), and genetically encoded calcium indicators, like GCaMP3 (37). Whereas the examples provided here concern neural development, GEEM is applicable to the study of nonautonomous, as well as autonomous, regulations of any cell type.
The transgene design used in Hemmar vectors provides a general strategy for high expression of transgenes, as the UAS-CD4-tdGFP cloned with a modified pUAST vector, pACU2, which contains the HIH cassette, shows reliably higher expression than the UAS-CD8-GFP cloned with pUAST. It is possible to modify the Hemmar vectors to express other functional proteins, such as Gal4, Gal80, QS, QF, and LexA. Improvements of current tools based on these unique designs will likely provide researchers with more freedom in probing challenging biological questions.
Methods
See SI Methods for details of methods for plasmid construction, fly strains, live imaging, quantitative analysis, and immunohistochemistry. See Table S1 for PCR fragments used for plasmid construction. The pACU2 and pDEST-Hemmar vectors are available from Addgene. UAS-CD4-tdGFP and UAS-CD4-tdTom lines are available from the Bloomington Stock Center.
Supplementary Material
Acknowledgments
We thank J. W. Posakony, J. Bischof, C. Bargmann, G. Rubin, and the Vienna Drosophila RNAi Center for plasmids; K. Venken, H. J. Bellen, G. Rubin, E. Buchner, C. H. Lee, W Song, and the Bloomington Drosophila Stock Center for fly lines; the Developmehtal Studies Hybridoma Bank for antibodies; J. Smyth, R. Shaw, T. Chen, and Q. Yuan for technical help; and M. Klassen, P. Soba, J. Wildonger, Q. Yuan, and S. Zhu for critical reading of the manuscript. This work was supported by a postdoctoral fellowship from the Jane Coffin Childs Memorial Fund (to C.H.) and by National Institutes of Health Grant 2R37NS040929 (to Y.-N.J.). L.Y.J. and Y.-N.J. are investigators of The Howard Hughes Medical Institute.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1106386108/-/DCSupplemental.
References
- 1.Guillemot F. Cellular and molecular control of neurogenesis in the mammalian telencephalon. Curr Opin Cell Biol. 2005;17:639–647. doi: 10.1016/j.ceb.2005.09.006. [DOI] [PubMed] [Google Scholar]
- 2.Marmigère F, Ernfors P. Specification and connectivity of neuronal subtypes in the sensory lineage. Nat Rev Neurosci. 2007;8(2):114–127. doi: 10.1038/nrn2057. [DOI] [PubMed] [Google Scholar]
- 3.Chotard C, Salecker I. Neurons and glia: Team players in axon guidance. Trends Neurosci. 2004;27:655–661. doi: 10.1016/j.tins.2004.09.001. [DOI] [PubMed] [Google Scholar]
- 4.Jan YN, Jan LY. Branching out: Mechanisms of dendritic arborization. Nat Rev Neurosci. 2010;11:316–328. doi: 10.1038/nrn2836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Margeta MA, Shen K. Molecular mechanisms of synaptic specificity. Mol Cell Neurosci. 2010;43:261–267. doi: 10.1016/j.mcn.2009.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brand AH, Perrimon N. Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development. 1993;118:401–415. doi: 10.1242/dev.118.2.401. [DOI] [PubMed] [Google Scholar]
- 7.Lai SL, Lee T. Genetic mosaic with dual binary transcriptional systems in Drosophila. Nat Neurosci. 2006;9:703–709. doi: 10.1038/nn1681. [DOI] [PubMed] [Google Scholar]
- 8.Gordon MD, Scott K. Motor control in a Drosophila taste circuit. Neuron. 2009;61:373–384. doi: 10.1016/j.neuron.2008.12.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Potter CJ, Tasic B, Russler EV, Liang L, Luo L. The Q system: A repressible binary system for transgene expression, lineage tracing, and mosaic analysis. Cell. 2010;141:536–548. doi: 10.1016/j.cell.2010.02.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Grueber WB, Ye B, Moore AW, Jan LY, Jan YN. Dendrites of distinct classes of Drosophila sensory neurons show different capacities for homotypic repulsion. Curr Biol. 2003;13:618–626. doi: 10.1016/s0960-9822(03)00207-0. [DOI] [PubMed] [Google Scholar]
- 11.Yamamoto M, Ueda R, Takahashi K, Saigo K, Uemura T. Control of axonal sprouting and dendrite branching by the Nrg-Ank complex at the neuron-glia interface. Curr Biol. 2006;16:1678–1683. doi: 10.1016/j.cub.2006.06.061. [DOI] [PubMed] [Google Scholar]
- 12.Williams DW, Truman JW. Cellular mechanisms of dendrite pruning in Drosophila: Insights from in vivo time-lapse of remodeling dendritic arborizing sensory neurons. Development. 2005;132:3631–3642. doi: 10.1242/dev.01928. [DOI] [PubMed] [Google Scholar]
- 13.Kuo CT, Jan LY, Jan YN. Dendrite-specific remodeling of Drosophila sensory neurons requires matrix metalloproteases, ubiquitin-proteasome, and ecdysone signaling. Proc Natl Acad Sci USA. 2005;102:15230–15235. doi: 10.1073/pnas.0507393102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Williams DW, Kondo S, Krzyzanowska A, Hiromi Y, Truman JW. Local caspase activity directs engulfment of dendrites during pruning. Nat Neurosci. 2006;9:1234–1236. doi: 10.1038/nn1774. [DOI] [PubMed] [Google Scholar]
