Abstract
The main consequence of the Chernobyl accident has been an increase in papillary thyroid carcinomas (PTCs) in those exposed to radioactive fallout as young children. Our aim was to identify genomic alterations that are associated with exposure to radiation. We used array comparative genomic hybridization to analyze a main (n = 52) and a validation cohort (n = 28) of PTC from patients aged <25 y at operation and matched for age at diagnosis and residency. Both cohorts consisted of patients exposed and not exposed to radioiodine fallout. The study showed association of a gain on chromosome 7 (7q11.22–11.23) with exposure (false discovery rate = 0.035). Thirty-nine percent of the exposed group showed the alteration; however, it was not found in a single case from the unexposed group. This was confirmed in the validation set. Because only a subgroup of cases in the exposed groups showed gain of 7q11.22–11.23, it is likely that different molecular subgroups and routes of radiation-induced carcinogenesis exist. The candidate gene CLIP2 was specifically overexpressed in the exposed cases. In addition, the expression of the genes PMS2L11, PMS2L3, and STAG3L3 correlated with gain of 7q11.22–11.23. An enrichment of Gene Ontology terms “DNA repair” (PMS2L3, PMS2L5), “response to DNA damage stimulus” (BAZ1B, PMS2L3, PMS2L5, RFC2), and “cell–cell adhesion” (CLDN3, CLDN4) was found. This study, using matched exposed and unexposed cohorts, provides insights into the radiation-related carcinogenesis of young-onset PTC and, with the exposure-specific gain of 7q11 and overexpression of the CLIP2 gene, radiation-specific molecular markers.
Keywords: FISH, post-Chernobyl
Ionizing radiation is a major known risk factor for thyroid cancer and particularly for papillary thyroid carcinoma (PTC), which is the most common cancer type of the thyroid gland. Both external and internal radiation exposures due to medical therapeutic schemes and accidental events, such as nuclear reactor accidents, have been shown to lead to an increased thyroid cancer risk (1). The most recent evidence for such an association between radiation exposure and an increased thyroid cancer risk comes from the Chernobyl power plant accident in 1986, which resulted in an excess of childhood PTC in regions of Ukraine, Belarus, and Russia contaminated by the radioiodine fallout (2). To date, PTC has developed in more than 4,000 individuals who were children or adolescents at the time of exposure. Thus, young age at exposure is a significant risk factor for the development of radiation-induced PTC. The Chernobyl Tissue Bank (CTB) has provided a systematic collection of thyroid tumors from those resident in the contaminated regions of Ukraine and Russia since 1998 (www.chernobyltissuebank.com). The CTB collection now also includes a substantial number of thyroid tumors from those who were conceived more than 3 mo after the accident and were therefore not exposed to radioiodine either in utero or in early childhood. It is crucial for any study of genetic factors that might be involved in the development of radiation-associated PTC that appropriate tumor, sex, and age-matched cohorts with exposed cases and nonexposed individuals for comparison are used. This approach minimizes variability in the study that results particularly with respect to age at diagnosis. Studies of early Chernobyl-related tumors by several groups found that a very high proportion showed RET rearrangements, predominantly RET/PTC3 (3–6). It was speculated that this rearrangement might be a marker for radiation-induced tumors (7). However, more recent papers have suggested that this may be related more to the young age of the patients in the study, because frequent RET/PTC rearrangements have also been observed, with similar prevalence, in sporadic papillary carcinomas from children and young adults (8, 9). These findings may therefore reflect more the association between the solid morphological subtype with RET/PTC3 rearrangement and the age of the patient at diagnosis, rather than the etiology of the tumor (5, 10). In addition, the level of RET/PTC expression in PTC has been shown to be highly variable as a result of heterogeneous distribution of RET/PTC within a particular tumor (11–14). This suggests that rearrangement of the RET oncogene may not be an initiating event and, therefore, the presence of a high frequency of this alteration is not directly linked to radiation exposure. Further studies have shown that PTCs diagnosed in childhood also show a much lower frequency of mutation of the BRAF oncogene, the most common molecular feature of adult-onset PTC (8, 15, 16). Taken together, the data from the previous studies suggest that childhood PTCs differ significantly from their adult counterparts and provide a strong motivation to search for gene alterations as alternative drivers of thyroid tumorigenesis in general and for radiation-specific DNA changes in Chernobyl-related PTC in particular. For this purpose, a genome-wide screening approach for the detection of somatic copy number alterations (CNAs) was used. PTCs of differing etiology and occurring at different ages have already been analyzed using array comparative genomic hybridization (CGH) to identify additional genomic changes in RET/PTC-positive tumors (17). We applied the same strategy to a cohort of 52 PTCs from patients who were matched for age at diagnosis and ethnicity, consisting of 33 PTCs from patients who were young at the time of exposure (median 1.3 y) and 19 PTCs from unexposed patients, who were born when the environment was no longer contaminated with radioiodine (i.e., more than 9 mo (equal to ≈33 half-lives of radioiodine) after the accident). A second, independent cohort of 28 PTC cases (16 exposed and 12 unexposed cases) was used to validate the previous findings of CNAs. The overall aim of this study was to identify genomic CNAs that are associated with exposure to ionizing radiation on an age- and residence-matched cohort of radiation-exposed and unexposed PTC.
Results
RET/PTC Status, Lymph Node Status, and Tumor Size Separate Copy Number Profiles After Unsupervised Hierarchical Clustering.
