Abstract
Methyl-sensitive cut counting (MSCC) with the HpaII methylation-sensitive restriction enzyme is a cost-effective method to pinpoint unmethylated CpGs at single base-pair resolution. However, it has the drawback of addressing only CpGs in the context of the CCGG site, leaving out the remainder of the possible 16 XCGX tetranucleotides in which CpGs are found. We expanded MSCC to include three additional enzymes to address a total of 5 of the 16 XCGX combinations. This allowed us to survey methylation at about one-third of all a mammalian genome's CpGs. Applied to mouse liver DNA, we correctly confirmed data reported with other methods showing hypomethylation to be concentrated at promoters and in CpG islands (CGIs), with gene bodies and intergenic regions being mostly methylated. Grouping unmethylated CpGs, characterized by high MSCC scores (7% false discovery rate), we found a large number of unmethylated regions not qualifying as CGIs located in intergenic and intronic regions, which are highly enriched in functional DNA sequences (open regulatory annotation database) as well as in noncoding yet highly conserved mammalian sequences thought to be important but with as yet unknown function. About 50% of MSCC-defined unmethylated regions do not overlap algorithm-defined CGIs and offer a novel search space in which new functionalities of DNA may be found in health and disease.
Keywords: methylome, single CpG, genome-wide, functional annotation, repetitive
Methylation of DNA at position five of the cytosine ring is a widespread modification in the vertebrate genomes (1). A family of DNA methyltransferases whose primary targets are the cytosines located at CpG dinucleotides catalyzes this chemical modification (2). Many CpGs are not distributed at random, because a significant proportion of them have coalesced into what has been called CpG islands (CGIs), where they are mostly hypomethylated, whereas CpGs outside of CGIs are mostly methylated (1, 3–5). CGIs are identified with computer algorithms that search for shared distinctive properties; traditionally CpG and (G + C) richness (5–8). A different approach selects CGIs between clusters of CpGs whose maximum inter-CpG distances are below a threshold (e.g., median genomic inter-CpG distance) (9). The filtering criteria used by all these programs seek to optimize the possibility that the selected CGIs are not the product of chance but the result of evolutionary processes. The most widely accepted explanation for the origin of CGIs is based in the tendency of 5-methylcytosines to undergo spontaneous deamination to uracil producing C-to-T mutations. This process drove a nonselective purge of CpGs from the broadly methylated genomic sequences with no evolutionary constraints (10, 11) (SI Appendix, Tables S1, S2, and S3). However, this purge did not occur in regions rich in regulatory elements that have been protected from being methylated. According to this simplified and generally accepted hypothesis, the functionality of a CGI is measured by the probability of finding it unmethylated. For example, the program called CpGCluster uses a statistical criterion (P value) to select for clusters with low probability of having been formed by chance (9). This means that these loci have retained their CpG density during evolution, presumably because of their prevalence in an unmethylated state. CGIs, including CpGCluster CGIs with the lowest P values, are more frequently found overlapping promoters (12), which supports connections between evolutionary origin, unmethylated state, and functionality. The idea that active promoters protect their CpGs from being methylated is supported by site-specific mutagenesis experiments. For example, mutations that prevent the transcription factor Sp1 from recognizing and binding its target sequences in a particular CGI remove the protection of that CGI from DNA methylation (13).
Research efforts focused on improving the prediction of locations of CGIs aim to identify functionally relevant epigenetic loci in development and disease; as a consequence, CGIs still constitute the framework on which the majority of researchers base their high-throughput methylation analyses. However, the filtering criteria used by these programs frequently fail to identify a large percentage of subsequences having the potential to encode regulatory functions that can be disrupted or activated by changes in methylation.
Inspection of the mammalian genome shows it to be divided into two classes of subsequences. In one class (85% of the genome), CpGs are sparse (one every 250 bp). The other class (15% of the genome) concentrates half of the total genomic CpGs at an average inter-CpG distance of 40 bp. At this density, methylation has been shown to have a deleterious effect on the functionality of the DNA elements (14). These relative CpG-rich subsequences accommodate the totality of the CGIs, regardless of the algorithms used to define them, and overlap with 95% of the RefSeq-defined transcription start site (TSS) regions (−3 Kb to +2 Kb from the TSS).
Our goal was to design a method that allows us to identify targets of methylation-mediated epigenetic processes throughout the genome without having to select a priori candidate subsequences. We developed a high-throughput sequencing-based DNA methylation analysis, which consists of an expanded (more comprehensive) version of the methyl-sensitive cut counting assay (MSCC) (15, 16). Applied to mouse, our method identifies the methylation status of 6 million CpGs (one-third of all existing CpGs) and covers 58% of the CpG-rich subsequences.
We found that a surprising proportion (50%) of our unmethylated regions (UMRs) do not meet the traditional CGI criteria. Interestingly, most of these non-CGI UMRs are located in noncoding DNA outside of promoters and are more enriched in experimentally determined regulatory sequences than CGI-like UMRs (UMRs that overlap predicted CGIs). At least 10% of the UMRs identified through MSCC contain literature-published liver-specific and liver-related regulatory sequences.
Results and Discussion
Coverage.
