Abstract
Although protein glycosylation systems are becoming widely recognized in bacteria, little is known about the mechanisms and evolutionary forces shaping glycan composition. Species within the genus Neisseria display remarkable glycoform variability associated with their O-linked protein glycosylation (pgl) systems and provide a well developed model system to study these phenomena. By examining the potential influence of two ORFs linked to the core pgl gene locus, we discovered that one of these, previously designated as pglH, encodes a glucosyltransferase that generates unique disaccharide products by using polyprenyl diphosphate-linked monosaccharide substrates. By defining the function of PglH in the glycosylation pathway, we identified a metabolic conflict related to competition for a shared substrate between the opposing glycosyltransferases PglA and PglH. Accordingly, we propose that the presence of a stereotypic, conserved deletion mutation inactivating pglH in strains of Neisseria gonorrhoeae, Neisseria meningitidis, and related commensals, reflects a resolution of this conflict with the consequence of reduced glycan diversity. This model of genetic détente is supported by the characterization of pglH “missense” alleles encoding proteins devoid of activity or reduced in activity such that they cannot exert their effect in the presence of PglA. Thus, glucose-containing glycans appear to be a trait undergoing regression at the genus level. Together, these findings document a role for intrinsic genetic interactions in shaping glycan evolution in protein glycosylation systems.
Keywords: epistasis, oligosaccharide biosynthesis, type IV pili
The surfaces of microbial species are dominated by diverse arrays of glycoconjugates that vary in structure and function. Despite their predominance and importance, there remain significant gaps in understanding the origins of polysaccharide diversity at the genetic level and what forces drive glycan diversification. Protein glycosylation systems based on both N- and O-linked modifications occur in many bacterial species, and among the substrates, surface localized proteins are well represented (1). The broad distribution of these systems strongly suggests that they are advantageous and affect fitness. Detecting selection at the genetic level in these systems is complicated because complex carbohydrates are not primary gene products and because, in many cases, the specific functions of the biosynthetic components and the glycans themselves are incompletely defined. Some systems display robust inter- and intrastrain glycan variability, suggesting that glycoform diversification is adaptive. For example, flagellar protein glycosylation systems in Campylobacter jejuni and Clostridium species display remarkable plasticity in glycoform expression (2–4). However, precise correlations between glycosylation gene content and glycoform phenotypes have been difficult to establish. Similarly, attempts at reconciling the glycosylation gene repertoire with particular populations and ecotypes are problematic. As such, the long-term evolutionary trends and dynamics of protein glycosylation systems remain poorly understood.
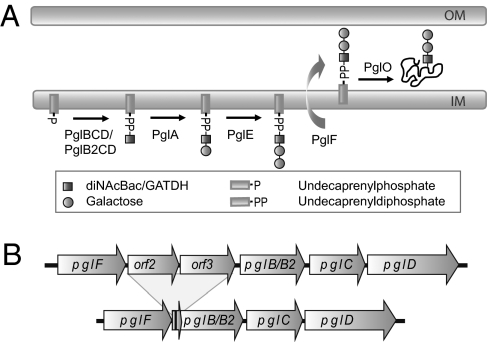
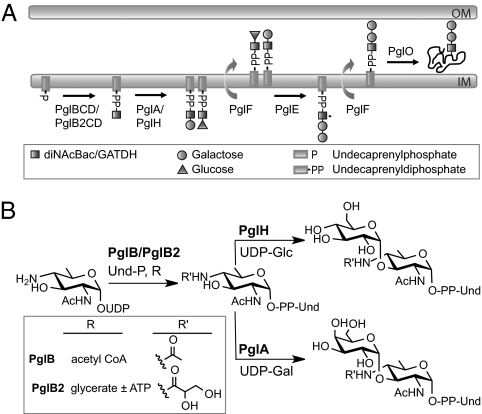
The broad-spectrum O-linked protein glycosylation (pgl) system expressed by species within the genus Neisseria represents a unique model system in which to study bacterial glycoconjugate biology and evolution. Three highly related neisserial species of importance to human health and disease are Neisseria gonorrhoeae (the agent of gonorrhea), Neisseria meningitidis (an agent of epidemic meningitis), and Neisseria lactamica (a commensal colonizing the oropharynx of young children). The genetics of glycan biosynthesis, modification, and transfer to protein have begun to be well characterized in these species (5, 6). Interestingly, some members of these species are capable of undergoing glycan variation as a result of phase-variable, slipped-strand mispairing events within the pglA and pglE genes (7). These genes are not part of the pgl core locus, the products of which function in the synthesis of undecaprenyl diphosphate (UndPP) monosaccharides (PglB, PglC, and PglD) and translocation into the periplasm (PglF) (Fig. 1A). PglA is a galactosyltransferase that acts on the UndPP-monosaccharide, whereas PglE is a galactosyltransferase that extends the PglA-generated UndPP-linked disaccharide to a trisaccharide (Fig. 1A) (8). Both the di- and trisaccharide forms can be further modified via O-acetylation mediated by PglI, the gene for which is also capable of undergoing phase variation (8). The pglB gene encodes a bifunctional protein with an acetyltransferase domain and a phospho-glycosyltransferase domain responsible for synthesis of 2,4-diacetamido-2,4,6-trideoxyhexose (DATDH). In addition, nearly one half of N. meningitidis isolates are reported to have a variant allele of pglB, designated pglB2, with an ATP grasp domain and a phospho-glycosyltransferase domain responsible for synthesis of glyceramido-acetamido trideoxyhexose (GATDH) rather than DATDH at the reducing end of the glycan (7, 9, 10). Recent studies have demonstrated that the DATDH moiety corresponds to N,N′-diacetylbacillosamine (diNAcBac) (11). UndPP-linked GATDH precursors generated by PglB2 can be further elaborated by PglA, PglE, and PglI, resulting in at least nine unique protein-associated glycans that can be generated from the neisserial pgl pan genome (6). Alleles of pglB2 appear to have been imported into a pglB background from an as yet unidentified source outside of the genus Neisseria (9). Given that each neisserial protein-associated glycan displays unique immunogenic and antigenic properties, these data strongly suggest that neisserial pgl systems are subject to selective pressures similar to those exerted on other bacterial surface glycoconjugates and adapt to such pressures by mechanisms analogous to those seen in these other systems (12, 13).