We analyzed genomic CNAs in PTCs from 52 patients. The RET/PTC expression status, BRAF mutation status, and the patients’ demographic and clinical data are briefly summarized in Table 1 and in full in Table S1. The overall copy number changes of the 52 PTC cases are shown in Fig. 1. In general, DNA gains (n = 81) were more frequent than DNA losses (n = 63). Five regions on chromosomes 1, 3, 4, and 12 were lost, and six regions on chromosomes 12, 19, 20, and 22 were gained in all cases investigated. Hierarchical cluster analysis (Fig. S1) separated the array CGH profiles into two main clusters, 1 (n = 23) and 2 (n = 29). Cluster 2 was subdivided into two subclusters, 2-1 (n = 14) and 2-2 (n = 15). Cluster 1 contained significantly more RET/PTC-positive cases compared with cluster 2 (P = 0.0096). Large tumors (pT2 and pT3) were associated with cluster 1 (P = 0.00087). Tumors that have metastasized to local lymph nodes (N1) were predominant in cluster 1 and cluster 2-2 (P = 0.04).
Table 1.
Characteristics of the discovery and validation set
| Exploratory set |
Validation set |
|||
| Characteristic | Exposed | Not exposed | Exposed | Not exposed |
| No. of cases | 33 | 19 | 16 | 12 |
| Age at exposure (y) | 1.26 (0.095–4.04) | NA | 2.55 (0.29–8.33) | NA |
| Age at operation (y) | 16.87 (13.99–18.99) | 14.15 (7.70–18.66) | 19.83 (16.37–24.55) | 16.82 (11.01–20.65) |
| Tumor size (cm) | 2.0 (0.7–5.0) | 1.8 (0.6–7.5) | 1.5 (0.5–3.8) | 2.45 (0.8–3.8) |
| BRAF V600E | 5/27 (18.5) | 3/15 (20) | 5/15 (33.33) | 3/10 (30) |
| RET/PTC | 15/28 (53.6) | 13/19 (68.4) | 5/12 (41.67) | 1/8 (12.5) |
| Lymph node positive | 20/33 (60.60) | 11/19 (57.89) | 7/16 (43.75) | 7/12 (58.33) |
Values are median (range) or n (%).
Fig. 1.
Frequency plots of copy number changes in the unexposed (Upper) and exposed (Lower) cases. The green bars (starting from the top) represent the frequencies (proportion of cases) showing a copy number gain at the according position in the genome, whereas the red bars (starting from the bottom) represent the frequencies of copy number losses. The blue frame points out the copy number gain on chromosome 7 that is only present in the exposed cases and that is associated with radiation exposure.
Estimation of Irradiation Dose and Proportion of Radiation-Induced Tumors.
Estimated dosimetry data were used because no individual dose measurements of the patients were available. The average thyroid dose of the patients was calculated using county-specific doses that were estimated for inhabitants that were born between 1968 and 1985 (18). Because the patients included in this study were considerably younger (age at the time of exposure, ≈2 y) than the birth cohort 1968–1985, the estimated thyroid dose was corrected by a factor of 2 (19). The resulting average dose for the patients was ≈150 mGy. An excess relative thyroid cancer risk of 6.5 for these patients was calculated using an estimation curve published in a study by Jacob et al. (19). Consequently, the estimated proportion of patients with radiation-induced PTC in our study was ≈85%.
CNAs Associated with Tumor Size and Sex.
Univariate supervised analysis showed associations [false discovery rate (FDR) <0.05; Table S2] of CNAs with tumor size (gain of 1q21.1–23.3, 7q22.1, 9p24.3, 10p15.3–15.1, 10q26.13–26.3, 11p11.12-cen, 12q24.11–24.23, and 16q22.1–23.3) and sex of patients (loss of 5q23.3–31.3). Candidate genes located in the altered regions were selected on the basis of their reported functional role or association with cancer risk in the literature and are summarized in Table S3. Correlation analysis (Spearman) showed that tumor size and age at operation are independent variables (P = 0.58 and ρ = −0.05). Therefore, age can be excluded as an underlying factor for the CNAs that were found to be associated with tumor size.
CNA Associated with Exposure to Radioiodine.
A DNA gain on chromosome 7 (7p14.1-q11.23, 32.1 Mb in size) was exclusively associated with a subgroup of patients exposed to the radioiodine fallout (univariate supervised analysis; FDR <0.05; Fig. 1 and Table S2). The alteration was present in 13 out of 33 cases from the exposed group and in none of the cases from the unexposed group (P = 0.0015, FDR = 0.035). This finding was verified by array CGH analysis of an independent validation set consisting of 28 PTC cases (16 exposed and 12 unexposed; Table 1 and Table S4). Supervised analysis found a smaller proportion of the alteration (7q11.22–11.23, 4.3 Mb in size; Table S2) associated with exposure to radioiodine fallout (P = 0.024; 6 out of 16 exposed cases, none out of 12 unexposed cases) in the validation set. LIMK1, RFC2, CLIP2, CLDN4, and CLDN3 were identified as tumor associated-candidate genes located on chromosome 7q11.22–11.23. Analysis of the genes mapped on chromosome 7q11.22–11.23 (68 Ensembl annotated genes) revealed significant enrichment of Gene Ontology (GO) terms associated with DNA repair (PMS2L3 and PMS2L5; P = 0.047), response to DNA damage stimulus (BAZ1B, PMS2L3, PMS2L5, and RFC2; P = 0.034), and cell–cell adhesion (CLDN3 and CLDN4; P = 0.041).