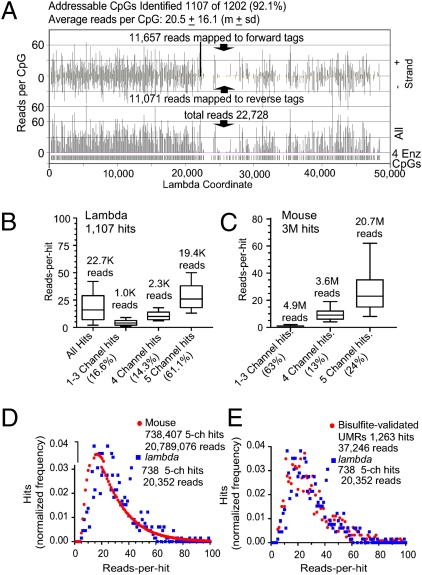
CpG tag libraries prepared from DNA digested with four methylation-sensitive restriction enzymes (4Enz; SI Appendix, SI Materials and Methods) were sequenced by Illumina Genome Analyzers reporting the sequences returned by individual sequencing channels of their flow cells. The data analyzed here were obtained from sequencing three CpG tag libraries, yielding three datasets that we called Slax1, Slxa2, and Slxa3 (SI Appendix, Table S4). The majority of the data analyzed below are from the Slxa3 dataset returned to us from five sequencing channels, which, when pooled, rendered 29 million reads that mapped to unique nonrandom CpGs (SI Appendix, Tables S4 and S5). The number of reads per identified CpG (MSCC score, see below) was found to vary from site to site because of disparities in the level of methylation, but a minimum coverage is required to be able to perceive these differences. We used the reads recovered from lambda phage (lambda) CpGs to gauge the coverage and followed the behavior of the 1,202 unmethylated lambda CpGs addressable by 4Enz during the preparation of our CpG tag libraries. A read frequency map of the 22,728 reads that mapped to the lambda genome is shown in Fig. 1A. We identified 92% (1,107 hits) of the 4Enz CpGs with at least 1 read and an average reads-per-hit ratio of 20.5 ± 16.1 (mean ± SD). Thus, although the measures come from equally unmethylated sites, not all were identified with similar frequency. The read values were scattered between a minimum of 0 (8%) and a maximum of 99. For the mouse genome, whose methylation status is unknown, it is necessary to discriminate between highly methylated CpGs and CpGs that were poorly covered. Because we aligned the reads returned by each of the five channels separately, we classified the CpGs according to the number of channels in which they were identified, revealing a relation between the experimental coverage and the number of channels in which each CpG was identified. The hits were grouped into three classes: one- to three-channel hits, four-channel hits, and five-channel hits (Fig. 1 B and C). We compared the number of CpGs identified in the mouse and lambda genomes according to these criteria. At a sequencing depth of about 30 million mapped reads from five channels, for lambda (100% unmethylated), the majority of the CpGs (61%) were identified by reads that came from all five channels and only a small portion (16%) from one to three channels. The proportion of lambda one- to three-channel hits plus the proportion of not identified CpGs estimate the failure rate of the method. In contrast to lambda, we found only 24% of mouse hits in five channels. The majority of which (63%) were in one to three channels at an average of 2.5 reads per hit, which reflects the widely methylated status of the genome and the sensitivity of the method to detect CpGs even when they are heavily methylated (Fig. 2). The box-and-whisker plots in Fig. 1 B and C show that CpGs located in the same class were identified with a similar level of reads. Interestingly, the MSCC scores (reads per hit) recorded for mouse or lambda five-channel hits show a similar distribution of values (median MSCC score of 24.5 for lambda and 23.2 for mouse). The distributions of MSCC scores obtained from the mouse and lambda genomes are compared in Fig. 1D, which reflect the difference between a completely unmethylated genome and one with different levels of hypomethylation. For the mouse genome, we obtained higher frequencies of CpGs identified with lower numbers of reads. However, when we compared lambda CpGs against those located in the newly discovered mouse UMRs, the MSCC score distributions showed no difference (Fig. 1E). By comparing different CpG tag libraries in which the same amount of lambda DNA was introduced as an internal standard, we found that certain sites systematically perform better or worse than others (SI Appendix, Fig. S2). Although this systematic bias increases the variability above that expected from the Poisson distribution, it is shown below that the level of methylation of a CpG under study is the primary parameter determining the final number of reads.
Fig. 1.
Analysis of reads that mapped to lambda DNA: comparison with mouse. (A) Plots of reads per hit (identified CpGs) along the lambda genome. (Upper) Reads-per-hit plot of identified forward and reverse tags. (Lower) Combined forward and reverse reads per hit. (B) Box-and-whisker plots (median, quartiles, and fifth and 95th percentiles) representing the reads-per-hit distribution as a function of the number of channels in which the CpG-identifying reads were found in the lambda genome. (C) Same as in B but for CpGs detected in the mouse genome. (D and E) Frequency histograms of the reads per hit recovered for CpGs identified with 4Enz in the five sequencing channels. ch, channel. (D) Distribution of read recovery from the lambda genome is compared with that of reads recovered from the whole-mouse genome. (E) Distribution of read recovery from the lambda genome is compared with that of the reads per hit found in CpGs that were located in UMRs of the mouse genome validated by bisulfite sequencing analysis.
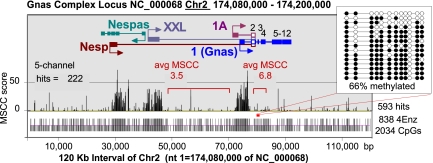
Fig. 2.