Fig. 1.
Glycosylation pathway and core pgl locus in Neisseria. (A) Current model of the broad spectrum O-linked glycosylation systems in Neisseria. (B) Gross polymorphisms in the neisserial core pgl locus associated with ORFs 2 and 3. Shown are the two states documented in strains of Neisseria (7, 10) and in a survey of currently available genome sequences (Table S1): the top configuration represents the ancestral form and the bottom corresponds to the deleted form. The stereotypic deletion retains the first 40 bp of ORF2 and the last 100 bp of ORF3. A 30-bp sequence of unknown origin that spans the ORF2 and 3 sequences is shown in black. Ancestral and deleted forms are found in combination with both pglB and pglB2 allele variants.
In addition to the distinctive reorganization involving pglB/B2, a second form of gross polymorphism at pgl locus involves the variable presence of two ORFs linked at the pgl locus consisting of the pglF, pglB, pglC, and pglD genes (Fig. 1B) (7, 10). These ORFs are arrayed in tandem between pglF and pglB/pglB2 (in the same orientation as these flanking genes) and are annotated as glycosyltransferases of the CAZy family 4. Each ORF also encompasses monotonous runs of cytosines, suggesting that, if expressed, they might be subject to phase variation. Gonococcal, meningococcal, and commensal strains lacking intact alleles of ORFs 2 and 3 still bear conserved remnants of the 5′ end of ORF2 and 3′ end of ORF3. This observation indicates that the intact state is ancestral and that a deletion event likely occurred once and then radiated through the metapopulations (Fig. 1B). Intact ORFs were reported in 67% of strains in a predominantly meningococcal strain collection (7), whereas available neisserial genome sequences show their presence in 81% of gonococcal strains, 65% of meningococcal strains, and 94% of commensal strains (Table S1). To date, however, no connections have been established between either ORF2 or ORF3 genotype and glycosylation phenotype (10).
Herein, we examine the potential contribution of ORF3 to protein glycosylation by using a systematic approach with genetically defined recombinant backgrounds, MS analyses, biochemical characterization, and glycan serotyping. Our findings unequivocally demonstrate that PglH (from ORF3) acts as a glucosyltransferase, which uses both UndPP-linked diNAcBac and GATDH. These findings thus connect major changes in pgl gene content to alterations in glycan repertoire and provide the infrastructure for assessing glycan evolution in this model system.
Results
A Previously Uncharacterized Glycosyltransferase Is Associated with Altered Disaccharide Glycan Composition.
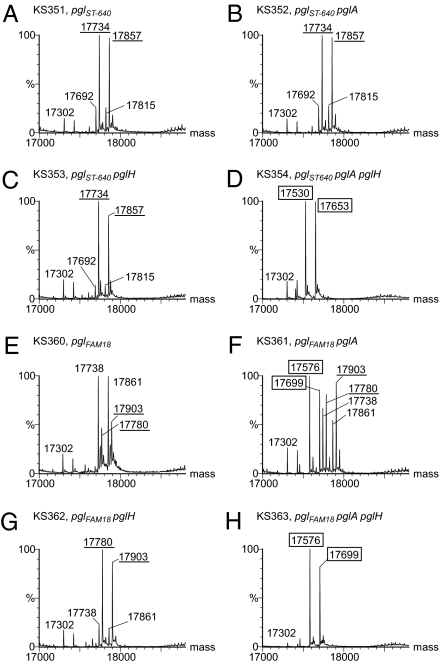
To address the potential contributions of ORFs 2 and 3 on neisserial O-linked protein glycosylation, loci encompassing them were introduced from diverse neisserial strains into N. gonorrhoeae strain N400 in which the pgl gene function and associated glycan structures have been defined (8). As ORFs 2 and 3 map between pglF and pglB, we used a procedure designed to exchange alleles of pglB/pglB2 by using a counter-selectable marker that allows gene replacement without the presence of selectable markers in the resulting recombinants (6). Specifically, we replaced the N400 locus (carrying the deleted form of ORFs 2 and 3) with those derived from the N. gonorrhoeae strain FA1090, N. meningitidis strains FAM18 and Z2491, and N. lactamica strain ST-640 (Fig. S1). These strains were chosen because they all possess in-frame variants of ORF3. Whereas FAM18 bears the pglB2 allele predicted to result in GATDH expression, the other strains carry the pglB allele. To assess the impact of the different pgl loci on protein glycosylation patterns, mature pilin (PilE) protein was purified from the surface of the recombinant strains. In the N. gonorrhoeae background strain N400, the PilE protein subunit is glycosylated with a single disaccharide at serine 63 (14). The intact PilE protein was examined from recombinant strains by using a top-down electrospray ionization (ESI)-MS approach. For recombinants carrying the pglFA1090, pglST-640, and pglZ2491 loci, the reconstructed molecular mass profiles of the corresponding spectra revealed a distribution of species identical to that seen for the parental background. These species corresponded to PilE modified with either the 432-Da glycan moiety [diNAcBac-O-acetylated hexose (diNAcBac-AcHex)] or a 390-Da glycan moiety [diNAcBac-hexose (diNAcBac-Hex)] and carrying one or two phosphoethanolamine (PE) moieties (Fig. 2A and Table 1). In the case of the strain carrying the pglFAM18 locus with a pglB2 allele, the predominant species corresponded to PilE with either a 436-Da glycan moiety [GATDH-hexose (GATDH-Hex)] or a 478-Da glycan moiety [GATDH-O-acetylated hexose (GATDH-AcHex)] and carrying one or two PE moieties (Fig. 2E). These signals were indistinguishable from those found previously for a recombinant with the meningococcal pgl8013 locus that carries a pglB2 allele and the deleted conformation of ORFs 2 and 3 (6).