- 15.Groth AC, Fish M, Nusse R, Calos MP. Construction of transgenic Drosophila by using the site-specific integrase from phage phiC31. Genetics. 2004;166:1775–1782. doi: 10.1534/genetics.166.4.1775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Venken KJ, He Y, Hoskins RA, Bellen HJ. P[acman]: A BAC transgenic platform for targeted insertion of large DNA fragments in D. melanogaster. Science. 2006;314:1747–1751. doi: 10.1126/science.1134426. [DOI] [PubMed] [Google Scholar]
- 17.Roseman RR, Pirrotta V, Geyer PK. The su(Hw) protein insulates expression of the Drosophila melanogaster white gene from chromosomal position-effects. EMBO J. 1993;12:435–442. doi: 10.1002/j.1460-2075.1993.tb05675.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Markstein M, Pitsouli C, Villalta C, Celniker SE, Perrimon N. Exploiting position effects and the gypsy retrovirus insulator to engineer precisely expressed transgenes. Nat Genet. 2008;40:476–483. doi: 10.1038/ng.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Grueber WB, Jan LY, Jan YN. Tiling of the Drosophila epidermis by multidendritic sensory neurons. Development. 2002;129:2867–2878. doi: 10.1242/dev.129.12.2867. [DOI] [PubMed] [Google Scholar]
- 20.Letourneur F, Cosson P. Targeting to the endoplasmic reticulum in yeast cells by determinants present in transmembrane domains. J Biol Chem. 1998;273:33273–33278. doi: 10.1074/jbc.273.50.33273. [DOI] [PubMed] [Google Scholar]
- 21.Feinberg EH, et al. GFP Reconstitution Across Synaptic Partners (GRASP) defines cell contacts and synapses in living nervous systems. Neuron. 2008;57:353–363. doi: 10.1016/j.neuron.2007.11.030. [DOI] [PubMed] [Google Scholar]
- 22.Ma D, et al. Role of ER export signals in controlling surface potassium channel numbers. Science. 2001;291:316–319. doi: 10.1126/science.291.5502.316. [DOI] [PubMed] [Google Scholar]
- 23.Vomel M, Wegener C. Neuroarchitecture of aminergic systems in the larval ventral ganglion of Drosophila melanogaster. PLoS One. 2008;3 doi: 10.1371/journal.pone.0001848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cohen M, Georgiou M, Stevenson NL, Miodownik M, Baum B. Dynamic filopodia transmit intermittent Delta-Notch signaling to drive pattern refinement during lateral inhibition. Dev Cell. 2010;19(1):78–89. doi: 10.1016/j.devcel.2010.06.006. [DOI] [PubMed] [Google Scholar]
- 25.Ramírez-Weber FA, Kornberg TB. Cytonemes: Cellular processes that project to the principal signaling center in Drosophila imaginal discs. Cell. 1999;97:599–607. doi: 10.1016/s0092-8674(00)80771-0. [DOI] [PubMed] [Google Scholar]
- 26.Shaner NC, et al. Improved monomeric red, orange and yellow fluorescent proteins derived from Discosoma sp. red fluorescent protein. Nat Biotechnol. 2004;22:1567–1572. doi: 10.1038/nbt1037. [DOI] [PubMed] [Google Scholar]
- 27.Pfeiffer BD, et al. Tools for neuroanatomy and neurogenetics in Drosophila. Proc Natl Acad Sci USA. 2008;105:9715–9720. doi: 10.1073/pnas.0803697105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gao S, et al. The neural substrate of spectral preference in Drosophila. Neuron. 2008;60:328–342. doi: 10.1016/j.neuron.2008.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Truman JW. Metamorphosis of the central nervous system of Drosophila. J Neurobiol. 1990;21:1072–1084. doi: 10.1002/neu.480210711. [DOI] [PubMed] [Google Scholar]
- 30.Lee HH, Jan LY, Jan YN. Drosophila IKK-related kinase Ik2 and Katanin p60-like 1 regulate dendrite pruning of sensory neuron during metamorphosis. Proc Natl Acad Sci USA. 2009;106:6363–6368. doi: 10.1073/pnas.0902051106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kuo CT, Zhu S, Younger S, Jan LY, Jan YN. Identification of E2/E3 ubiquitinating enzymes and caspase activity regulating Drosophila sensory neuron dendrite pruning. Neuron. 2006;51:283–290. doi: 10.1016/j.neuron.2006.07.014. [DOI] [PubMed] [Google Scholar]
- 32.Kitamoto T. Conditional modification of behavior in Drosophila by targeted expression of a temperature-sensitive shibire allele in defined neurons. J Neurobiol. 2001;47(2):81–92. doi: 10.1002/neu.1018. [DOI] [PubMed] [Google Scholar]
- 33.Cherbas L, Hu X, Zhimulev I, Belyaeva E, Cherbas P. EcR isoforms in Drosophila: Testing tissue-specific requirements by targeted blockade and rescue. Development. 2003;130:271–284. doi: 10.1242/dev.00205. [DOI] [PubMed] [Google Scholar]
- 34.Yu HH, Chen CH, Shi L, Huang Y, Lee T. Twin-spot MARCM to reveal the developmental origin and identity of neurons. Nat Neurosci. 2009;12:947–953. doi: 10.1038/nn.2345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Griffin R, et al. The twin spot generator for differential Drosophila lineage analysis. Nat Methods. 2009;6:600–602. doi: 10.1038/nmeth.1349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bishop DL, Misgeld T, Walsh MK, Gan WB, Lichtman JW. Axon branch removal at developing synapses by axosome shedding. Neuron. 2004;44:651–661. doi: 10.1016/j.neuron.2004.10.026. [DOI] [PubMed] [Google Scholar]
- 37.Tian L, et al. Imaging neural activity in worms, flies and mice with improved GCaMP calcium indicators. Nat Methods. 2009;6:875–881. doi: 10.1038/nmeth.1398. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.