Abstract
We describe a simple method of using rolling circle amplification to amplify vector DNA such as M13 or plasmid DNA from single colonies or plaques. Using random primers and φ29 DNA polymerase, circular DNA templates can be amplified 10,000-fold in a few hours. This procedure removes the need for lengthy growth periods and traditional DNA isolation methods. Reaction products can be used directly for DNA sequencing after phosphatase treatment to inactivate unincorporated nucleotides. Amplified products can also be used for in vitro cloning, library construction, and other molecular biology applications.
A fundamental requirement for molecular biology is the isolation and amplification of specific DNA sequences. Target sequences are typically inserted into circular vectors, propagated in a biological host, and isolated by physical methods (Sambrook et al. 1989). However, such methods are laborious, costly, and not amenable to high-density formats. PCR is also used to amplify defined sequences, but can introduce sequence errors and is limited to amplification of short DNA segments (Innis et al. 1990).
In nature, the replication of circular DNA molecules such as plasmids or viruses frequently occurs via a rolling circle mechanism (Kornberg and Baker 1992). As a laboratory method, linear rolling circle amplification (RCA) (Fire and Xu 1995; Liu et al. 1996; Lizardi et al. 1998) is the prolonged extension of an oligonucleotide primer annealed to a circular template DNA. A continuous sequence of tandem copies of the circle is synthesized. RCA has the advantage of not requiring a thermal cycling instrument. Two primers are used to perform exponential (or hyperbranched) RCA, one for each strand (Lizardi et al. 1998). A cascade of strand displacement reactions results in an exponential amplification.
Previously, RCA had been used to amplify small DNA circles approximately 100 nt in length. However, the rate for plasmid-sized targets is only about 20 copies per hour, limiting the usefulness with plasmids or other circles larger than 0.2 kb. We describe here a technique called multiply-primed rolling circle amplification (multiply-primed RCA) that uses the unique properties of φ29 DNA polymerase and random primers to achieve a 10,000-fold amplification. This robust process allows amplification of circular DNA directly from cells or plaques, generating high-quality template for use in DNA sequencing, probe generation, or cloning. The method is simple and is optimally performed at 30°C, making it suitable for a variety of applications. This will make it attractive for high-throughput processes and 384-well formats.
RESULTS
Increased Yield in RCA Using Random Hexamer Primers
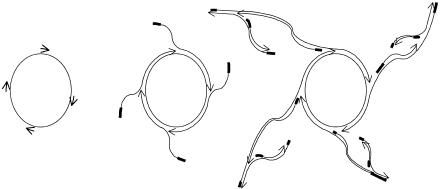
In multiply-primed RCA, the use of multiple primers annealed to a circular template DNA generates multiple replication forks (Fig. 1). RCA proceeds by displacing the nontemplate strand. In this way, product strands are “rolled off” of the template as tandem copies of the circle. Random priming allows synthesis of both strands, resulting in double-stranded product. A cascade of priming events results in exponential (or hyperbranched) amplification.
Figure 1.
Scheme for multiply-primed rolling circle amplification. Oligonucleotide primers complementary to the amplification target circle are hybridized to the circle. The 3′ ends of the DNA strands are indicated by arrowheads to show the polarity of polymerization. Thickened lines indicate the location of the original primer sequences within the product strands. The addition of DNA polymerase and deoxynucleoside triphosphates (dNTPs) to the primed circle results in the extension of each primer, and displacement of each newly synthesized strand results from elongation of the primer behind it. Secondary priming events can subsequently occur on the displaced product strands of the initial rolling circle amplification step. It is possible that the last intermediate shown in the figure makes only a small contribution to the overall yield of amplified DNA.