Fig. 2.
ORF3/pglH is associated with altered disaccharide glycan composition. ESI MS analysis of intact PilE with pili from either the N400 pglST-640 or N400 pglFAM18 strains with different pgl backgrounds were carried out to characterize the glycan structure. (A–D) N400 pglST-640 has the pglB allele responsible for synthesizing diNAcBac glycans. The strains KS351, KS352, and KS353 all produced two major signals that represent PilE carrying the diNAcBac-AcHex disaccharide with one (17,734 Da) or two (17,857 Da) PE modifications. For KS354, the two major signals represent PilE with diNAcBac monosaccharide with one (17,530 Da) or two (17,653 Da) PE modifications. (E–H) N400 pglFAM18 has the pglB2 allele accountable for synthesizing GATDH glycans. The strains KS360, KS361, and KS362, all produced signals that represent PilE carrying GATDH-AcHex disaccharides with one (17,780 Da) or two (17,903 Da) PE modifications and also GATDH-Hex disaccharides with one (17,738 Da) or two (17,861 Da) PE modifications. For KS363, the two major signals represent PilE with GATDH monosaccharides with one (17,576 Da) or two (17,699 Da) PE modifications. Also, KS361 produced two major signals that represent PilE carrying GATDH monosaccharide with one or two PE modifications. Monosaccharides are indicated with molecular weights (in Da) boxed. All acetylated disaccharides are underlined. Table S2 shows all the ion species, m/z values, and corresponding molecular weights.
Table 1.
Pgl genotypes and phenotypes of strains
| Strain | Relevant genotype | Glycoform* | mAb† | pAb‡ | |
| N. gonorrhoeae N400 strains | |||||
| KS100 | recA6 (22) | diNAcBac-AcHex | npg2 | — | |
| KS141 | pglA (8) | diNAcBac | npg1 | — | |
| KS442 | galE | nd | npg1 | — | |
| KS351 | pglST-640 | diNAcBac-AcHex | npg2 | pDAb2 | |
| KS352 | pglST-640 pglA | diNAcBac-AcHex | — | pDAb2 | |
| KS353 | pglST-640 pglH | diNAcBac-AcHex | npg2 | — | |
| KS354 | pglST-640 pglA pglH | diNAcBac | npg1 | — | |
| KS355 | pglST-640 galE | nd | — | pDAb2 | |
| KS356 | pglST-640 pglA galE | diNAcBac-AcHex | — | pDAb2 | |
| KS357 | pglST-640 pglH galE | diNAcBac | npg1 | — | |
| KS358 | pglST-640 pglEon | diNAcBac-AcHexHex | npg3 | pDAb2 | |
| KS359 | pglST-640 pglEonpglA | diNAcBac-AcHex | — | pDAb2 | |
| KS419 | pglST-640 pglA pglI | diNAcBac-Hex | — | pDAb2 | |
| KS416 | pglST-640 pglH pglI | diNAcBac-Hex | npg2 | — | |
| KS387 | pglST-640 pilE | KS351 | npg2 | pDAb2 | |
| KS388 | pglST-640 pglA pilE | KS352 | — | pDAb2 | |
| KS389 | pglST-640 pglH pilE | KS353 | npg2 | — | |
| KS390 | pglST-640 pglA pglH pilE | KS354 | npg1 | — | |
| KS360 | pglFAM18 | GATDH-AcHex | — | — | |
| KS361 | pglFAM18 pglA | GATDH-AcHex | — | pDAb2 | |
| GATDH | pGAb2 | ||||
| KS362 | pglFAM18 pglH | GATDH-AcHex | — | — | |
| KS363 | pglFAM18 pglA pglH | GATDH | — | — | |
| KS364 | pglFAM18 galE | nd | — | — | |
| KS365 | pglFAM18 pglA galE | GATDH-AcHex | — | pDAb2 | |
| GATDH | pGAb2 | ||||
| KS366 | pglFAM18 pglH galE | GATDH | — | — | |
| KS367 | pglFAM18 pglEon | GATDH-AcHexHex | npg3 | — | |
| KS368 | pglFAM18 pglEonpglA | GATDH-AcHex | — | pDAb2 | |
| pGAb2 | |||||
| KS420 | pglFAM18 pglA pglI | GATDH-Hex | — | pDAb2 | |
| pGAb2 | |||||
| KS417 | pglFAM18 pglH pglI | GATDH-Hex | — | — | |
| KS393 | pglFAM18 pilE | KS360 | — | — | |
| KS394 | pglFAM18 pglA pilE | KS361 | — | pDAb2 | |
| pGAb2 | |||||
| KS395 | pglFAM18 pglH pilE | KS362 | — | — | |
| KS396 | pglFAM18 pglA pglH pilE | KS363 | — | — | |
| KS369 | pglZ2491 | diNAcBac-AcHex | npg2 | pDAb2 | |
| KS370 | pglZ2491 pglA | diNAcBac-AcHex | — | pDAb2 | |
| KS371 | pglZ2491 pglH | diNAcBac-AcHex | npg2 | — | |
| KS372 | pglZ2491 pglA pglH | diNAcBac | npg1 | — | |
| KS373 | pglZ2491 galE | nd | — | pDAb2 | |
| KS374 | pglZ2491 pglA galE | diNAcBac-AcHex | — | pDAb2 | |
| KS375 | pglZ2491 pglH galE | diNAcBac | npg1 | — | |
| KS376 | pglZ2491 pglEon | diNAcBac-AcHexHex | npg3 | pDAb2 | |
| KS377 | pglZ2491 pglEonpglA | diNAcBac-AcHex | — | pDAb2 | |
| KS421 | pglZ2491 pglA pglI | diNAcBac-Hex | — | pDAb2 | |
| KS418 | pglZ2491 pglH pglI | diNAcBac-Hex | npg2 | — | |
| KS399 | pglZ2491 pilE | KS369 | npg2 | pDAb2 | |
| KS400 | pglZ2491 pglA pilE | KS370 | — | pDAb2 | |
| KS401 | pglZ2491 pglH pilE | KS371 | npg2 | — | |