Validation of the Gain on Chromosome Band 7q11 by FISH.
The copy number gain of chromosome 7q11.22–11.23 was investigated further by FISH on formalin-fixed, paraffin-embedded tissue sections of the same PTC cases used for array CGH analysis and was confirmed in all cases analyzed. FISH confirmed gain of 7q11.22–11.23 that was found by array CGH in 17 of the exposed cases (signal to reference ratio: 3/2; Fig. S2), whereas cases with normal copy number of the region in array CGH (n = 11, 6 exposed and 5 unexposed cases) also showed normal copy number of the region after FISH (signal to reference ratio: 2/2).
Assessment of Expression of the Candidate Genes of the Chromosome Region 7q11.22–11.23 by Quantitative RT-PCR.
To assess whether the gain in copy number of 7q11.22–11.23 also resulted in an increased expression at the mRNA level, quantitative RT-PCR (qRT-PCR) was performed on the key genes located in the gained region (CLDN3, CLDN4, CLIP2, LIMK1, PMS2L2, PMS2L3, PMS2L11, RFC2, and STAG3L3). Expression levels in tumors harboring the gain were compared with those with a normal copy number at this region. The mRNA expression of three out of the nine genes (PMS2L11, PMS2L3, and STAG3L3) was significantly elevated in cases showing the gain (P < 0.05). The genes LIMK1 and PMS2L2 showed twofold increased expression in the gained cases compared with those not showing the gain (Fig. S3 and Table 2). CLIP2 was the only gene to show significantly elevated mRNA expression between the exposed and nonexposed groups, irrespective of the gain of 7q11 (P = 0.04; Fig. S4 and Table 2).
Table 2.
Summary of results of qRT-PCR analysis of the candidate genes from the gained region on 7q11.22–11.23
| Gene | Function | mRNA gene expression | P value |
| Comparison, exposed/gain* with exposed/no gain† | |||
| CLDN3‡ CLDN4‡ | Integral membrane proteins, components of tight junction strands; overexpressed in various human tumors (32–36) | CLDN3: 0.9; CLDN4: 1.5 | 0.733; 0.078 |
| LIMK1 | LIM kinase, regulates actin dynamics (accurate spindle orientation); role in tumor-cell invasion and metastasis (47–49) | LIMK1: 2.4 | 0.078 |
| PMS2L2, PMS2L3§¶, PMS2L11 | Belong to family of PMS2-related genes; highly conserved motifs among MutL proteins, suggesting a role in DNA mismatch repair (40–42, 57) | PMS2L2: 4.8; PMS2L3: 2.1; PMS2L11: 4.9 | 0.256; 0.027; 0.037 |
| STAG3L3 | Stromal antigen 3-like 3, encodes a protein of as yet unknown function | STAG3L3: 1.8 | 0.042 |
| Comparison, exposed with nonexposed | |||
| RFC2¶ | Part of protein complex required for DNA replication and repair; amplified in glioblastomas and colorectal carcinomas (54, 55, 58) | RFC2: 1 | 0.886 |
| CLIP2 | Cytoplasmic linker protein, links organelles to microtubules; amplified in glioblastomas and colorectal carcinomas (50, 54, 55) | CLIP2: 1.5 | 0.039 |
Fold-changes greater than 1.5 and significant P values (<0.5) are in bold.
*Exposed/gain: cases from the exposed group that showed DNA gain on chromosome 7q11.22–11.23 on array CGH.
†Exposed/no gain: cases from the exposed group that showed no DNA gain on chromosome 7q11.22–11.23 on array CGH.
‡, §, ¶Genes linked to significantly enriched GO terms (P < 0.05) ‡”cell-cell adhesion,” §”DNA repair,” and ¶”response to DNA damage stimulus.”
Characterization of the Gain on Chromosome Band 7q11 by High-Resolution Array CGH.
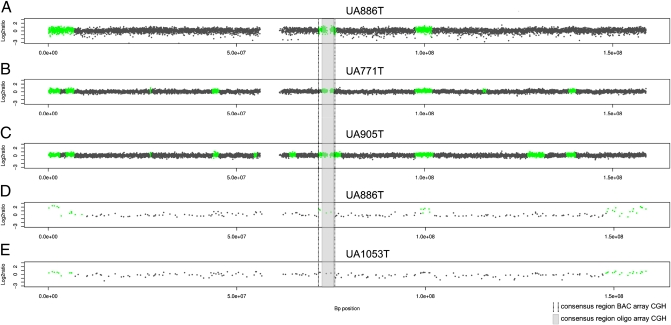
To characterize the radiation-associated gain of 7q11.22–11.23 in more detail, three of the cases that showed the copy number gain were analyzed using high-resolution oligo array CGH. The Agilent 180-k arrays that were used give a theoretical resolution of 17 kb, whereas 1-Mb bacterial artificial chromosome (BAC) arrays only allow copy number typing at a resolution greater than 1000 kb. Fig. 2 illustrates the results of the oligo array CGH analysis. The gain of 7q11.22–11.23 was confirmed in all three cases analyzed. The position of breakpoints flanking the gained region varied slightly between the three cases, whereas the consensus region of the gain (7:73096427–76001049, 2.9 Mb) is approximately 1.4 Mb smaller than the consensus region found by BAC array CGH. This is due to an uncertainty interval of approximately 1 Mb in BAC array CGH, whereas that of high-resolution array CGH profiles is much narrower. However, the consensus region found by high-resolution array CGH covers the core of the BAC array CGH consensus region and hence confirms the BAC array results at a much higher resolution.