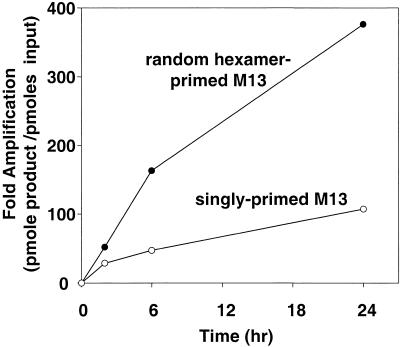
φ29 DNA polymerase was chosen because of its capacity to perform strand displacement DNA synthesis for more than 70,000 nt without dissociating from the template (Blanco et al. 1989) and its stability, which allows efficient DNA synthesis to continue for many hours. Random priming of M13 single-stranded DNA gave significantly more DNA synthesis compared with a single specific primer (Fig. 2). Pyrophosphatase was added to the reaction to eliminate the inhibitory accumulation of pyrophosphate. The M13 DNA was amplified 375-fold in 24 h by using random hexamers.
Figure 2.
Comparison of amplification efficiency between singly- and randomly-primed M13 single-strand circular DNA. Rolling circle reactions were performed as described and contained either 1 ng of singly-primed single-strand M13 DNA or 1 ng of random hexamer-primed single-strand M13 DNA. Reactions were incubated at 34°C for 24 h and aliquots were taken to measure DNA synthesis as indicated.
Amplification of Plasmid and Bacteriophage DNA from Colonies or Plaques
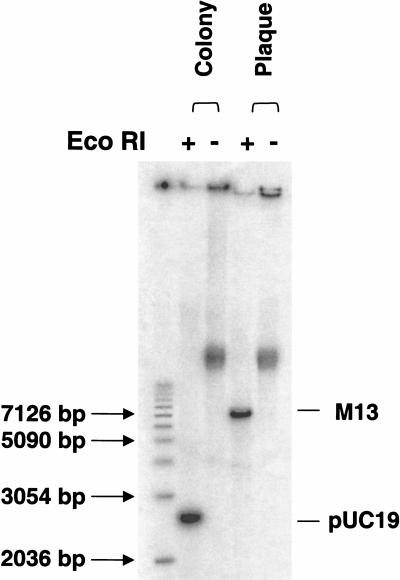
A small amount of material from bacterial colonies or plaques was picked and heated in the presence of random hexamer primers, as described. The heating step inactivates nucleases, releases the plasmid or phage DNA from cells or phage particles, and denatures the DNA, allowing primer annealing. Amplification was performed in the presence of radioactively labeled deoxycytidine triphosphate (dCTP) to quantify DNA synthesis and to visualize the reaction products after gel electrophoresis. Cleavage of the amplification products by restriction endonuclease EcoRI gave linear 7.2-kb (M13) and 2.7-kb double-stranded (pUC19) DNA fragments, demonstrating that the amplification product was indeed tandem repeats of the target (Fig. 3). No DNA products were observed by using colonies that did not contain plasmid DNA (not shown).
Figure 3.
Amplification of pUC19 from colonies and M13mp19 from plaques. Amplification reactions were performed as indicated directly on material picked from a colony or a plaque. Half of the radioactively labeled reaction products were cleaved with EcoRI and both cleaved and uncleaved samples were analyzed by agarose gel electrophoresis. The positions of linear, duplex M13, and pUC19 DNA are indicated.
Approximately 80%of the multiply-primed RCA products on the gel were converted to the linear form by EcoRI digestion. This high yield of specific product occurred in spite of the presence of an excess of bacterial genomic DNA over plasmid DNA. For 200 copies of pUC19, genomic DNA would be in an eightfold excess in the cell. Therefore, in addition to being efficient, amplification of the circular vector DNA was highly selective. The short heating step may not release the bacterial DNA from its association with the bacterial membrane (Kornberg and Baker 1992), leaving it less accessible for amplification than the more easily released pUC19 DNA. In addition, a plasmid DNA would be copied with orders of magnitude greater frequency than the bacterial chromosome during the linear RCA phase of the amplification. Finally, the large chromosomal DNA would have a greater likelihood of acquiring RCA-terminating nicks than would a small vector DNA.