| KS402 | pglZ2491 pglA pglH pilE | KS372 | npg1 | — | |
| KS378 | pglFA1090 | diNAcBac-AcHex | npg2 | — | |
| KS379 | pglFA1090, pglA | diNAcBac | npg1 | — | |
| KS380 | pglFA1090, pglH | diNAcBac-AcHex | npg2 | — | |
| KS381 | pglFA1090 pglA pglH | diNAcBac | npg1 | — | |
| KS382 | pglFA1090 galE | nd | npg1 | — | |
| KS383 | pglFA1090 pglA galE | nd | npg1 | — | |
| KS384 | pglFA1090 pglH galE | nd | npg1 | — | |
| KS385 | pglFA1090 pglEon | diNAcBac-AcHexHex | npg3 | — | |
| KS386 | pglFA1090 pglEonpglA | nd | npg1 | — | |
| KS405 | pglFA1090 pilE | KS378 | npg2 | — | |
| KS406 | pglFA1090 pglA pilE | KS379 | npg1 | — | |
| KS407 | pglFA1090 pglH pilE | KS380 | npg2 | — | |
| KS408 | pglFA1090 pglA pglH pilE | KS381 | npg1 | — | |
| N. gonorrhoeae 4/3/1 (pilEind) strains | |||||
| KS101 | pilEind (23) | nd | npg2 | — | |
| KS105 | pglC (5) | nd | — | — | |
| KS122 | pglA (5) | nd | npg1 | — | |
| KS127 | pglEon (5) | nd | npg3 | — | |
| KS310 | pglB28013 pglEon (6) | nd | npg3 | — | |
| KS311 | pglB28013 (6) | nd | — | — | |
| KS312 | pglB28013 pglA (6) | nd | — | — | |
| 4/3/1 strains, ectopic lctP::pglH expression | |||||
| KS443 | pglHST-640 | nd | npg2 | pDAb2 | |
| KS444 | pglHST-640 pglA | nd | — | pDAb2 | |
| KS454 | pglHST-640 R373H pglA | nd | — | pDAb2 | |
| KS445 | pglHZ2491 | nd | npg2 | pDAb2 | |
| KS446 | pglHZ2491 pglA | nd | — | pDAb2 | |
| KS456 | pglHZ2491 R371H pglA | nd | — | pDAb2 | |
| KS447 | pglHFAM18 | nd | npg2 | — | |
| KS448 | pglHFAM18 pglA | nd | npg1 | pDAb2 | |
| KS458 | pglHFAM18 R373H pglA | nd | npg1 | — | |
| KS449 | pglHFA1090 | nd | npg2 | — | |
| KS450 | pglHFA1090 pglA | nd | npg1 | — | |
| KS451 | pglHFA1090 H371R | nd | npg2 | — | |
| KS452 | pglHFA1090 H371R pglA | nd | npg1 | pDAb2 | |
| N. gonorrhoeae FA1090 strains | |||||
| KS300 | recA6 (22) | diNAcBac, | npg1 | — | |
| diNAcBac- | npg3 | ||||
| AcHexHex | |||||
| KS422 | pglH | diNAcBac, | npg1 | — | |
| diNAcBac- | npg3 | ||||
| AcHexHex | |||||
| KS423 | pglA pglH | diNAcBac | npg1 | — | |
| KS459 | pglHH371R | nd | npg1 | pDAb2 | |
| npg3 | |||||
| KS460 | pglA pglHH371R | nd | npg1 | pDAb2 | |
* Glycoform (qualitative MS) or parental strain in which MS was performed. Hex, hexose; Ac, acetyl-group; nd, not determined.
†mAb reactivity, npg1 recognizes the monosaccharide diNAcBac; npg2 recognizes the disaccharide diNAcBac-Hex where galactose is added by PglA (i.e., diNAcBac-Gal); npg3 recognizes the trisaccharides diNAcBac-GalGal and GATDH-GalGal; −, no reactivity.
‡pAb reactivity, pDAb2 and pGAb2 recognize the disaccharides diNAcBac-Hex and GATDH-Hex, respectively, where glucose is added by PglH (i.e., diNAcBac-Glc and GATDH-Glc, respectively); −, no reactivity.
Next, we examined the effects of a pglA null mutation (that precludes the addition of galactose to UndPP-linked diNAcBac and GATDH sugars) and an ORF3 disrupting mutation. Surprisingly, inactivating pglA or ORF3 independently in the N400 pglST-640, N400 pglZ2491, and N400 pglFAM18 backgrounds failed to dramatically alter the MS-based glycoform profiles (Fig. 2 B, C, F, and G and Table 1). Specifically, MS analyses demonstrated that N400 pglST-640 pglA and N400 pglZ2491 pglA continued to generate the diNAcBac-AcHex disaccharide whereas N400 pglFAM18 pglA afforded a mixture of GATDH-AcHex disaccharides and GATDH monosaccharide. The latter strain also showed an alteration in microheterogeneity attributable to reduced O-acetylation (Fig. 2F). As ORF2 was in an out-of-frame configuration in these three backgrounds, ORF3 was implicated as encoding a glycosyltransferase equivalent to that of PglA (i.e., addition of a hexose moiety to UndPP-linked diNAcBac/GATDH sugars). In contrast, mutating pglA in the N400 pglFA1090 background resulted in detection of only diNAcBac monosaccharide glycoform (Table 1).
To examine the influence of ORF3 in more detail, an orf3 null mutation was introduced into the pglA recombinant strains. This resulted in strains expressing only the monosaccharide diNAcBac and GATDH glycoforms, confirming that ORF3 was responsible for the disaccharide observed in the absence of PglA (Fig. 2 D and H and Table 1). Based on these data (summarized in Fig. S2A) and subsequent findings, we designated ORF3 as pglH.