Fig. 2.
High-resolution oligo array CGH profiles of PTCs UA0886T (A), UA0771T (B), and UA0905T (C), which were positive for gain of 7q11.22–11.23. (D and E) Copy number profiles (1 Mb) of case UA886T, which is positive for gain of 7q11.22–11.23, and of UA1053T, which is not gained in 7q11.22–11.23. The black dotted lines mark the gain on chromosome 7q11.22–11.23, as identified after 1-Mb array CGH and the gray area the consensus region of the gain after oligo array CGH. The high-resolution array CGH profiles show varying positions of breakpoints flanking the gain, whereas the consensus region covers the core of the gained region identified by BAC array CGH. The gap (≈280 kb) within the gained region of the 180-k profiles relates to the array design, which does not cover this particular region.
Association of the Gain on Chromosome Band 7q11 with Clinical Characteristics.
The gain of chromosome band 7q11 was not associated with any clinical characteristics of the patients such as sex, lymph node status, tumor size, morphological subtype, RET/PTC, or BRAF mutation status. The only association that was identified was gain of the region with exposure to radiation.
Discussion
In this study we describe genomic CNAs in two age-matched cohorts of PTC from young patients who were either exposed or not exposed to the radioiodine contaminated fallout from the Chernobyl accident. There is evidence from several studies that radiation exposure alters gene expression during the carcinogenic process by the induction of CNAs, even at low doses of radiation (20). However, direct linkage of specific genetic changes to radiation exposure in cancers that have arisen many years after the exposure to radiation is a challenge, because although radiation-induced effects such as an increased incidence of cancer can be clearly detected in an exposed population, in an individual cancer they may be hidden among a number of non–radiation-associated changes. The only way to tease out the radiation-induced changes is to use cohorts of closely matched cancer cases to minimize the number of variables for which genetic effects are known (e.g., age, sex, and ethnicity). For thyroid cancer, it is well established that age has a significant influence on the molecular biology of the tumors. This is evidenced by data showing that the prevalence of the type of RET rearrangement varies with the average age of patients, with RET/PTC3 being more common in children and RET/PTC1 being more common in adults (5, 9, 21). There are also age-specific changes in the predominant pathomorphology of PTCs; hence the solid subtype of PTC is more common in younger cohorts and the follicular and papillary subtypes in older cohorts (22, 23). Previously published studies suggesting a radiation gene expression signature in post-Chernobyl cases (24, 25) therefore need careful interpretation, because the nonirradiated comparator population is usually drawn from an adult series of PTCs instead of childhood cases. We therefore aimed to identify radiation-specific CNAs in PTCs by directly comparing age-matched tumor cohorts, for which the main difference between them was exposure to radioiodine fallout. The systematic collection of thyroid cancers through the CTB project has enabled us to select appropriately matched cohorts of patients.
Unsupervised clustering of copy number profiles separated the cases into two main groups that correlated with the RET/PTC status of tumors, tumor size, and lymph node status. These findings indicate that the CNAs correlate with the morphological and molecular phenotypes of the investigated cases. The various copy number changes that are present in addition to the RET/PTC rearrangement in RET/PTC positive cases concur with the findings of previous studies reporting that RET/PTC only occurs in a subset of tumor cells and therefore is unlikely to be the only driver of PTCs (12, 13, 26). Many of the CNAs described in this study have already been described in PTCs from children in one of our previous studies (17) (Table S5).
The main finding of the study is that, in addition to the association of CNAs with the above-mentioned tumor phenotypes, a chromosomal gain of 7q11.22–11.23 is associated with tumors from patients exposed to radioiodine in fallout from the Chernobyl accident. This result was confirmed in a second independent set of tumors by array CGH and by FISH in a subset of cases in which up to 24% of tumor epithelial cells of individual cases showed the alteration. The gain on 7p14.1-q11.23, which was originally found to be associated with exposure to radiation in the main data set, was narrowed down to a smaller region on 7q11.22–11.23 after analysis of CNAs in the validation set. The gain was shown to be absent in all unexposed cases from both tumor sets and was exclusively present in approximately one third of the exposed cases from both tumor sets and thus seemed to be very robust. Our results suggest that the group of exposed cases is, in fact, composed of a number of molecular subgroups. These subgroups show both inter- and intratumor heterogeneity with respect to copy number gains and losses, and therefore it is likely there is more than one molecular route associated with radiation-induced carcinogenesis. It is assumed that a critical event that is required to initiate genomic amplification is a DNA double-strand break, which is instantly followed by the formation of a large so-called DNA palindrome (27). Although double-strand breaks are induced even at very low doses of radiation (28), the subsequent misrepair of primary DNA lesions may vary in a dose-dependent manner, thus leading to different genomic phenotypes defined by CNAs and chromosomal rearrangements.
The frequency of sporadically occurring PTCs in young people is on the order of 1.5 per million per year (29), and the increase in this age group after Chernobyl is on the region of 100-fold (2). Moreover, the estimated proportion of radiation-induced PTCs in our study is ≈85% (18, 19). Thus, it is unlikely that there were a significant number of sporadic PTCs among the exposed group of our study. Moreover, exposed and nonexposed tumors were matched, as far as possible, for as many parameters that could potentially bias the results, such as age, residency, and RET/PTC status, and thus it seems reasonable to propose that copy number gain of 7q11.22–11.23 represents a marker for exposure to radiation. To substantiate this observation, this study needs to be validated by typing of cases from a larger independent validation set, matched for the same criteria as used in the present study, and which, in addition, incorporates detailed data on individually received dose of irradiation and on latency of tumors.