Exonuclease-Resistant Random Primers Increase Amplification by φ29 DNA Polymerase
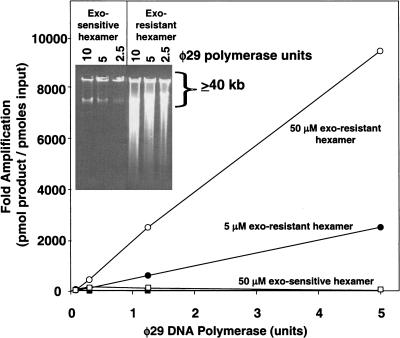
The degradation of primers by the proofreading, 3′-5′ exonuclease activity of φ29 DNA polymerase reduces yields. Primers resistant to degradation were used to prolong the reaction and allow the use of higher concentrations of DNA polymerase. Exonuclease-resistant (exo-resistant) random-hexamer primers were made by using thiophosphate linkages for the two 3′ terminal nucleotides (5′-NpNpNpNpsNpsN-3′). With exo-resistant primers, RCA yield increased linearly when using up to five units of φ29 DNA polymerase (Fig. 4). Up to a 10,000-fold amplification was achieved starting with 1 ng of M13 template. This compared favorably with the 375-fold amplification observed by using exo-sensitive primers (Fig. 2). It corresponded to an amplification rate of ∼800 copies per hour, a 40-fold improvement over the rate of 20 copies per hour achieved with linear RCA. No amplification was seen in the absence of added primer. Analysis by agarose gel electrophoresis (Fig. 4, inset) confirmed the improvement in yield using exo-resistant primers and showed the average product length to be greater than 40 kb. These results are consistent with the observation that, in assays containing more than 0.3 units of φ29 DNA polymerase per nanogram of DNA using unmodified primers, the amplification yield was decreased (Fig. 4) and primers are degraded (data not shown).
Figure 4.
Effect of exonuclease-resistant random primers on amplification. Annealing reactions (20 μL) were performed as described except that the template DNA was M13 double-strand RF DNA and the primers were either exo-resistant or exo-sensitive hexamers. Reactions contained 1 ng of M13 DNA with exo-resistant or exo-sensitive primers and φ29 DNA polymerase as indicated. Reactions were incubated at 34°C for 13 h. DNA synthesis was quantitated by the incorporation of radioactive nucleotide.
DNA Sequencing by Using Template Amplified by RCA with Random Hexamer Primers
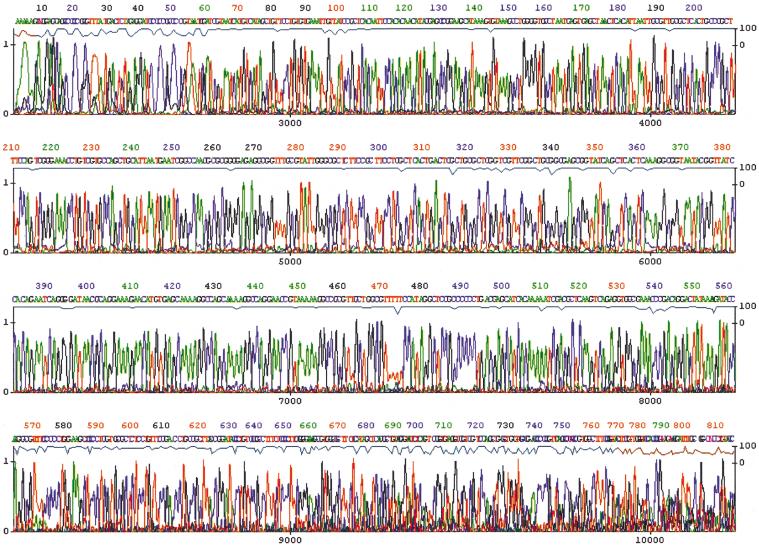
DNA from a saturated culture of XL1-blue transformed with a plasmid from a DNA library was amplified directly by using thiophosphate-modified random hexamers and φ29 DNA polymerase. Amplification products were treated with calf intestinal alkaline phosphatase to dephosphorylate remaining dNTPs. The sample was heated to inactivate the enzymes and used directly as a template for DNA sequencing (Fig. 5). The random hexamer used in the amplification does not need to be removed before sequencing, presumably because it does not anneal at the elevated temperatures used in cycle-sequencing reactions. The quality and read length of the sequence was indistinguishable from that obtained with ∼100 ng DNA template purified by standard methods. Similar results were obtained by using bacterial colonies in place of saturated cultures (data not shown).