We also used monoclonal antibodies recognizing specific epitopes associated with the diNAcBac monosaccharide and PglA-dependent diNAcBac-Gal forms of protein-associated glycans (6) to assess the impact of PglH on glycan antigenicity in the diNAcBac-expressing recombinants. Immunoblotting demonstrated that glycoproteins from all backgrounds with active PglA reacted with npg2 (recognizing diNAcBac-Gal), whereas those with PglH alone did not (Fig. S2B). Thus, the disaccharide glycoform associated with PglH activity is an antigenically distinct disaccharide glycan form. In addition, the presence of active PglH in the absence of PglA correlated with the failure to react with the npg1 mAb (recognizing diNAcBac monosaccharide) (Fig. S2B). This effect was specific, as npg1 mAb reactivity was restored by introduction of a pglH null mutation into the pglA backgrounds. These associations were not observed in the strain with the pglFA1090 locus, consistent with the other data that it carries a defective pglH allele.
PglH Is a Glucosyltransferase Synthesizing an UndPP-Disaccharide.
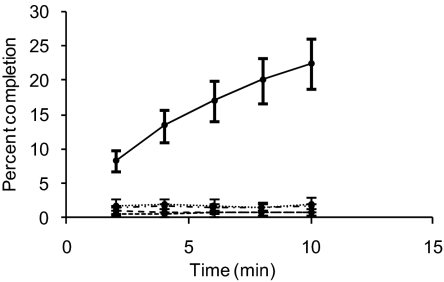
To determine the glycosyltransferase specificity of PglH, the allele from N. meningitidis strain Z2491 was expressed and purified as a maltose binding protein (MBP) fusion. Purified MBP-PglH was incubated with UndPP-diNAcBac and radiolabeled versions of five different NDP-sugars: UDP-Glc, UDP-Gal, UDP-GlcNAc, UDP-GalNAc, and GDP-Man. All five of these activated sugars are found endogenously in neisserial species and could serve as the native substrate for PglH. As shown in Fig. 3, PglH is specific for transfer of glucose, showing that UDP-Glc is the preferred substrate of this enzyme and that UndPP-diNAcBac-Glc is the product of the reaction. To further confirm the identity of the radiolabeled product observed in the assay, the PglH product UndPP-diNAcBac-Glc was generated and treated under acidic conditions to hydrolyze the glycosyl diphosphate, and the resultant disaccharide was labeled with 2-aminobenzamide (2-AB). The fluorescently labeled sugar was purified by using normal-phase HPLC and the identity of the disaccharide peak was confirmed by MALDI MS, providing definitive evidence that PglH produces a disaccharide product comprising diNAcBac-Glc (Fig. S3).
Fig. 3.
PglH is a glucosyltransferase synthesizing disaccharide glycans. In vitro radioactivity-based assay using recombinantly expressed PglH from N. meningitidis strain Z2491 shows that it specifically transfers UDP-Glc (solid line) to Und-PP-diNAcBac. In comparison, PglH shows no transferase activity in the presence of UDP-Gal, (round dot) UDP-GlcNAc (square dot), UDP-GalNAc (dash), and GDP-Man (dash/dot). Assays were performed in triplicate, and error bars indicate SD.
PglB2 has not yet been biochemically characterized; therefore, the UndPP-GATDH substrate cannot be synthesized for the parallel assay with PglH. Instead, we examined the glycan monosaccharide composition by using purified pilin derived from different backgrounds by GC after methanolysis and trimethylsilyl derivatization. This confirmed that the hexose transferred by PglH is glucose in the context of both diNAcBac and GATDH substrates (Fig. S4). We also examined the effects of a galE null mutation on glycoform expression. GalE catalyzes the interconversion between UDP-glucose and UDP-galactose, and neisserial galE mutants express galactose-deficient LPS (15) and galactose-deficient PilE glycan (16). We confirmed the role of GalE in PglA-mediated disaccharide biosynthesis, as the null mutation creates a phenocopy of the pglA mutant as seen by generation of the diNAcBac and GATDH-based glycoforms in MS analysis (strains KS354/KS357, KS363/KS366, and KS372/KS375; Table 1). In strains expressing pglB alleles (synthesizing diNAcBac monosaccharide), these effects were detected by reactivity with the npg1 mAb (Fig. S5A) in addition to MS analysis (Table 1). In contrast, the galE null mutation had no effect on disaccharide glycoform biosynthesis mediated by PglH as evidenced by MS analysis (strains KS356, KS365, and KS374; Table 1).
Immunochemical Analysis of Glycans Synthesized in the Presence of PglH.
To examine the immunogenicity and antigenicity of the glycans synthesized by PglH, PilE (in the form of purified pili) bearing the diNAcBac- and GATDH-based disaccharides were used to immunize rabbits, and the responses were monitored by immunoblotting with the N. gonorrhoeae strains and recombinants. As previously detailed, reactivity specific to other glycoproteins was used as a readout (6). The antibody response engendered by immunization with PilE modified with the diNAcBac-AcGlc (from strain KS370, pglZ2491 pglA) was specific to both diNAcBac- and GATDH-based PglH-derived glycans, whereas that following immunization with PilE modified with the PglH-derived GATDH-AcGlc (from strain KS361, pglFAM18 pglA) was more restricted, as it reacted with only the GATDH-AcGlc–expressing strains (Table 1 and Fig. S5 B and C). Taken together, these results show that protein-linked PglH-derived disaccharides are immunogenic and antigenic, and that both diNAcBac- and GATDH-based PglH-derived glycans act as glycosyl donors in general, broad-spectrum protein glycosylation. Moreover, these findings demonstrate that strains bearing active alleles of both pglA and pglH simultaneously express mixtures of glycoproteins with two distinct glycoforms (Table 1 and Figs. S2B and S5).
Elaboration of UndPP-Disaccharides Generated in the Presence of PglH.