Single CpG resolution profile of hypomethylation on a genome-wide scale. The MSCC data from the genomic region spanning the Gnas complex locus are shown. The promoters for Nesp, Nespas, Gnasxl, and Exon1A lie within differentially methylated regions (DMRs) that have been identified in the locus. Exon 1 of Gnas is located in a biallelic UMR contiguous to the DMR containing exon 1A. Tick bars at the bottom of the figure indicate the positions of CpGs and 4Enz CpGs. The majority of the sites are largely resistant to 4Enz, except for those located in three regions colocalizing with the described promoters. Low avgMSCC scores for two regions outside the three major UMRs indicate that these regions are heavily methylated (Fig. 3). (Inset) Bisulfite sequencing analysis confirms this conclusion.
Assigning Methylation Status at Single-CpG Resolution.
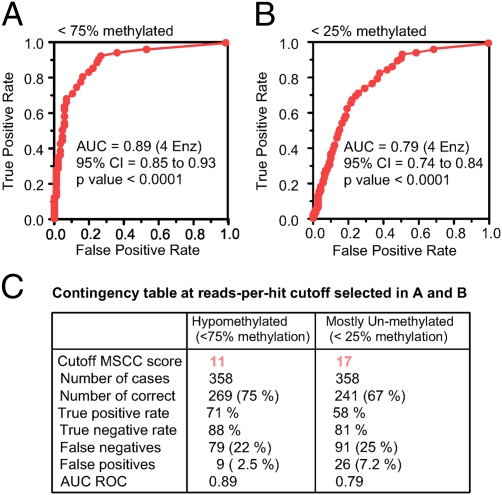
The 4Enz set of methylation-sensitive restriction enzymes accurately targets 6 million CpGs located in the context of five different patterns (CCGG, ACGT, GCGC, CCGC, and GCGG) and collects information about their methylation state. The number of CpGs and the genomic regions that can be studied (addressable CpGs) depend not only on the number of restriction enzymes used but on the number of CpGs with unique tags (SI Appendix, Tables S1, S2, S3, S4, and S5). Although the abundance of any individual tag in the CpG tag library is expected to be inversely proportional to the methylation state of the addressed CpG (15, 16), the demonstrated local bias impairs the usefulness of the method to perceive moderate variations in levels of methylation within a genome (SI Appendix, Fig. S2). Despite this limitation, the method is sensitive to detect the small fraction of hypomethylated CpGs in the genome (Fig. 2). We used receiver operating characteristic (ROC) analysis to visualize and compare the performance of the method to classify CpGs in different categories of methylation according to their MSCC scores. ROC curves (Fig. 3) were built following the strategy described in SI Appendix, SI Materials and Methods using the MSCC scores and methylation status of a panel of 358 CpGs validated by bisulfite sequencing analysis.
Fig. 3.
ROC analysis generated from MSCC scores and bisulfite sequencing data. (A) MSCC scores are used to classify CpGs as hypomethylated (<75% methylation) or heavily methylated (>75% methylation). (B) Same as in A but classifying CpGs as mostly unmethylated (<25% methylation) or not. (C) Summary of the results when an MSCC score of 11 or 17 is selected as the optimal cutoff for the classification process. The true-positive rate indicates CpGs with a methylation rate <75% (25%) and an MSCC score >11 (or 17)/CpGs with a methylation rate <75% (or <25%). The true-negative rate indicates CpGs with a methylation rate >75% (or 25%) and an MSCC score <11 (or 17)/total CpGs with a methylation rate >75% (or 25%). The false-negative rate indicates CpGs with a methylation rate <75% (or 25%) and an MSCC score <11 (or 17). The false-positive rate indicates CpGs with a methylation rate >75% (or 25%) and an MSCC score >11 (or 17).
Fig. 3 shows the area under the curve (AUC) plots for two situations: detection of sites <75% methylated and detection of sites <25% methylated. The AUC in Fig. 3A is 0.89 ± 0.02 and represents the probability that a randomly selected hypomethylated CpG (<75% methylation) will score a higher number of reads than a randomly selected heavily methylated CpG (>75% methylation). We found a MSCC score of 11 as the optimal cutoff for this classification (SI Appendix, Fig. S3A). A similar analysis to detect mostly unmethylated sites (<25% methylation) (Fig. 3B and SI Appendix, Fig. S3B) showed that using an MSCC score of 17 as a cutoff allows us to classify 67% of CpGs correctly as mostly unmethylated with a false discovery rate (FDR) of 7.2% (Fig. 3C). Single-CpG MSCC scores are affected by local systematic bias (SI Appendix, Fig. S2); however, CpGs with a tendency to be overestimated or underestimated are randomly distributed in the genome. When the level of methylation of a discrete region of the genome is measured by averaging the counts from the individual CpGs, the bias tends to cancel. For this reason, the larger the number of CpGs that can be covered in the MSCC analysis, the higher is the accuracy with which the method evaluates the level of unmethylation of a given region.
The average MSCC (avgMSCC) score was thus used to quantify the level of hypomethylation of two regions known to differ by 50% in their methylation status as a result of imprinting (17) (SI Appendix, Fig. S4). As recorded in three independent experiments, the differentially methylated 1A domain of the Gnas locus produced an avgMSCC score that was one-half of that scored for the neighboring completely unmethylated exon 1 domain (SI Appendix, Fig. S4). We concluded that avgMSCC scores reflect the hypomethylation status of a region accurately and reproducibly.
Distribution of Hypomethylation in the Genome.
