Abstract
Convergent gene pairs with head-to-head configurations are widespread in both eukaryotic and prokaryotic genomes and are speculated to be involved in gene regulation. Here we present a unique mechanism of gene regulation due to convergent transcription from the antagonistic prgX/prgQ operon in Enterococcus faecalis controlling conjugative transfer of the antibiotic resistance plasmid pCF10 from donor cells to recipient cells. Using mathematical modeling and experimentation, we demonstrate that convergent transcription in the prgX/prgQ operon endows the system with the properties of a robust genetic switch through premature termination of elongating transcripts due to collisions between RNA polymerases (RNAPs) transcribing from opposite directions and antisense regulation between complementary counter-transcripts. Evidence is provided for the presence of truncated RNAs resulting from convergent transcription from both the promoters that are capable of sense–antisense interactions. A mathematical model predicts that both RNAP collision and antisense regulation are essential for a robust bistable switch behavior in the control of conjugation initiation by prgX/prgQ operons. Moreover, given that convergent transcription is conserved across species, the mechanism of coupling RNAP collision and antisense interaction is likely to have a significant regulatory role in gene expression.
Keywords: inverse expression, overlapping DNA, gene-regulatory network
Convergent transcription from two opposing promoters of partially overlapping genes on opposite strands of DNA gives rise to transcripts with potential sense–antisense interactions in the overlapping region. Such convergent transcription is widespread in eukaryotes including the mouse (1), human (2), Drosophila melanogaster (3), and Arabidopsis thaliana (4) genomes and a number of prokaryotes such as Listeria monocytogenes (5), Mycoplasma pneumoniae (6), and Escherichia coli (7). We postulate that convergent, overlapping gene organization provides two-layered regulation by a combination of transcriptional interference and antisense regulation through RNA: RNA interactions between complementary transcripts (8).
Transcription from promoters of convergent overlapping genes gives rise to a finite probability that opposing elongating RNA polymerases (RNAPs) collide head-on, thus exerting a suppressive effect on transcription. Such suppressive influence of one transcriptional activity on a second transcriptional activity occurring in cis is referred to as transcriptional interference (9), as has been reported in prokaryotic (10, 11) and eukaryotic systems (12, 13). Furthermore, the two transcripts, having complementary sequence in the overlapping region, may exert antisense regulation through RNA: RNA interactions (8, 14). Given the frequent occurrence of sense–antisense transcripts in both prokaryotic and eukaryotic genomes, convergent transcription may play a significant role in gene regulation by both mechanisms. However, there have been few attempts to document the relative effect of each mechanism in control of transcription of overlapping, convergent genes.
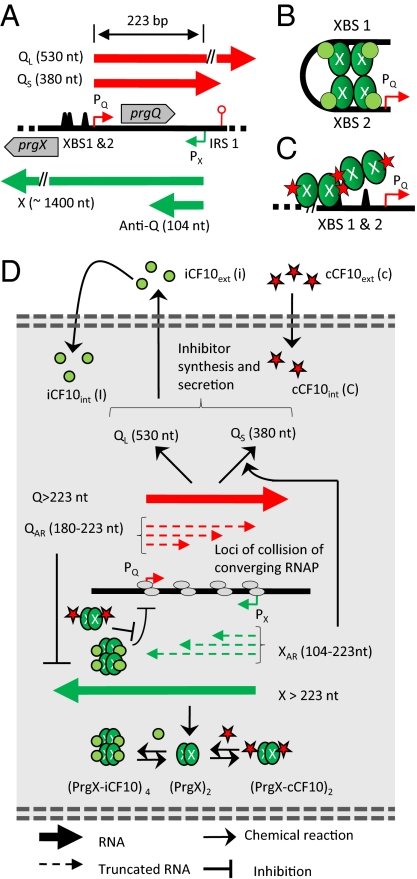
The present study investigates the role of transcriptional interference arising due to RNAP collision and antisense regulation due to expression of complementary transcripts from operons prgQ and prgX, in plasmid pCF10 that regulates the conjugative transfer of antibiotic resistance between Enterococcus faecalis donor and recipient cells (15). Promoter PQ drives expression of the prgQ operon, which encodes pheromone inhibitor peptide, iCF10 (AITLIFI) as well as the pCF10 conjugation machinery (16). Approximately 223 bp downstream of the prgQ start site, the convergent promoter PX drives expression of the prgX operon which encodes repressor PrgX regulating PQ transcription by binding to the operators XBS 1 and 2 of promoter PQ (Fig. 1 A–C and Fig. S1 A and B). In addition to the full-length prgX transcript (denoted X), transcription from PX also produces a 104-nt noncoding RNA, Anti-Q, by processing or termination within the 5′ end of the prgX transcript (17). The interaction of Anti-Q with nascent prgQ transcripts affects folding of prgQ RNA and results in a short 380-nt terminated QS transcript (at IRS1, Fig. 1A) that encodes iCF10, but is incapable of inducing conjugation (18).