We next sought to examine whether PglE can further elaborate the PglH-generated disaccharide. MS analyses of PilE purified from strains carrying pglST-640, pglZ2491, or pglFAM18 in an otherwise N400 pglA pglEon background showed the presence of only the PglH-generated disaccharide (results for KS359, KS377, and KS368 are summarized in Table 1). PglE is therefore unable to modify the PglH-generated disaccharide. The galactose residues in the previously characterized di- and trisaccharide glycoforms (with the exception of the GATDH-based trisaccharide) undergo O-acetylation by PglI (6, 8, 17). To explore whether PglI is responsible for acetylation of glucose in the PglH disaccharides, a pglI null mutation was introduced into the recombinant backgrounds. These strains synthesized nonacetylated disaccharides as demonstrated by MS analysis of purified PilE (results for KS419, KS416, KS420, KS417, KS421, and KS418 are summarized in Table 1).
Polymorphisms in PglH Are Associated with Diminished Glycosyltransferase Activity.
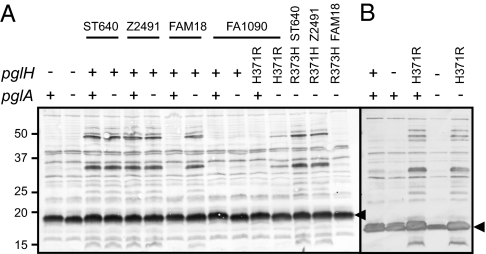
To confirm that the effects seen in strains carrying different pgl loci were attributable to pglH and not indirect effects associated with strain construction, recombinants were created in which pglH alone was expressed from an ectopic site in otherwise unaltered backgrounds. By using immunoblot-based glycan serotyping, we confirmed that pglH was both necessary and sufficient for expression of the alternative disaccharide by using both UndPP-linked diNAcBac and GATDH as precursors (Fig. 4A). In addition, we found that the atypical glycosylation phenotypes observed for the FA1090- and FAM18-based recombinants could be directly attributed to their pglH genotypes.
Fig. 4.
Identification of pglH polymorphisms associated with diminished glucosyltransferase activity. Expression of the diNAcBac-Glc disaccharide was monitored by reactivity of endogenous glycoproteins following immunoblotting with pDAb2 polyclonal antibody. (A) Variant alleles of pglH were expressed ectopically in the strain N400 derivative 4/3/1. In the strain expressing the pglHFAM18 allele, glycoform-specific immunoreactivity was detected only in the pglA-null background, and in the strain expressing the pglHFA1090 allele, no immunoreactivity was seen in either background. Conversion of H371R in the pglHFA1090 allele partially restored activity but only in the pglA null background. Introduction of his in place of arg 371/373 in the other alleles had varying effects. Minus signs denote absence of the pglH gene or pglA::kan. Plus signs denote ectopically expressed pglH or intact pglA. Strains used are KS101, KS122, KS443, KS444, KS445, KS446, KS447, KS448, KS449, KS450, KS451, KS452, KS454, KS456, and KS458. (B) Mutation H371R in the endogenous pglH locus of strain FA1090 restores diNAcBac-Glc expression, and activity was seen in the presence of an intact pglA allele. Minus signs denote pglA::kan or pglH::ermC/rpsL; plus signs denote intact pglA or pglH. Strains used are KS300, KS422, KS459, KS423, and KS460. Arrow denotes the position of the major glycoprotein PilE.
To determine the nature of the defect manifest in the pglHFA1090 ORF, a comparative alignment of PglH from several strains was examined (Fig. S6). This revealed one residue unique to PglHFA1090 (and an identical allele in the gonococcal strain F62) at position 371 as a histidine versus a conserved arginine. A pglHFA1090 allele in which the histidine codon at residue 371 was substituted with the arginine codon (H371R) restored PglH protein glycosylation activity as assessed by glycan serotyping. However, the reactivity was observed only in a pglA background, and the level of glycosylation was lower than that seen in strains expressing the other pglH alleles (Fig. 4A). Conversely, introducing the histidine 371 substitution into the pglHFAM18 allele abolished the disaccharide glycosylation activity seen in the pglA background. However, making the equivalent mutations in the pglHZ2491 and pglHST-640 alleles did not diminish glycosylation.
To corroborate the finding that polymorphisms at residue 371 influence PglH activity, the H371R mutation was introduced into the endogenous pglH gene of strain FA1090. As shown in Fig. 4B, the single amino acid substitution was sufficient to restore PglH activity in the WT FA1090 strain. Moreover, this effect was seen even in the presence of PglA, in contrast to observations in the N400 background. The disparate effect of the arginine substitution in the two backgrounds is not attributable to differences in PglH expression. Rather, it stems from polymorphisms in pglA, as pglAFA1090 has previously been shown to be associated with reduced activity relative to that of the allele in strain N400 (6). Together, these findings document epistatic interactions involving pglH at the intragenic and intergenic levels.
Discussion
Although the presence of a widely distributed deletion within the pgl core locus of Neisseria species was established more than 10 years ago, the biological consequences of this gross polymorphism remained elusive. Here, we establish that PglH, corresponding to one of two ORFs encompassed within the deletion, encodes a glucosyltransferase that acts to synthesize disaccharides by using UndPP-linked forms of diNAcBac and GATDH as substrates. Together with the protein-associated glycans previously defined, these two disaccharides in their unmodified and O-acetylated forms expand the pan-neisserial glycome to include 13 distinct glycoforms. These findings also definitively link a pervasive genome rearrangement disseminated at the genus level to a reduction in protein glycoform repertoire.
We also observed surprisingly high levels of polymorphism within pglH, and, in two cases, we were able to demonstrate corresponding functional alterations indicative of altered selection. The first case is that exemplified by the allele found in gonococcal strains FA1090 and F62. Here, the gene product is devoid of detectable glycosyltransferase activity as measured in vivo. The basis for this defect was mapped to residue 371, as restoration of a consensus arginine residue (in place of histidine) resurrected activity both in the ectopic locus in N400 as well as in the native locus of FA1090. The allele carried in gonococcal strain SK-92–679 may also encode a defective transferase, as it encodes a proline rather than the consensus arginine at this site (Fig. S6). The second case is that associated with the pglHFAM18 allele, which encodes a functional product but whose activity is diminished as seen by an admixture of both monosaccharide and disaccharide forms in its presence. Furthermore, pglHFAM18 is unable to exert its activity in the presence of PglA.