We mapped 29 million reads to 3 million unique CpG tags (SI Appendix, Table S5) and built a read frequency table (all-CpGs-all-hits frequency table; SI Appendix, Table S6) listing all addressable CpG tags (forward and reverse) and the number of times that each unique tag was identified. The MSCC threshold values derived from ROC analysis were used to assign methylation status to each CpG listed in this table, giving a distribution of hypomethylation throughout the entire genome.
The landscape that emerged from this analysis is in complete agreement with the known genome-wide mosaic pattern showing heavily methylated sites sharply separated from hypomethylated sites (Fig. 2 and SI Appendix, Fig. S4). However, the hypomethylated regions corresponded poorly with predicted CGIs. We analyzed the degree of hypomethylation in four different sets of CGIs: Gardiner-Garden and Frommer (GG&F), Takai and Jones (T&J), CpGCluster CGIs, and Epi-CGIs, with the last being based on an algorithm that combines the GG&F criterion with information gathered from epigenetic marks (6–9) (Fig. 4).
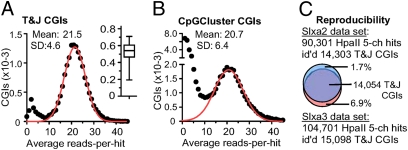
Fig. 4.
Analysis of methylation at CGIs. (A) T&J CGIs, wherein addressable CpGs represent one-third of the total CpGs, were selected for this analysis. At each CGI, the average reads-per-hit ratio was calculated, and the distribution of these ratios is represented as a frequency histogram (black dots). The red curve is the result of fitting a Gaussian model to the data. Means and SDs were calculated from this model. (Inset) Distribution of the fractional addressability of CpGs among T&J CGIs. The box-and-whisker plot depicts the median, first and third quartiles, and fifth and 95th percentiles. (B) Same as in A but analyzing CGIs predicted by CpGCluster. (C) Reproducibility in the identification of unmethylated T&J CGIs with HpaII hits in two experiments (SI Appendix, Table S4). The 5-ch HpaII hits of the Slxa2 experiment collected 4,396,101 reads; those of the Slxa3 experiment collected 5,904,634 reads. 5-ch, five-channel.
Although our method can interrogate, on average, one of every two CpGs located in CGI-like loci, not all CGIs are interrogated to the same extent (Fig. 4 A and Inset). We included in our analysis those CGIs in which we can address at least one-third of the total CpGs (79% of T&J, 52% of GG&F, 56% of CpGCluster CGIs, and 60% of Epi-CGIs; SI Appendix, Table S8). Fig. 4A shows the distribution of the degree of unmethylation (avgMSCC score) for 16,731 of the mouse genome's T&J CGIs. According to the scores, the T&J set partitions into 14,993 (∼90%) hypomethylated islands [MSCC score: 21.5 ± 4.6 (mean ± SD)] and 1,738 (∼10%) heavily methylated islands. Bisulfite sequencing confirmed this partition of CGIs (SI Appendix, Fig. S5). The majority of T&J CGIs are located in promoter regions (SI Appendix, Table S7). This bias is probably what makes the T&J set very specific in terms of predicting UMRs; however, sensitivity is a concern for this algorithm. We performed the same analysis for the methylation status of CGIs originated with the CpGCluster algorithm (Fig. 4B). This set is three times larger than the T&J set, but 58% (39,022) of them were found to be heavily methylated, whereas only 32% (21,386) were found to be unmethylated [avgMSCC score: 20.7 ± 6.4 (mean ± SD)]. Notice that whereas the T&J algorithm was more effective in predicting UMRs, it missed classifying a substantial number of CGIs compared with CpGCluster (8,926 CGIs). We found that 70% of these 8,926 unmethylated loci are located in introns or intergenic regions. We extended this analysis to GG&F CGIs and Epi-CGIs (SI Appendix, Table S8). The methylation profile of the GG&F CGIs resembled that of CpGCluster CGIs (Fig. 4B); however, the number of unmethylated loci increased to 23,318 (SI Appendix, Table S8). The Epi-CGI algorithm appears to be as effective as the T&J algorithm in predicting unmethylated islands (67% of the Epi-CGIs analyzed); even though it finds the smallest number of unmethylated loci (11,191), these are less biased toward TSS regions compared with the T&J set (SI Appendix, Tables S7 and S8).
Reproducibility.
The avgMSCC score of five-channel hits in our dataset (Slxa3) was 28.3. Thus, five-channel hits identify unmethylated CpGs. We found that of 199,618 HpaII five-channel hits with an avgMSCC score of 29.5, 104,701 hits mapped to 15,098 (unmethylated) T&J CGIs. In an independent experiment sequenced to a similar depth (Slxa2), we found that 90,301 HpaII five-channel hits mapped to 14,303 T&J CGIs. A common set of 14,054 T&J CGIs was identified in the two experiments (Fig. 4C), indicating that the MSCC approach is a highly reproducible tool to identify UMRs.
Genome-Wide Annotation of Experimentally Determined Hypomethylated Regions.
We made the CpGCluster program (9) specific for 4Enz CpGs (GCGC, CCGG, GCGG, CCGC, and ACGT) and used it to cluster all CpGs identified in five channels. This group comprises 738,407 CpGs with a median MSCC score of 23, of which 90% scored more than 10 reads (Fig. 1C). The modified CpGCluster program creates clusters of addressed CpGs with specific inter-CpG distances (SI Appendix, Fig. S6). If all 6 million addressable CpGs were randomly distributed, the distances between neighboring sites should follow the geometrical distribution with a mean intersite separation of 311 bp. We set the distance to 300 bp to search for hypomethylated CpG clusters (w300 clusters) and found 559,901 hypomethylated CpGs grouped in 64,266 clusters (SI Appendix, Table S9). Although the five-channel CpGs included in these clusters have an avgMSCC score of 29, we also found 287,456 CpGs with an avgMSCC score of 6.9. This finding is an indication that certain clusters could have a considerable number of CpGs with high rates of methylation (SI Appendix, Fig. S7B and SI Appendix, Table S10).