Figure 5.
Sequencing of DNA amplified from a saturated bacterial culture. A saturated culture of XL1-blue (Stratagene) containing a random library plasmid (2- to 3-kb inserts in pUC18, 2 μL) was amplified as described for 12 h (see Methods). The amplified DNA was treated with calf intestine phosphatase, heated to 95° for 3 min, and sequenced by using the DYEnamic ET terminator sequencing kit and run on a MegaBACE 1000 DNA sequencer.
DISCUSSION
For genome sequencing, there is a need for methods to amplify circular templates of variable insert size, in a high-density, automated format. We describe a rapid, scalable method for the in vitro amplification of circular DNA molecules that uses multiply-primed RCA. An important factor for the success of this method is the unique nature of φ29 DNA polymerase: This single subunit, proofreading DNA polymerase, is able to incorporate >70,000 nt per binding event (Blanco et al. 1989). It has excellent strand displacement activity and is very stable, with linear reaction kinetics at 30°C for over 12 h. The use of random hexamer primers with 3′ thiophosphate-protected ends is also important, allowing circular DNA molecules to be amplified at least 10,000-fold by protecting the primers from the 3′ exonuclease activity of φ29 DNA polymerase. To achieve amplification, φ29 DNA polymerase appears to initiate multiple replication forks on each circle and to perform an exponentially cascading strand displacement amplification.
Multiply-primed RCA is an improvement over linear RCA in allowing an increased rate of synthesis and yield. In practice, conventional linear RCA and exponential RCA have been limited to small circles of <200 nt in circumference. The single primers used in linear RCA can only provide for DNA synthesis at a rate of ∼50 nt/sec from each circle, requiring several hours to achieve maximal 50-fold amplification for a 7250-nt M13 circle (Fig. 2). In contrast, the method described here yields several thousand-fold amplification (Fig. 4). Multiply-primed RCA with φ29 DNA polymerase also has the benefit of generating double-stranded products, allowing subsequent DNA sequencing of either strand, restriction endonuclease digestion, and other methods used in cloning, labeling, and detection.
Of note, multiply-primed RCA with φ29 DNA polymerase effectively amplified even large DNA circles such as bacterial artificial chromosomes (BACs) and cosmids (not shown). Unlike PCR, this method does not appear to be limited by target length. φ29 DNA polymerase readily synthesizes DNA strands of ∼0.5 Mb in length (Baner et al. 1998). Additional studies are underway with circular microbial genomes to define the upper limit of circle amplification with this method.
A simple amplification protocol was developed to use multiply-primed RCA with φ29 DNA polymerase to prepare templates for DNA sequencing directly from colonies, plaques, or liquid cultures. This involves heat lysis of the organism containing the DNA of interest, an isothermal amplification step, and either dilution or phosphatase treatment to reduce or eliminate remaining dNTPs. We used this method to generate sequence data in <6 h starting with 4 μL of saturated bacterial culture. Because amplification from colonies and plaques is efficient, overnight liquid cultures can be eliminated. Of immediate utility for genome sequencing centers would be compatibility of this method with high-density robotic formats, potentially eliminating a significant bottleneck in production sequencing. Initial results using this technique in a 96-well, high-throughput format on saturated cultures from a plasmid library gave an average read length of 553 bases in at least 80% of the samples by using the DYEnamic ET terminator reagent kit with a MegaBACE 1000.