The disseminated distribution of the deleted ORF2/pglH variant strongly suggests that this polymorphism is linked to altered fitness related to protein glycosylation. However, it is difficult to exclude that this situation reflects selection at a tightly linked locus and that the prevalence of the deletion is caused by unrelated effects. The existence of pglH missense alleles associated with nondetectable and reduced transferase activities argues against this scenario, and thus strengthens the idea that the glucose-containing disaccharide itself currently represents a trait in the active process of regression. If this is the case, to what can we ascribe this regressive nature? We propose that genetic interactions between pglA and pglH are likely to play a major role. As both glycosyltransferases result in the synthesis of disaccharides by using shared monosaccharide substrates (Fig. 5 and Fig. S7), redundancy may have allowed the passive accumulation of pglH mutations. Alternatively, incompatibility or conflict mediated by competition between the two pathways may have led to active selection. Although the activities of PglA and PglH are not mutually exclusive of one another (in the laboratory), the PglH activity precludes synthesis of the trisaccharide glycoform mediated by PglE. It is also conceivable that genetic antagonism imposed by the bifurcated pathway might be resolved by regressive alleles of pglA as suggested by the low activity associated with the FA1090 gene (6). An interesting facet of the “active-selection hypothesis” is that both pglA and pglH are capable of high-frequency, on/off expression as a result of hotspots for frame-shifting events in the ORFs. Theoretically, such a mechanism could significantly ameliorate the potential for pathway incompatibility. However, some pglA and pglH alleles lack the sequences driving phase variation, and strains carrying them will invariably express galactose- or glucose-containing oligosaccharides, respectively. Further studies of connections between the phase variability status and genotypes are ongoing.
Fig. 5.
PglA and PglH compete for substrates generated by PglB/PglB2 in a branched pathway for protein glycan biosynthesis. (A) PglH is involved in the broad-spectrum O-linked glycosylation systems in Neisseria by adding glucose onto diNacBac or GATDH UndPP monosaccharide. (B) The figure displays the chemical reactions of the phospho-glycosyltransferases, PglB/PglB2, and the glycosyltransferases, PglA and PglH. Boxed are the substrates (R) for the two glycosyltransferases, PglB and PglB2, and side groups (R′) on the glycans. Und, undecaprenol; UDP, uridine diphosphate.
As the roles of O-linked protein glycosylation in neisserial species remain poorly understood, attempts to define the functional consequences of an altered glycan repertoire are somewhat limited. Glycan diversification may be driven by selection at the level of the adaptive and innate immune systems. Hamadeh and colleagues reported that human serum and IgA1 polyclonal antibodies directed toward terminal galactose residues bound to meningococcal pili and blocked complement-mediated lysis (18). Although the mechanistic details behind those observations remain unknown, pglH expression would likely preclude or diminish recognition of glycoproteins by anti-Gal antibodies. Banerjee and colleagues reported that phase variable alleles of pglA were overrepresented in isolates associated with disseminated forms of disease (19). This finding was refuted by a subsequent study that failed to find an absolute correlation between disease manifestation and pglA phase-variable status (20). Our findings on pglH and its interactions with pglA indicate that broader studies are needed to assess the potential influence of O-linked protein glycosylation on the propensity of strains to promote disseminated gonococcal disease.
In conclusion, the prevalent loss of pglH mediated by deletion, together with the presence of alleles encoding defective products, corresponds to a significant reduction in glycan diversity. The pervasive deletion of pglH represents the second major alteration of pgl gene content impacting at the genus level, following that documented for the introgression of the pglB2 allele (9). Thus, the work clearly demonstrates that gene loss as well as gene acquisition can drive protein glycan macroevolution. Finally, we found strong evidence that intrinsic genetic interactions may contribute to the evolutionary trajectory of protein glycosylation systems.
Materials and Methods
Bacterial Strains and Culture Conditions.
The bacterial strains used in this study are described in Table 1 and in SI Materials and Methods.
SDS/PAGE and Immunoblotting.
Procedures for SDS/PAGE and immunoblotting have been previously described (21). Immunoreactive proteins were detected by immunoblotting by using the glycan-specific monoclonal antibodies npg1, npg2, and npg3 (6), rabbit polyclonal antibodies pDAb2 and pGAb2 (SI Materials and Methods), and alkaline phosphatase-coupled goat anti-rabbit secondary antibodies (Sigma).
Sample Preparation and ESI MS Analysis of Intact PilE.
Type IV pili were isolated and treated with a methanol/chloroform wash/precipitation procedure as described (8). Data were acquired on a quadrupole time-of-flight mass spectrometer (Q-Tof micro; Waters) equipped with the standard Z-spray ESI source as previously described (6, 8).
In Vitro Radioactivity-Based Assay.
The glycosyltransferase PglH was heterologously overexpressed in Escherichia coli and purified as a MBP fusion protein as described in SI Materials and Methods. The ability of PglH to transfer UDP-Glc, UDP-Gal, UDP-GlcNAc, UDP-GalNAc, and GDP-Man was analyzed by using a radioactivity-based assay, and the identity of the PglH product was verified via characterization of the 2-AB–labeled glycan. Further details are described in SI Materials and Methods.
Supplementary Material
Acknowledgments
The authors thank Jos van Putten for providing the N. gonorrhoeae galE mutant and Warren Wakarchuk for helpful discussions on the structure–function relationships of PglH. This research was supported in part by Research Council of Norway Grants 166931, 183613, and 183814; by funds from the Department of Molecular Biosciences and Center for Molecular Biology and Neurosciences of the University of Oslo; and by National Institutes of Health Grant GM039334.
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1103321108/-/DCSupplemental.