Isolation and Analysis of UMRs.
We showed that single CpGs with MSCC scores ≥17 can be classified as mostly unmethylated (<25% methylation), generating a low number of false-positive results (7% FDR). We reasoned that a hypomethylated region with an avgMSCC score ≥17 will have the majority of its CpGs in an unmethylated state. After calculating and applying this cutoff, we ended with a set of 46,804 UMRs that span 22 million bp of the mouse genome and include 1.3 million unmethylated CpGs (SI Appendix, Tables S9 and S11). A number of studies have shown that CpGs located in close proximity tend to share a common methylation state (15, 18, 19). The penetrance of this correlation increases with the proximity of neighboring CpGs. We found that 97% of nonaddressable CpGs located in UMRs have at least 1 addressable CpG within a distance of 100 bp. The correlation between methylation states for CpGs within a distance of 100 bp has been estimated to be ∼75% (18). Bisulfite sequencing analysis of randomly selected UMRs confirms the comethylation phenomenon (SI Appendix, Figs. S7 and S8). We conclude that the UMRs with avgMSCC scores ≥17 constitute a set of mostly unmethylated sequences.
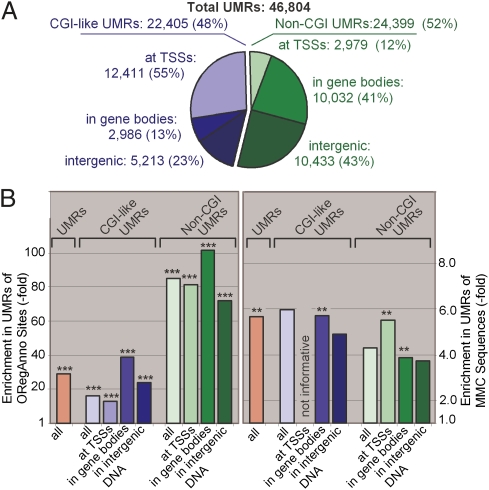
We analyzed the colocalization of UMRs with predicted CGIs and classified them in non-CGI UMRs (do not overlap) and CGI-like UMRs (overlap). The result showed a poor correspondence. For example the T&J and CpGCluster CGIs fail to detect 75% and 60%, respectively, of the experimentally determined UMRs (SI Appendix, Table S9). Indeed, 52% of the UMRs could not be detected by any of the CGI-defining algorithms (Fig. 5A). The high failure rate, even after combining different CGI sets, suggests that the algorithms are failing to include one or more critical features. We hypothesize that protein-binding DNA elements must be a ubiquitous feature shared by all the UMRs.
Fig. 5.
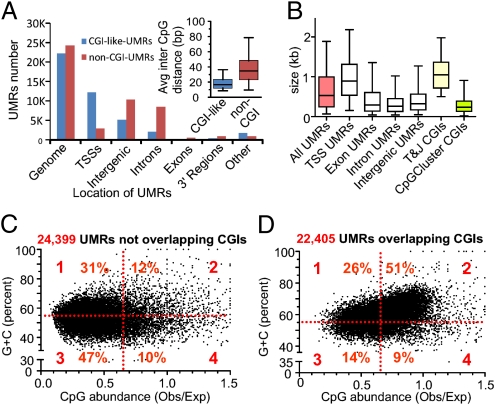
CGI-like and non-CGI UMRs as detected genome-wide in the mouse liver genome. (A) Distribution of UMRs in the major genomic compartments: TSS regions (−3 Kb to +2 Kb of the TSS) and 3′ region of genes (3 Kb). For this segmentation, the RefSeq definition of genes was used. If a UMR is not completely included in one of the five categories, it was labeled as “other.” UMRs were classified as overlapping or not in the combined set of CGIs (T&J, GG&F, or CpGCluster). (Inset) Distributions of the average inter-CpG distance calculated for each UMR, represented as box-and-whisker plots depicting the median, quartiles, and fifth and 95th percentiles. (B) Size distribution of UMRs and comparison with CGIs. Box-and-whisker plots (median, quartiles, and fifth and 95th percentiles) represent the distribution of sizes for UMRs that were completely included in the indicated genomic regions. (C and D) Scatter plots represent the (C + G) content vs. Obs/Exp CpG ratio in UMRs that overlap and do not overlap CGIs. Note the large proportion of UMRs that do not meet CGI criteria. Obs/Exp, observed/expected.