Fig. 1.
The prgQ–prgX genetic locus controls conjugation of pCF10 plasmid in Enterococcus faecalis. (A) Convergent promoters PQ (red) and PX (green) drive expression of the antagonistic prgQ and prgX operons, respectively, with a 223-bp overlap. Transcripts (indicated by thick arrows) from the inducible PQ promoter include 380 nt QS RNA terminating at an inverted repeat sequence (IRS1) and 530 nt QL RNA that results in expression of downstream conjugation-related genes. Transcripts from constitutive antisense PX promoter include noncoding Anti-Q and full-length X RNA coding for PrgX protein. (B and C) Promoter PQ in the repressed state (B) and the derepressed state (C). (D) Model for the prgQ–prgX genetic switch. Successful transcription gives rise to full-length Q and X RNA (>223 nt). Failed transcription, upon collision of converging RNAP, gives rise to various sizes of truncated RNA (<223 nt) indicated by dashed arrows. Some truncated RNAs (QAR and XAR) have the potential to exert antisense interaction with full-length counter-transcripts (not to scale).
PQ is normally in a repressed state whereas PX is constitutive (Fig. 1B). The switch of PQ between the repressed state (conjugation off, Fig. 1B) and the derepressed state (conjugation on, Fig. 1C) is controlled by competing interactions of PrgX with inhibitor iCF10 and an inducer, cCF10, a chromosomally coded heptapeptide (LVTLVFV) that is produced predominantly by recipient cells (16) because in donor cells, the pCF10-encoded PrgY protein sequesters endogenous cCF10 (19, 20). In the absence of recipient cells and thus low ratios of cCF10 to iCF10, iCF10-bound PrgX tetramers cause a DNA loop between XBS 1 and 2 sites, stabilizing a repressing complex at PQ (Fig. 1B) (21, 22). In the derepressed state, the cCF10 imported from the environment via membrane protein PrgZ (23) displaces iCF10, resulting in the disruption of PrgX-iCF10 tetramers and the DNA loop (Fig. 1C). This disruption leads to increased levels of prgQ transcription, which serves to titrate the Anti-Q RNA, and results in expression of a longer 530-nt transcript called QL (Fig. 1A), capable of inducing conjugation by a post-transcriptional mechanism (24).
In this report we used a mathematical model to demonstrate that convergent transcription of the prgX/prgQ system plays a regulatory role in endowing the system with a bistable switch-like behavior, a characteristic of many robust biological switches, including regulation of competence in Bacillus subtilis (25), lysogeny and lytic state in bacteriophage λ (26), and during cell fate determination in Xenopus laevis oocytes (27). We provide experimental evidence that RNAP collision gives rise to shorter truncated transcripts that exert an antisense effect on transcripts of prgX and prgQ. Both transcriptional interference as a result of RNAP collision and antisense regulation are necessary for the prgX/prgQ genetic switch to function in a robust manner.
Results
Modeling the prgQ–prgX Genetic Network: Coupled Effect of RNAP Collision and Antisense Regulation.
A model for conjugation control by the prgX/prgQ operons is shown in Fig. 1D. The proposed model is based in part on previously described mechanisms (15, 18). We included additional regulatory effects contributed by transcriptional interference caused by RNAP collision and antisense effects caused by truncated RNA produced as a result of transcription interference. The underlying hypothesis is that RNAP collision causes failure in transcription, thus suppressing the transcription rate from both promoters. A second element is the generation of truncated transcripts that are released upon RNAP collision. We consider the hybridization of these short RNAs to their complementary counter-transcripts.