References
- 1.Nothaft H, Szymanski CM. Protein glycosylation in bacteria: Sweeter than ever. Nat Rev Microbiol. 2010;8:765–778. doi: 10.1038/nrmicro2383. [DOI] [PubMed] [Google Scholar]
- 2.Champion OL, et al. Comparative phylogenomics of the food-borne pathogen Campylobacter jejuni reveals genetic markers predictive of infection source. Proc Natl Acad Sci USA. 2005;102:16043–16048. doi: 10.1073/pnas.0503252102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Carter AT, et al. Independent evolution of neurotoxin and flagellar genetic loci in proteolytic Clostridium botulinum. BMC Genomics. 2009;10:115. doi: 10.1186/1471-2164-10-115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Twine SM, et al. Motility and flagellar glycosylation in Clostridium difficile. J Bacteriol. 2009;191:7050–7062. doi: 10.1128/JB.00861-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Vik A, et al. Broad spectrum O-linked protein glycosylation in the human pathogen Neisseria gonorrhoeae. Proc Natl Acad Sci USA. 2009;106:4447–4452. doi: 10.1073/pnas.0809504106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Børud B, et al. Genetic, structural, and antigenic analyses of glycan diversity in the O-linked protein glycosylation systems of human Neisseria species. J Bacteriol. 2010;192:2816–2829. doi: 10.1128/JB.00101-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Power PM, et al. Genetic characterization of pilin glycosylation and phase variation in Neisseria meningitidis. Mol Microbiol. 2003;49:833–847. doi: 10.1046/j.1365-2958.2003.03602.x. [DOI] [PubMed] [Google Scholar]
- 8.Aas FE, Vik A, Vedde J, Koomey M, Egge-Jacobsen W. Neisseria gonorrhoeae O-linked pilin glycosylation: functional analyses define both the biosynthetic pathway and glycan structure. Mol Microbiol. 2007;65:607–624. doi: 10.1111/j.1365-2958.2007.05806.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chamot-Rooke J, et al. Alternative Neisseria spp. type IV pilin glycosylation with a glyceramido acetamido trideoxyhexose residue. Proc Natl Acad Sci USA. 2007;104:14783–14788. doi: 10.1073/pnas.0705335104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kahler CM, et al. Polymorphisms in pilin glycosylation locus of Neisseria meningitidis expressing class II pili. Infect Immun. 2001;69:3597–3604. doi: 10.1128/IAI.69.6.3597-3604.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hartley MD, et al. Biochemical characterization of the O-linked glycosylation pathway in Neisseria gonorrhoeae responsible for biosynthesis of protein glycans containing N,N'-diacetylbacillosamine. Biochemistry. 2011 doi: 10.1021/bi2003372. May 12 (Epub ahead of print) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bentley SD, et al. Genetic analysis of the capsular biosynthetic locus from all 90 pneumococcal serotypes. PLoS Genet. 2006;2:e31. doi: 10.1371/journal.pgen.0020031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang L, Wang Q, Reeves PR. The variation of O antigens in gram-negative bacteria. Subcell Biochem. 2010;53:123–152. doi: 10.1007/978-90-481-9078-2_6. [DOI] [PubMed] [Google Scholar]
- 14.Hegge FT, et al. Unique modifications with phosphocholine and phosphoethanolamine define alternate antigenic forms of Neisseria gonorrhoeae type IV pili. Proc Natl Acad Sci USA. 2004;101:10798–10803. doi: 10.1073/pnas.0402397101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Robertson BD, Frosch M, van Putten JP. The role of galE in the biosynthesis and function of gonococcal lipopolysaccharide. Mol Microbiol. 1993;8:891–901. doi: 10.1111/j.1365-2958.1993.tb01635.x. [DOI] [PubMed] [Google Scholar]
- 16.Stimson E, et al. Meningococcal pilin: A glycoprotein substituted with digalactosyl 2,4-diacetamido-2,4,6-trideoxyhexose. Mol Microbiol. 1995;17:1201–1214. doi: 10.1111/j.1365-2958.1995.mmi_17061201.x. [DOI] [PubMed] [Google Scholar]
- 17.Warren MJ, Roddam LF, Power PM, Terry TD, Jennings MP. Analysis of the role of pglI in pilin glycosylation of Neisseria meningitidis. FEMS Immunol Med Microbiol. 2004;41:43–50. doi: 10.1016/j.femsim.2004.01.002. [DOI] [PubMed] [Google Scholar]
- 18.Hamadeh RM, Estabrook MM, Zhou P, Jarvis GA, Griffiss JM. Anti-Gal binds to pili of Neisseria meningitidis: the immunoglobulin A isotype blocks complement-mediated killing. Infect Immun. 1995;63:4900–4906. doi: 10.1128/iai.63.12.4900-4906.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Banerjee A, et al. Implications of phase variation of a gene (pgtA) encoding a pilin galactosyl transferase in gonococcal pathogenesis. J Exp Med. 2002;196:147–162. doi: 10.1084/jem.20012022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Power PM, et al. The phase-variable allele of the pilus glycosylation gene pglA is not strongly associated with strains of Neisseria gonorrhoeae isolated from patients with disseminated gonococcal infection. Infect Immun. 2007;75:3202–3204. doi: 10.1128/IAI.01501-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Freitag NE, Seifert HS, Koomey M. Characterization of the pilF-pilD pilus-assembly locus of Neisseria gonorrhoeae. Mol Microbiol. 1995;16:575–586. doi: 10.1111/j.1365-2958.1995.tb02420.x. [DOI] [PubMed] [Google Scholar]
- 22.Tonjum T, Freitag NE, Namork E, Koomey M. Identification and characterization of pilG, a highly conserved pilus-assembly gene in pathogenic Neisseria. Mol Microbiol. 1995;16:451–464. doi: 10.1111/j.1365-2958.1995.tb02410.x. [DOI] [PubMed] [Google Scholar]
- 23.Wolfgang M, van Putten JP, Hayes SF, Dorward D, Koomey M. Components and dynamics of fiber formation define a ubiquitous biogenesis pathway for bacterial pili. Embo J. 2000;19:6408–6418. doi: 10.1093/emboj/19.23.6408. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.