The original quantitative criteria used to define CGIs were based on a small set of sequences, likely biased by the limited size of the 1985 GenBank database (4, 7). Later programs readjusted thresholds and changed how the edges of CGIs are defined (5, 8). However, most of these new computational methods still rely on the three initially considered DNA features: Obs/Exp CpG ratio, G + C content, and length. We found these three parameters to be highly variable among the UMRs (Fig. 5). For example, many non-CGI UMRs meet the first two criteria mentioned, but all of them have sizes below the 500 bp required by the T&J algorithm (Fig. 5C, second quadrant). Also, many UMRs do not overlap CGIs but meet the size required by different algorithms (SI Appendix, Table S9). This shows that the size (base pairs) of a region rich in CpGs should not be used as a filter to select for putatively functionally important unmethylated sequences. The size of the UMRs was found to be primarily a function of their genomic location, with larger UMRs overlapping TSSs and smaller UMRs being located in intergenic regions (Fig. 5B). On the other hand, there are UMRs that overlap predicted CGIs but the CpG and G + C richness are below the CGI thresholds (Fig. 5D, quadrants 1, 3, and 4), suggesting that in addition to colocalizing, they cover substantially different sequences. The inability to predict the edges of CGIs seems not to be trivial. It has been reported that tissue- and cancer-specific differentially methylated regions map to the shores of CGIs (20). Recently, computational algorithms have incorporated hidden Markov models (HMMs) to improve the detection of CGI borders. Many of the HMM CGIs incorporated the shores of previously defined islands (5). Although this new strategy predicts a relative large number of CGIs, the majority (75%) do not overlap our experimentally determined UMRs. Our description of thousands of UMRs not qualifying as CGIs outside of promoter regions is in agreement with the previous discovery of nonpromoter UMRs subject to tissue-specific de novo methylation and suggests the existence of additional tissue-specific UMRs not evident in liver (21).
The relation between CpG density and methylation has been studied in promoter and nonpromoter regions. Whereas promoters with high CpG density (more than one CpG every 20 bp) are found to be primary unmethylated, the methylation rate at single-copy DNAs outside promoters was found to increase with the CpG density until a threshold value of 0.025 (one CpG every 40 bp) was reached; beyond this threshold, the methylation rates fell sharply (19). Interestingly, we found that CGI-like UMRs and non-CGI UMRs differ in their CpG density. CGI-like UMRs have a median inter-CpG distance of 18 bp, which is just above the expected distance for the CpG dinucleotide in a DNA sequence with identical base composition (A = T = C = G). In contrast, the median distance between CpGs in the non-CGI UMRs is 34 bp (Fig. 5A, Inset). Whether these differences have a relationship to the functionality of the underlying sequences has to be determined in future experiments. On the other hand, there is a clear pattern in how these two kinds of UMR partition among different noncoding compartments of the genome (Fig. 5A). Whereas 55% of CGI-like UMRs localize to TSS regions, only 12% of non-CGI UMRs do. Whereas less than 35% of CGI-like UMRs localize to intronic or intergenic sequences, more than 80% of non-CGI UMRs do. The finding that 70% of UMRs are located in exons, introns, and intergenic regions was unexpected. Introns and intergenic sequences account for almost 98% of mammalian genomes, and most of the CpGs located in these are heavily methylated (15). Mammalian DNA regulatory sequences are principally located in noncoding sequences but concentrated preferentially in the 5′-flanking regions of genes, leaving introns and intergenic regions virtually devoid of evolutionary constraints. However, the comparison of complete genomes has revealed a large number of conserved non–protein-coding DNA sequences mapping to intergenic and intronic regions, for most of which their biological function remains unknown (22).
We hypothesized that UMRs are highlighting functional sequences, which, if proven, emphasizes the usefulness of our method as a tool to identify loci at which epigenetic mechanisms could influence complex phenotypes and diseases. To provide genome-wide evidence in favor of our hypothesis, we evaluated the functional significance of our UMRs by asking if they overlap with highly conserved mammalian sequences and/or with experimentally determined protein-binding sites. To test the potential functionality of our UMRs, we used two tracks of the University of California, Santa Cruz Genome Browser (23). One is the mammal most conserved (MMC) track of conserved sequences or elements based on whole-genome alignments of different mammalian species. The other is the track for open regulatory annotation (ORegAnno), which includes literature-curated regulatory regions and transcription factor-binding sites (24). We reasoned that the sequences of these tracks could be used as probes that would allow us to follow the partitioning of regulatory sequences between the methylated regions and UMRs of the genome. We found that our UMRs are indeed enriched in both MMCs and ORegAnno sites (SI Appendix, Table S12). For ORegAnno sites, 25% populating 10% of the UMRs and non-CGI UMRs (with the majority in intronic and intergenic regions) reached enrichments of 100-fold compared with the concentration at which these regulatory elements are found in the whole genome (Fig. 6 and SI Appendix, Table S12). The likelihood that this enrichment occurred by chance is less than 4 in 100,000. Interestingly, 98% of all ORegAnno elements overlapping UMRs belong to binding sites for Esr1 and Foxa2. The Foxa 2 gene codes for the forkhead box protein A2, which is a transcriptional activator for liver-specific genes (25), and the Esr1 gene codes for the nuclear estrogen receptor α. This is the major estrogen receptor expressed in the liver, where it regulates glucose homeostasis as well as lipid metabolism (26). Among the remaining 2% of regulatory elements, we also found tissue-related transcription-binding sites (i.e., Hnf4A). The hepatocyte nuclear factor 4α is a transcription factor found upstream in the regulation pathways of several hepatic genes (27). Surprisingly, the non-CGI UMRs produced, on average, a threefold greater enrichment in regulatory elements than the CGI-like UMRs.
Fig. 6.
Genomic distribution of UMRs and enrichment of annotated features. (A) Distribution of CGI-like and non-CGI UMRs in genomic regions. UMRs at TSS regions (−3 kb to +2 kb of TSS), in gene bodies (non-TSS exons, introns, and +3 kb of 3′ not-transcribed regions), and in intergenic DNA do not add up to 100 because only those UMRs with >90% overlap were considered. (B) Enrichment of ORegAnno sites and MMC sequences in the UMRs located in the indicated genomic regions. Enrichments are compared with abundance in the undiluted genome (numerical values are provided in SI Appendix, Table S12). **P < 0.005; ***P < 0.0001.