Enrichment analysis of GO terms associated with the genes of the gained region on chromosome band 7q11 revealed the terms “DNA repair,” “response to DNA damage stimulus,” and “cell–cell adhesion” (Table 2). This suggests deregulation of DNA repair processes and subsequent chromosomal instability, which is one of the main features of cells after exposure to ionizing irradiation (30). Response to DNA damage stimulus includes any genes associated with DNA damage response. Therefore, those genes of the gained region associated with this GO term are likely to reflect response to the DNA-damaging radiation the patients were exposed to. Further, deregulated cell–cell adhesion is an important feature of malignant cells (31), leading to tumor invasion and metastasis. The genes CLDN3 and CLDN4, located on chromosome 7q11.22–11.23, are associated with the GO term cell–cell adhesion and have already been reported to be overexpressed in various human tumors (32–36). qRT-PCR analysis of nine genes located on chromosome 7q11.22–11.23 revealed significantly elevated mRNA expression of the three genes PMS2L11, PMS2L3, and STAG3L3 in the exposed cases that showed a gain on 7q11 compared with those that did not show the gain. This correlation strongly suggests an impact of the copy number gain of chromosome band 7q11 on mRNA expression of these genes. Although not statistically significant, the twofold increase in the level of mRNA expression of two additional genes, LIMK1 and PMS2L2, suggests that these may also be of biological relevance in the gain of this chromosomal region. The observation that there is no absolute correlation of mRNA expression with gain of 7q11.22–11.23 is not surprising because it is known from a study by Järvinen et al. (37) that the extent of correlation between copy number gains and mRNA expression is highly variable and does not exceed one third of genes located in the gained region. PMS2L2, PMS2L3, and PMS2L11 belong to the family of PMS2-related genes. PMS2 encodes a protein that acts as a heterodimer with MLH1 in DNA mismatch repair, and heterozygous germ line mutations of PMS2 are associated with hereditary nonpolyposis colon cancer (38, 39). PMS2-related genes share a high degree of identity, including several highly conserved motifs with PMS2, suggesting a functional role in the mismatch repair system (40–42). Because mismatch repair proteins are known to be recruited to the sites of various types of DNA damage including double-strand breaks (43), altered expression of PMS2L genes could interfere with repair processes and contribute to the tumor development. The gene STAG3L3 (Stromal antigen 3-like 3) encodes a protein of as yet unknown function. It is known that STAG3, a member of the synaptonemal complex, is involved in sister chromatid cohesion (44, 45). Barber et al. (46) suggest that chromatin cohesion defects as a result of mutations in STAG3, among other genes, may be a major cause of chromosome instability in human cancer. A similar function of STAG3L3 could link this gene to cancer. LIMK1, which also showed an elevated mRNA expression, is known to regulate actin dynamics and is critical for accurate spindle orientation (47). LIMK1 plays a role in tumor-cell invasion and metastasis (48, 49). CLIP2 mRNA was shown to be overexpressed in the exposed group compared with the unexposed group. CLIP2 (Cap-Gly domain containing linker protein 2) is known to link organelles with microtubules and is deleted in Williams-Beuren syndrome (50, 51). The CLIP2 protein contains a conserved Smc domain (structural maintenance of chromosomes) that is linked to chromosome segregation and cell division (52). Because accurate chromosome segregation during mitosis is required for chromosome stability, and both chromosome missegregation and chromosomal instability are hallmarks of cancer cells (53), this could potentially link CLIP2 to cancer. The CLIP2 gene has been shown to be amplified in glioblastomas and colorectal carcinomas (54, 55), and its potential role in radiation-associated PTC needs to be investigated in future studies.
This study provides a cytogenetic marker and a candidate gene associated with radiation-associated post-Chernobyl PTCs. The gain on 7q11.22–11.23 and the candidate genes the altered region harbors have not been reported in PTC so far. In particular, CLIP2, a gene with an as yet unknown cancer-related function, was shown to be overexpressed in the radiation-exposed cases, suggesting an important and hitherto unknown role in radiation-induced carcinogenesis.
Materials and Methods
Patient Data and Tumor Tissues.
For this study a frequency-matched case–control design (56) was used, and sequential cases of PTC who were born after first January 1987 (nonexposed) were identified from the CTB database. For inclusion in the Genrisk-T cohort, these cases must have frozen tissue available for extraction of RNA and DNA from both normal and tumor tissue and be resident in one of the following areas of Ukraine (receiving an average thyroid dose of 40–120 mGy): Zhytomir, Cherkasse, Chernigov, Kiev, Rovno, or Sumy. Cases were matched as closely as possible on residency, age at operation, and sex to cases of PTC born before April 26, 1986 (exposed).
Experimental Procedures.