A two-part mathematical model was formulated to describe the mechanism and to quantitatively evaluate the behavior of transcription from PQ and PX in response to inducer cCF10 (9). The first part is a discrete model that enumerates the frequency of RNAP collision and the generation of truncated RNA (Figs. S2 and S3). The second part is an ordinary differential equation (ODE)-based mathematical model on the mass balance of the components of the pCF10 system (Fig. 1D, Fig. S4, and Tables S1–S6).
The discrete model simulated RNAP moving along the overlapping DNA between PQ and PX (SI Text and Fig. S2). Upon RNAP binding to the promoter, a delay of 10 s is allowed for formation of the elongation complex (28) before RNAP moves at a velocity of elongation at 50 bp/s, which has been reported as the average velocity of RNAP in E. coli in the presence of pausing (29). The maximal RNAP binding rate at a derepressed PQ was set at 0.1/s on the basis of RNAP initiation rates estimated for the Pbla system in E. coli (30) and the PR-PRE system in bacteriophage λ (10). Using experimentally determined relative strengths of promoters PQ and PX under repressed and derepressed states in Fig. 2, the RNAP binding rates at PQ and PX under these two states were calculated. RNAP collision is considered to occur when converging RNAPs from PQ and PX are separated by 60 bp (RNAP footprint). This result takes into consideration occlusion of PQ due to passage of RNAP originating from PX and vice versa. Our experimental data suggested that RNAP collision did not result in complete abolition of transcription on the basis of a 10% level of X transcript observed upon derepression; we thus assume that 90% of RNAP collisions result in transcriptional termination (31), releasing short transcripts terminated at the point of collision (SI Text and Fig. S2). Movement of RNAP is not hindered by binding of PrgX to DNA (Fig. S5A). Successful passage of RNAP across the overlapping region (223 nt) without collision generates the full-length Q or X RNA. RNAP collision gives rise to a population of truncated RNA with varying length of <223 nt from PQ and PX (Fig. 1D). Previous work showed that stem loops at 46–51 nt and 80–83 nt of PX transcript respectively interact with stem loops at 173–178 nt and 156–161 nt of PQ transcript (Fig. S3) (32). On the basis of these data and in silico RNA structural analysis using Sfold (33), we postulate that truncated PX RNAs (including Anti-Q RNA) between 104 nt and 223 nt in length and truncated Q RNA between 180 nt and 223 nt are capable of RNA:RNA interaction with their counter-transcripts Q and X, respectively, due to the presence of stem loops (Fig. S3). These subpopulations of truncated RNAs capable of antisense effects are collectively denoted as QAR (180–223 nt) and XAR (104–223 nt) RNA from PQ and PX promoters, respectively (Fig. 1D).
Fig. 2.
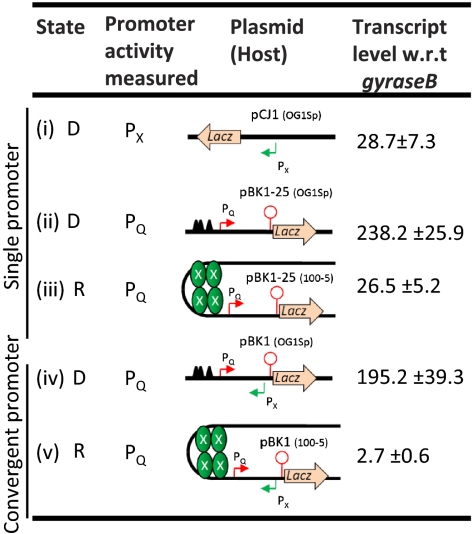
Interference exerted on PQ expression due to convergent transcription. (i) pCJ1 (OG1Sp), single PX promoter construct; (ii) pBK1-25 (OG1Sp), single PQ promoter construct in derepressed (D) state; (iii) pBK1-25 (100-5), single PQ promoter construct in repressed (R) state; (iv) pBK1 (OG1Sp), convergent promoter construct measuring expression from derepressed PQ in the presence of PX; and (v) pBK1 (100-5), convergent promoter construct measuring expression from repressed PQ in the presence of PX. The lacZ mRNA expression levels (normalized to gyraseB) shown are an average of three independent qPCR experiments.