Relying on the way the protein-binding sites were partitioned, we believe that most of the UMRs represent regions rich in regulatory elements, with many of them being liver-related and possibly liver-specific. We used the “David Bioinformatics Functional Annotation Tool” Web application to bin UMR-containing genes according to functional annotation and analyzed the enrichments in three selected categories: biological process, molecular function, and tissue specificity (28). We took the top 5% of our UMRs, ranked according to their avgMSCC score. In each functional category, we sorted the results according to significance (smaller P values on top). For “tissue specificity,” liver is the most significantly enriched tissue (P value of 2 × 10−12.). For “biological process,” genes related to system development are at the top (P value of 1 × 10−8). Finally, for “molecular function,” genes related to steroid hormone receptor activity are the most enriched class of function (P value of 1 × 10−5).
In conclusion, our screening for hypomethylated CpGs showed that CGIs are weak predictors of sensitive epigenetic loci and, in addition, revealed a large unexpected number of non-CGI UMRs with the highest enrichment in regulatory elements. The fact that 50% of the UMRs have one-half of the average CpG content of traditional CGIs prompts a rethinking of the relationship between CpG density, unmethylation, and functionality of the genomic subsequences. A mechanistic link between these variables is beginning to emerge. A recent report shows that the genomic insertion of a promoterless CpG cluster is able to recruit the Cfp1 protein, which can bind to unmethylated CpGs and attract the Setd1 histone H3K4 methyltransferase complex, which, in turn, creates a new focus of H3K4me3 modification (29). There is evidence that this modification repels the methyltransferase involved in de novo methylation. However, only one-half of the cells carry the insertion-acquired methylation, indicating that CpG density, per se, is insufficient to maintain the unmethylated state. Our hypomethylation map showing thousands of UMRs not qualifying as CGIs but with the highest enrichment in DNA regulatory motifs supports the idea that methylation is the default state of CpGs, except for those than are protected from de novo methylation, which may be mediated by the action of DNA-binding proteins.
The idea that DNA-containing regulatory elements are furnished with a critical CpG density working as a local signal to recruit proteins able to create epigenetic marks that highlight functional sequences is simple and attractive. In this scenario, the unmethylated genomic regions (UMRs) could reflect the footprint of regulatory DNA-binding proteins that protected local sequences from the activity of de novo methylases.
4Enz MSCC vs. HpaII MSCC.
The MSCC approach was first published in 2009 by Ball et al. (15) in a proof-of-principle report. The average reads-per-hit values (MSCC score) reported for unmethylated CCGGs was 5.0 ± 15.4, which identified 69% of the addressable sites with at least one read. In discussing the usefulness of the approach, the authors used statistical criteria to conclude that by increasing the sequencing depth, the method would allow for the identification of the UMRs of a genome. Having sequenced to a greater depth, which reached average MSCC scores for HpaII of 10.4 ± 10.0 for all CCGGs identified at least once, we sought to determine to what extent the HpaII hits are able to report on the hypomethylation measured using 4Enz five-channel hits and a window of 300 bp. Our all-channel HpaII hits allow for formation of 109,877 of 300-bp–based clusters, which overlapped with only 25,373 (54.2%) of 46,804 of our UMRs and with 7,207 (18%) of 24,399 of our non-CGI UMRs. We thus conclude that only the expanded 4Enz MSCC reports reliably on the genome's hypomethylation.
Materials and Methods
CpG-tag libraries were prepared according to strategies similar to those outlined in ref. 15 except that four instead of one methylation-sensitive restriction enzyme was used, and the tags were retrieved with EcoP15I and not MmeI. CpG tag libraries were sequenced by Illumina Inc. using Genome Analyzer's Solexa technology. Illumina Inc returned approximately 1 Gigabase of sequence partitioned into 36-nt long reads per library. After mapping to the mm9 Mus musculus reference genome, the data were analyzed by creating frequency histograms showing the number of times reads were found that identified any given CpG. For further details on methods and data analysis, including materials, acknowledgements, 12 tables, and 8 figures, see S1 Appendix, SI Materials and Methods.
Supplementary Material
Acknowledgments
This study was supported by the Intramural Research Program of the National Institutes of Health (2009 National Institutes of Health Director's Challenge Award and Project Z01 ES101643 to L.B.).
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1105713108/-/DCSupplemental. Tab-delimited file for Table S6 available upon request.