DNA from tumors of the discovery and validation cohort was analyzed by BAC array CGH. After primary data analysis the data were subjected to unsupervised hierarchical cluster analysis. To check whether CNAs are associated with radiation exposure or clinical features of the patients, supervised univariate testing was conducted. Selected alterations found by array CGH were validated by FISH on formalin-fixed paraffin-embedded sections of the same cases analyzed by array CGH. Three of the cases that showed a gain that was found to be associated with exposure to radiation and one case that did not show the gain were further characterized by high-resolution array CGH. Moreover candidate genes were tested for differential expression in the groups of cases. A detailed description of the experimental procedures can be found in SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank the International Pathology Panel of the Chernobyl Tissue Bank for confirmation of diagnosis; Dr. A. Abrosimov, Prof. T. I. Bogdanova, Prof. V. LiVolsi, Prof. M. Ito, Prof. J. Rosai, and Prof. E. D. Williams; Dr. Peter Jacob for discussion and recommendations on the estimation of thyroid doses and the proportion of radiation-induced tumors among the exposed cases of this study; and Prof. Bauke Ylstra for providing the array design used for the 180-k Agilent arrays. This study was supported by European Commission Grant FP6-36495–GENRISK-T.
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1017137108/-/DCSupplemental.
References
- 1.Ron E, et al. Thyroid cancer after exposure to external radiation: A pooled analysis of seven studies. Radiat Res. 1995;141:259–277. [PubMed] [Google Scholar]
- 2.Cardis E, et al. Cancer consequences of the Chernobyl accident: 20 years on. J Radiol Prot. 2006;26:127–140. doi: 10.1088/0952-4746/26/2/001. [DOI] [PubMed] [Google Scholar]
- 3.Fugazzola L, et al. Oncogenic rearrangements of the RET proto-oncogene in papillary thyroid carcinomas from children exposed to the Chernobyl nuclear accident. Cancer Res. 1995;55:5617–5620. [PubMed] [Google Scholar]
- 4.Klugbauer S, Lengfelder E, Demidchik EP, Rabes HM. High prevalence of RET rearrangement in thyroid tumors of children from Belarus after the Chernobyl reactor accident. Oncogene. 1995;11:2459–2467. [PubMed] [Google Scholar]
- 5.Thomas GA, et al. High prevalence of RET/PTC rearrangements in Ukrainian and Belarussian post-Chernobyl thyroid papillary carcinomas: A strong correlation between RET/PTC3 and the solid-follicular variant. J Clin Endocrinol Metab. 1999;84:4232–4238. doi: 10.1210/jcem.84.11.6129. [DOI] [PubMed] [Google Scholar]
- 6.Smida J, et al. Distinct frequency of ret rearrangements in papillary thyroid carcinomas of children and adults from Belarus. Int J Cancer. 1999;80:32–38. doi: 10.1002/(sici)1097-0215(19990105)80:1<32::aid-ijc7>3.0.co;2-l. [DOI] [PubMed] [Google Scholar]
- 7.Nikiforov YE, Rowland JM, Bove KE, Monforte-Munoz H, Fagin JA. Distinct pattern of ret oncogene rearrangements in morphological variants of radiation-induced and sporadic thyroid papillary carcinomas in children. Cancer Res. 1997;57:1690–1694. [PubMed] [Google Scholar]
- 8.Powell N, et al. Frequency of BRAF T1796A mutation in papillary thyroid carcinoma relates to age of patient at diagnosis and not to radiation exposure. J Pathol. 2005;205:558–564. doi: 10.1002/path.1736. [DOI] [PubMed] [Google Scholar]
- 9.Tuttle RM, et al. ret/PTC activation is not associated with individual radiation dose estimates in a pilot study of neoplastic thyroid nodules arising in Russian children and adults exposed to Chernobyl fallout. Thyroid. 2008;18:839–846. doi: 10.1089/thy.2008.0072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nikiforov YE. Genetic alterations involved in the transition from well-differentiated to poorly differentiated and anaplastic thyroid carcinomas. Endocr Pathol. 2004;15:319–327. doi: 10.1385/ep:15:4:319. [DOI] [PubMed] [Google Scholar]
- 11.Rhoden KJ, et al. Real-time quantitative RT-PCR identifies distinct c-RET, RET/PTC1 and RET/PTC3 expression patterns in papillary thyroid carcinoma. Lab Invest. 2004;84:1557–1570. doi: 10.1038/labinvest.3700198. [DOI] [PubMed] [Google Scholar]
- 12.Unger K, et al. Heterogeneity in the distribution of RET/PTC rearrangements within individual post-Chernobyl papillary thyroid carcinomas. J Clin Endocrinol Metab. 2004;89:4272–4279. doi: 10.1210/jc.2003-031870. [DOI] [PubMed] [Google Scholar]
- 13.Unger K, et al. RET rearrangements in post-Chernobyl papillary thyroid carcinomas with a short latency analysed by interphase FISH. Br J Cancer. 2006;94:1472–1477. doi: 10.1038/sj.bjc.6603109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ciampi R, Nikiforov YE. RET/PTC rearrangements and BRAF mutations in thyroid tumorigenesis. Endocrinology. 2007;148:936–941. doi: 10.1210/en.2006-0921. [DOI] [PubMed] [Google Scholar]