Multiply-primed RCA will have additional applications. With an error rate of 1 in 106–107 bases (Esteban et al. 1993), φ29 DNA polymerase will be useful for amplification of genomic DNA or cDNA. Amplification of genomic DNA may require some additional DNA preparation steps because simple boiling of bacterial cells did not effectively release the chromosomal DNA for use as a template (Fig. 3). Multiply-primed RCA is suitable for amplification of mitochondrial DNA and microbial genomes ranging up to 6.5 Mb in length (T. Hawkins, pers. comm.). Mitochondrial DNA was amplified directly from whole cells, bypassing the conventional approach of isolating mitochondrial DNA by CsCl gradient centrifugation. The amplification of microbial genomes offers the prospect of obtaining DNA sequencing templates from unculturable organisms. The use of RCA may eliminate the need for cellular hosts to propagate DNA or cDNA targets and libraries. This method also holds promise for in vitro propagation of unclonable circular templates.
METHODS
DNA and Enzymes
M13mp19 single-stranded viral (+) strand and double-stranded replicative form (RF) DNA were from Life Technologies; random hexamers were from Amersham Pharmacia Biotech; thiophosphate-modified random hexamer (5′-NpNpNpNpsNpsN-3′) was from Molecular Staging or the Keck oligonucleotide synthesis facility, Yale University School of Medicine; and φ29 DNA polymerase was from Amersham Pharmacia Biotech. Yeast pyrophosphatase was from Boehringer-Mannheim.
Primed M13 DNA Templates
For preparation of singly-primed M13 DNA, 50 pmoles primer (5′ TCT GTT TAT AGG GCC TCT TCG CTA TTA CGC CAG C 3′) and 2.75 pmoles (6.5 μg) of single-strand M13mp19 circles were annealed in 100 μL of 20 mM Tris-HCl (pH 7.5) and 40 mM NaCl. For preparation of random hexamer-primed M13 DNA, annealing reactions (60 μL) contained 20 mM Tris-HCl (pH 7.5), 20 mM KCl, 0.1 mM ethylenediaminetetraacetic acid (EDTA), 15 fmoles (35 ng) of single-strand M13 circles, and 6000 pmoles random hexamer. Reactions were heated to 95°C for 1 min and cooled slowly to room temperature over 30 min.
Rolling Circle Reactions
Twenty-microliter reactions at 34°C contained 50 mM Tris-HCl (pH 7.5); 10 mM MgCl2; 20 mM ammonium sulfate; 5% glycerol; 200 μg/mL bovine serum albumin; 1 mM each dNTP, α-[32P] dCTP, 67 cpm/pmol total dNTPs; 0.02 units yeast pyrophosphatase; and 0.3 units φ29 DNA polymerase unless otherwise indicated. Incorporation of acid precipitable radioactive deoxyribonucleotide was determined with cut glass fiber filters. Fold amplification equals pmoles dNTP incorporation/pmoles nucleotide input M13 DNA.
Amplification of pUC19 from Colonies and M13mp19 from Plaques
Polyethylene tubing (Intramedic, PE20, 1.09-mm outer diameter) 1 cm in length was stabbed into a colony of Escherichia coli transformed with plasmid pUC19 or a plaque of bacteriophage M13mp19 in a lawn of E. coli (XL1-blue, Stratagene). The tubing was then placed into a thermocycler tube (200 μL) containing 20 μL of 20 mM Tris-HCl (pH 7.5), 40 mM NaCl, and 1 mM EDTA. Random hexamer primer was added to a final concentration of 50μM, heated to 95°C for 3 min, and cooled slowly to room temperature over 30 min. Reactions were brought to a final volume of 40 μL containing 0.6 units φ29 DNA polymerase and 0.04 units yeast pyrophosphatase, with reaction conditions as described for RCA. Reactions were incubated at 37°C for 8 h. Reaction products were digested with EcoRI, electrophoresis through an agarose gel (1.0%, Tris-borate-EDTA buffer), and analyzed with a Storm 860 PhosphorImager (Amersham Pharmacia Biotech).