References
- 1.Suzuki MM, Bird A. DNA methylation landscapes: Provocative insights from epigenomics. Nat Rev Genet. 2008;9:465–476. doi: 10.1038/nrg2341. [DOI] [PubMed] [Google Scholar]
- 2.Leonhardt H, Bestor TH. Structure, function and regulation of mammalian DNA methyltransferase. EXS. 1993;64:109–119. doi: 10.1007/978-3-0348-9118-9_5. [DOI] [PubMed] [Google Scholar]
- 3.Antequera F, Bird A. Number of CpG islands and genes in human and mouse. Proc Natl Acad Sci USA. 1993;90:11995–11999. doi: 10.1073/pnas.90.24.11995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bird AP. CpG-rich islands and the function of DNA methylation. Nature. 1986;321:209–213. doi: 10.1038/321209a0. [DOI] [PubMed] [Google Scholar]
- 5.Irizarry RA, Wu H, Feinberg AP. A species-generalized probabilistic model-based definition of CpG islands. Mamm Genome. 2009;20:674–680. doi: 10.1007/s00335-009-9222-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bock C, Walter J, Paulsen M, Lengauer T. CpG island mapping by epigenome prediction. PLOS Comput Biol. 2007;3:e110. doi: 10.1371/journal.pcbi.0030110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gardiner-Garden M, Frommer M. CpG islands in vertebrate genomes. J Mol Biol. 1987;196:261–282. doi: 10.1016/0022-2836(87)90689-9. [DOI] [PubMed] [Google Scholar]
- 8.Takai D, Jones PA. Comprehensive analysis of CpG islands in human chromosomes 21 and 22. Proc Natl Acad Sci USA. 2002;99:3740–3745. doi: 10.1073/pnas.052410099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hackenberg M, et al. CpGcluster: A distance-based algorithm for CpG-island detection. BMC Bioinformatics. 2006 doi: 10.1186/1471-2105-7-446. 10.1186/1471-2105-7-446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shimizu TS, Takahashi K, Tomita M. CpG distribution patterns in methylated and non-methylated species. Gene. 1997;205:103–107. doi: 10.1016/s0378-1119(97)00542-8. [DOI] [PubMed] [Google Scholar]
- 11.Simmen MW. Genome-scale relationships between cytosine methylation and dinucleotide abundances in animals. Genomics. 2008;92:33–40. doi: 10.1016/j.ygeno.2008.03.009. [DOI] [PubMed] [Google Scholar]
- 12.Hackenberg M, et al. Prediction of CpG-island function: CpG clustering vs. sliding-window methods. BMC Genomics. 2010;11:327. doi: 10.1186/1471-2164-11-327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Macleod D, Charlton J, Mullins J, Bird AP. Sp1 sites in the mouse aprt gene promoter are required to prevent methylation of the CpG island. Genes Dev. 1994;8:2282–2292. doi: 10.1101/gad.8.19.2282. [DOI] [PubMed] [Google Scholar]
- 14.Weber M, et al. Distribution, silencing potential and evolutionary impact of promoter DNA methylation in the human genome. Nat Genet. 2007;39:457–466. doi: 10.1038/ng1990. [DOI] [PubMed] [Google Scholar]
- 15.Ball MP, et al. Targeted and genome-scale strategies reveal gene-body methylation signatures in human cells. Nat Biotechnol. 2009;27:361–368. doi: 10.1038/nbt.1533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Suzuki M, et al. Optimized design and data analysis of tag-based cytosine methylation assays. Genome Biol. 2010;11:R36. doi: 10.1186/gb-2010-11-4-r36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liu J, Yu S, Litman D, Chen W, Weinstein LS. Identification of a methylation imprint mark within the mouse Gnas locus. Mol Cell Biol. 2000;20:5808–5817. doi: 10.1128/mcb.20.16.5808-5817.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Down TA, et al. A Bayesian deconvolution strategy for immunoprecipitation-based DNA methylome analysis. Nat Biotechnol. 2008;26:779–785. doi: 10.1038/nbt1414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Edwards JR, et al. Chromatin and sequence features that define the fine and gross structure of genomic methylation patterns. Genome Res. 2010;20:972–980. doi: 10.1101/gr.101535.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Doi A, et al. Differential methylation of tissue- and cancer-specific CpG island shores distinguishes human induced pluripotent stem cells, embryonic stem cells and fibroblasts. Nat Genet. 2009;41:1350–1353. doi: 10.1038/ng.471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Straussman R, et al. Developmental programming of CpG island methylation profiles in the human genome. Nat Struct Mol Biol. 2009;16:564–571. doi: 10.1038/nsmb.1594. [DOI] [PubMed] [Google Scholar]
- 22.Retelska D, Beaudoing E, Notredame C, Jongeneel CV, Bucher P. Vertebrate conserved non coding DNA regions have a high persistence length and a short persistence time. BMC Genomics. 2007;8:398. doi: 10.1186/1471-2164-8-398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fujita PA, et al. The UCSC Genome Browser database: Update 2011. Nucleic Acids Res. 2011;39(Database issue):D876–D882. doi: 10.1093/nar/gkq963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Griffith OL, et al. Open Regulatory Annotation Consortium ORegAnno: An open-access community-driven resource for regulatory annotation. Nucleic Acids Res. 2008;36(Database issue):D107–D113. doi: 10.1093/nar/gkm967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wederell ED, et al. Global analysis of in vivo Foxa2-binding sites in mouse adult liver using massively parallel sequencing. Nucleic Acids Res. 2008;36:4549–4564. doi: 10.1093/nar/gkn382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gao H, Fält S, Sandelin A, Gustafsson JA, Dahlman-Wright K. Genome-wide identification of estrogen receptor alpha-binding sites in mouse liver. Mol Endocrinol. 2008;22:10–22. doi: 10.1210/me.2007-0121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hoffman BG, et al. Locus co-occupancy, nucleosome positioning, and H3K4me1 regulate the functionality of FOXA2-, HNF4A-, and PDX1-bound loci in islets and liver. Genome Res. 2010;20:1037–1051. doi: 10.1101/gr.104356.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Huang W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 29.Thomson JP, et al. CpG islands influence chromatin structure via the CpG-binding protein Cfp1. Nature. 2010;464:1082–1086. doi: 10.1038/nature08924. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.