- 15.Lima J, et al. BRAF mutations are not a major event in post-Chernobyl childhood thyroid carcinomas. J Clin Endocrinol Metab. 2004;89:4267–4271. doi: 10.1210/jc.2003-032224. [DOI] [PubMed] [Google Scholar]
- 16.Nikiforova MN, et al. Low prevalence of BRAF mutations in radiation-induced thyroid tumors in contrast to sporadic papillary carcinomas. Cancer Lett. 2004;209:1–6. doi: 10.1016/j.canlet.2003.12.004. [DOI] [PubMed] [Google Scholar]
- 17.Unger K, et al. Array CGH demonstrates characteristic aberration signatures in human papillary thyroid carcinomas governed by RET/PTC. Oncogene. 2008;27:4592–4602. doi: 10.1038/onc.2008.99. [DOI] [PubMed] [Google Scholar]
- 18.Jacob P, et al. Thyroid cancer among Ukrainians and Belarusians who were children or adolescents at the time of the Chernobyl accident. J Radiol Prot. 2006;26:51–67. doi: 10.1088/0952-4746/26/1/003. [DOI] [PubMed] [Google Scholar]
- 19.Jacob P, et al. Thyroid cancer risk in areas of Ukraine and Belarus affected by the Chernobyl accident. Radiat Res. 2006;165:1–8. doi: 10.1667/rr3479.1. [DOI] [PubMed] [Google Scholar]
- 20.Mullenders L, Atkinson M, Paretzke H, Sabatier L, Bouffler S. Assessing cancer risks of low-dose radiation. Nat Rev Cancer. 2009;9:596–604. doi: 10.1038/nrc2677. [DOI] [PubMed] [Google Scholar]
- 21.Zitzelsberger H, Bauer V, Thomas G, Unger K. Molecular rearrangements in papillary thyroid carcinomas. Clin Chim Acta. 2010;411:301–308. doi: 10.1016/j.cca.2009.11.028. [DOI] [PubMed] [Google Scholar]
- 22.Williams D. Radiation carcinogenesis: lessons from Chernobyl. Oncogene. 2008;27(Suppl 2):S9–S18. doi: 10.1038/onc.2009.349. [DOI] [PubMed] [Google Scholar]
- 23.Harach HR, Williams ED. Childhood thyroid cancer in England and Wales. Br J Cancer. 1995;72:777–783. doi: 10.1038/bjc.1995.410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Port M, et al. A radiation-induced gene signature distinguishes post-Chernobyl from sporadic papillary thyroid cancers. Radiat Res. 2007;168:639–649. doi: 10.1667/RR0968.1. [DOI] [PubMed] [Google Scholar]
- 25.Detours V, et al. Genome-wide gene expression profiling suggests distinct radiation susceptibilities in sporadic and post-Chernobyl papillary thyroid cancers. Br J Cancer. 2007;97:818–825. doi: 10.1038/sj.bjc.6603938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhu Z, Ciampi R, Nikiforova MN, Gandhi M, Nikiforov YE. Prevalence of RET/PTC rearrangements in thyroid papillary carcinomas: Effects of the detection methods and genetic heterogeneity. J Clin Endocrinol Metab. 2006;91:3603–3610. doi: 10.1210/jc.2006-1006. [DOI] [PubMed] [Google Scholar]
- 27.Tanaka H, et al. Intrastrand annealing leads to the formation of a large DNA palindrome and determines the boundaries of genomic amplification in human cancer. Mol Cell Biol. 2007;27:1993–2002. doi: 10.1128/MCB.01313-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Löbrich M, et al. In vivo formation and repair of DNA double-strand breaks after computed tomography examinations. Proc Natl Acad Sci USA. 2005;102:8984–8989. doi: 10.1073/pnas.0501895102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Muir CS, Waterhouse J, Mack T, Powell J, Whelan S, editors. Cancer Incidence in Five Continents: Volume V. Lyon, France: IARC Scientific Publications; 1987. [Google Scholar]
- 30.Mahaney BL, Meek K, Lees-Miller SP. Repair of ionizing radiation-induced DNA double-strand breaks by non-homologous end-joining. Biochem J. 2009;417:639–650. doi: 10.1042/BJ20080413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Moh MC, Shen S. The roles of cell adhesion molecules in tumor suppression and cell migration: A new paradox. Cell Adh Migr. 2009;3:334–336. doi: 10.4161/cam.3.4.9246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rangel LB, et al. Tight junction proteins claudin-3 and claudin-4 are frequently overexpressed in ovarian cancer but not in ovarian cystadenomas. Clin Cancer Res. 2003;9:2567–2575. [PubMed] [Google Scholar]
- 33.Kominsky SL, et al. Clostridium perfringens enterotoxin elicits rapid and specific cytolysis of breast carcinoma cells mediated through tight junction proteins claudin 3 and 4. Am J Pathol. 2004;164:1627–1633. doi: 10.1016/S0002-9440(10)63721-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.de Oliveira SS, de Oliveira IM, De Souza W, Morgado-Díaz JA. Claudins upregulation in human colorectal cancer. FEBS Lett. 2005;579:6179–6185. doi: 10.1016/j.febslet.2005.09.091. [DOI] [PubMed] [Google Scholar]
- 35.Santin AD, et al. Overexpression of Clostridium perfringens enterotoxin receptors claudin-3 and claudin-4 in uterine carcinosarcomas. Clin Cancer Res. 2007;13:3339–3346. doi: 10.1158/1078-0432.CCR-06-3037. [DOI] [PubMed] [Google Scholar]
- 36.Mees ST, et al. Expression of tight and adherens junction proteins in ulcerative colitis associated colorectal carcinoma: Upregulation of claudin-1, claudin-3, claudin-4, and beta-catenin. Int J Colorectal Dis. 2009;24:361–368. doi: 10.1007/s00384-009-0653-y. [DOI] [PubMed] [Google Scholar]