Amplification of DNA for Use in DNA Sequencing
A saturated culture of XL1-blue, containing a random library plasmid (2- to 3-kb inserts in pUC18, 2 μL) was added to 8 μL of Tris-EDTA buffer and heated to 95°C for 3 min. Amplification premix (10μL) was added, yielding a final concentration of 50 mM Tris-HCl (pH 8.2), 5 mM MgCl2, 75 mM KCl, 0.1 mM dithiothreitol, 100 pmoles (5μM final concentration) thiophosphate protected random hexamer, 5 units φ29 DNA polymerase, 0.03 units yeast pyrophosphatase, and 0.1 mM dNTPs. Reactions were 30°C for 12 h. Reaction products were treated with calf intestine alkaline phosphatase (Amersham Pharmacia Biotech) at 37°C for 30 min and then heated to 95°C for 3 min. A 4-μL aliquot was used as a template for sequencing by using 5 pmoles of universal primer and a DYEnamic ET terminator sequencing kit (Amersham Pharmacia Biotech). The reaction was then ethanol precipitated and run on a MegaBACE 1000 DNA sequencer (Amersham Pharmacia Biotech) at 9 kV for 110 min.
Acknowledgments
We thank Rajanikanta Bandaru and Debra Itzkowitz for synthesis of oligonucleotides, the United States Department of Energy for BAC library clones, Qiuling Zong, Zhenyu Sun, and Alvaro Gordon-Escobar for technical assistance, and Carl Fuller, Trevor Hawkins, Stephen Kingsmore, and the reviewers for improvements to the manuscript.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL rogerl@molecularstaging.com; FAX (203) 776-5276.
Article and publication are at www.genome.org/cgi/doi/10.1101/gr.180501.
REFERENCES
- Baner J, Nilsson M, Mendel-Hartvig M, Landegren U. Signal amplification of padlock probes by rolling circle replication. Nucleic Acids Res. 1998;26:5073–5078. doi: 10.1093/nar/26.22.5073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanco L, Bernad A, Lazaro JM, Martin G, Garmendia C, Salas M. Highly efficient DNA synthesis by the phage phi 29 DNA polymerase. Symmetrical mode of DNA replication. J Biol Chem. 1989;264:8935–8940. [PubMed] [Google Scholar]
- Esteban JA, Salas M, Blanco L. Fidelity of Phi29 DNA polymerase. Comparison between protein-primed initiation and DNA polymerization. J Biol Chem. 1993;268:2719–2726. [PubMed] [Google Scholar]
- Fire A, Xu SQ. Rolling replication of short DNA circles. Proc Natl Acad Sci. 1995;92:4641–4645. doi: 10.1073/pnas.92.10.4641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Innis MA, Gelfand DH, Sninsky JJ, editors. PCR protocols. San Diego: Academic Press; 1990. [Google Scholar]
- Kornberg A, Baker TA. DNA replication. San Francisco: W.H. Freeman and Company; 1992. [Google Scholar]
- Liu D, Daubendiek SL, Zillman MA, Ryan K, Kool ET. Rolling circle DNA synthesis: Small circular oligonucleotides as efficient templates for DNA polymerases. J Am Chem Soc. 1996;118:1587–1594. doi: 10.1021/ja952786k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lizardi PM, Huang X, Zhu Z, Bray-Ward P, Thomas DC, Ward DC. Mutation detection and single-molecule counting using isothermal rolling-circle amplification. Nat Genet. 1998;19:225–232. doi: 10.1038/898. [DOI] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T. Molecular cloning: A laboratory manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]