- 37.Järvinen AK, et al. High-resolution copy number and gene expression microarray analyses of head and neck squamous cell carcinoma cell lines of tongue and larynx. Genes Chromosomes Cancer. 2008;47:500–509. doi: 10.1002/gcc.20551. [DOI] [PubMed] [Google Scholar]
- 38.Li GM, Modrich P. Restoration of mismatch repair to nuclear extracts of H6 colorectal tumor cells by a heterodimer of human MutL homologs. Proc Natl Acad Sci USA. 1995;92:1950–1954. doi: 10.1073/pnas.92.6.1950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lynch HT, de la Chapelle A. Genetic susceptibility to non-polyposis colorectal cancer. J Med Genet. 1999;36:801–818. [PMC free article] [PubMed] [Google Scholar]
- 40.Nicolaides NC, et al. Genomic organization of the human PMS2 gene family. Genomics. 1995;30:195–206. doi: 10.1006/geno.1995.9885. [DOI] [PubMed] [Google Scholar]
- 41.Osborne LR, et al. PMS2-related genes flank the rearrangement breakpoints associated with Williams syndrome and other diseases on human chromosome 7. Genomics. 1997;45:402–406. doi: 10.1006/geno.1997.4923. [DOI] [PubMed] [Google Scholar]
- 42.Kondo E, Horii A, Fukushige S. The human PMS2L proteins do not interact with hMLH1, a major DNA mismatch repair protein. J Biochem. 1999;125:818–825. doi: 10.1093/oxfordjournals.jbchem.a022354. [DOI] [PubMed] [Google Scholar]
- 43.Hong Z, et al. Recruitment of mismatch repair proteins to the site of DNA damage in human cells. J Cell Sci. 2008;121:3146–3154. doi: 10.1242/jcs.026393. [DOI] [PubMed] [Google Scholar]
- 44.Pezzi N, et al. STAG3, a novel gene encoding a protein involved in meiotic chromosome pairing and location of STAG3-related genes flanking the Williams-Beuren syndrome deletion. FASEB J. 2000;14:581–592. doi: 10.1096/fasebj.14.3.581. [DOI] [PubMed] [Google Scholar]
- 45.Prieto I, et al. Mammalian STAG3 is a cohesin specific to sister chromatid arms in meiosis I. Nat Cell Biol. 2001;3:761–766. doi: 10.1038/35087082. [DOI] [PubMed] [Google Scholar]
- 46.Barber TD, et al. Chromatid cohesion defects may underlie chromosome instability in human colorectal cancers. Proc Natl Acad Sci USA. 2008;105:3443–3448. doi: 10.1073/pnas.0712384105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kaji N, Muramoto A, Mizuno K. LIM kinase-mediated cofilin phosphorylation during mitosis is required for precise spindle positioning. J Biol Chem. 2008;283:4983–4992. doi: 10.1074/jbc.M708644200. [DOI] [PubMed] [Google Scholar]
- 48.Yoshioka K, Foletta V, Bernard O, Itoh K. A role for LIM kinase in cancer invasion. Proc Natl Acad Sci USA. 2003;100:7247–7252. doi: 10.1073/pnas.1232344100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Davila M, Frost AR, Grizzle WE, Chakrabarti R. LIM kinase 1 is essential for the invasive growth of prostate epithelial cells: Implications in prostate cancer. J Biol Chem. 2003;278:36868–36875. doi: 10.1074/jbc.M306196200. [DOI] [PubMed] [Google Scholar]
- 50.Hoogenraad CC, Akhmanova A, Galjart N, De Zeeuw CI. LIMK1 and CLIP-115: Linking cytoskeletal defects to Williams syndrome. Bioessays. 2004;26:141–150. doi: 10.1002/bies.10402. [DOI] [PubMed] [Google Scholar]
- 51.Ferrero GB, et al. An atypical 7q11.23 deletion in a normal IQ Williams-Beuren syndrome patient. Eur J Hum Genet. 2010;18:33–38. doi: 10.1038/ejhg.2009.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Marchler-Bauer A, et al. CDD: specific functional annotation with the Conserved Domain Database. Nucleic Acids Res. 2009;37(Database issue):D205–D210. doi: 10.1093/nar/gkn845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ricke RM, van Ree JH, van Deursen JM. Whole chromosome instability and cancer: A complex relationship. Trends Genet. 2008;24:457–466. doi: 10.1016/j.tig.2008.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Suzuki T, et al. Genetic analysis of human glioblastomas using a genomic microarray system. Brain Tumor Pathol. 2004;21:27–34. doi: 10.1007/BF02482174. [DOI] [PubMed] [Google Scholar]
- 55.Lassmann S, et al. Array CGH identifies distinct DNA copy number profiles of oncogenes and tumor suppressor genes in chromosomal- and microsatellite-unstable sporadic colorectal carcinomas. J Mol Med. 2007;85:293–304. doi: 10.1007/s00109-006-0126-5. [DOI] [PubMed] [Google Scholar]
- 56.Katz MH. Study Design and Statistical Analysis: A Practical Guide for Clinicians. Cambridge, UK: Cambridge Univ Press; 2006. [Google Scholar]
- 57.Horii A, Han HJ, Sasaki S, Shimada M, Nakamura Y. Cloning, characterization and chromosomal assignment of the human genes homologous to yeast PMS1, a member of mismatch repair genes. Biochem Biophys Res Commun. 1994;204:1257–1264. doi: 10.1006/bbrc.1994.2598. [DOI] [PubMed] [Google Scholar]
- 58.Tomida J, et al. DNA damage-induced ubiquitylation of RFC2 subunit of replication factor C complex. J Biol Chem. 2008;283:9071–9079. doi: 10.1074/jbc.M709835200. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.