From the discrete model simulation the rates of generation of Q, X, QAR, and XAR species were calculated (Fig. 3 A and B) for simulation using the ODE model (SI Text and Fig. S2). A set of eight ODEs describes the balance of RNA species QS, QL, X, XAR and QAR, intracellular iCF10 and cCF10, and extracellular iCF10 (Eqs. S25–S32), considering the rate of production, degradation, and dilution due to volume expansion caused by growth as well as interactions among these components (SI Text). For RNA species, interaction between sense transcripts (Q and QAR) and antisense transcripts (X and XAR) following second-order kinetics (32) was also considered (SI Text). Hybrid RNA duplex between X RNA and QAR is considered to be unavailable for translation (Fig. 1D) (8). The formation of RNA duplex between XAR and nascent Q RNA gives rise to formation of QS RNA (18, 32). Nascent Q transcripts that do not terminate proceed to elongate and form QL. Translation of QS and QL leads to production of the secreted inhibitor peptide iCF10 (15). Transport of iCF10 and cCF10 across the membrane protein PrgZ is considered to follow first-order kinetics (23). The model assumes that due to cCF10-sequestering activity of PrgY protein, the effect of endogenous cCF10 in donor cells is negligible (20).
Fig. 3.
Mathematical modeling predicts RNAP collision and antisense regulation are required for bistability in the prgQ–prgX switch. (A and B) Frequency of various sizes of truncated and full-length RNA as a result of RNAP collision in the repressed (RNAP firing rates fQ,R = 0.011 s−1 from PQ, and fX= 0.012 s−1 from PX) and derepressed (fQ,D = 0.1 s−1 and fX= 0.012 s−1) state from PQ (A) and PX (B). (C) Steady-state level of QL at different cCF10 concentrations shows characteristic bistable switch behavior. (D) Effect of decoupling RNAP collision (RC) and antisense regulation (AR) on the steady-state response of QL to cCF10.
The ODE model was solved to obtain a steady-state solution to a fixed concentration of extracellular cCF10 (Fig. 3 C and D). The parameter values were obtained from the literature or experimental data (Tables S1 and S2). To test the sensitivity of the steady-state behavior to the parameter values, the parameters in the model were lumped into six dimensionless parameters (Tables S3 and S4) to reduce the model to a set of six nondimensional ODEs (Eqs. S48–S53). The values of three of these dimensionless numbers were determined from experimental data (SI Text), whereas the remaining three were obtained from literature and subjected to sensitivity analysis (Table S6). In the dimensionless model, the term corresponding to RNA interaction between truncated RNAs XAR and QAR was eliminated as it did not affect the steady-state behavior of the system. Simultaneous parameter space search for the remaining three dimensionless parameters demonstrated that steady-state behavior shown in Fig. 3 C and D existed over at least two orders of magnitude (Fig. S4A and Table S6). The details of model derivation are described in SI Text.
PQ Activity in Presence and Absence of Convergent Transcription.
The strength of PQ in the presence (repressed state) and absence (derepressed state) of repressing PrgX complexes was quantified by measuring steady-state expression levels of the lacZ transcript from plasmids containing either a single PQ promoter or both convergent promoters with the lacZ reporter fused downstream of IRS1 (Fig. 2). Two kinds of host strains were used: (i) OG1Sp, a wild-type host strain; and (ii) 100-5, a derivative of OG1Sp that provides PrgX in trans from the chromosome at levels similar to wild-type pCF10 plasmid (Fig. S1B). By combining the plasmids and host strains a set of five conditions was created as shown in Fig. 2. The intrinsic promoter strength of PQ (derepressed state) in the absence of convergent transcription is 8.3-fold higher than that of the constitutive PX (Fig. 2, ii vs. i). The repression of PQ by supplying PrgX in trans caused a 9-fold reduction in expression in the absence of convergent transcription (Fig. 2, iii vs. ii). In the repressed state, PQ has almost identical strength to PX (Fig. 2, i vs. iii). In the derepressed state PQ becomes the stronger promoter (Fig. 2, i vs. ii). Whereas convergent transcription from PX caused only a modest (∼15%) reduction of transcripts from PQ in the derepressed state (stronger PQ than PX; Fig. 2, ii vs. iv), it reduced PQ transcript levels by ∼90-fold in the repressed state (similar strength of PQ and PX; Fig. 2, ii vs. v). These results show that convergent transcription greatly enhances the effect of repression on PQ.
Model Simulation and Bistable Behavior.
The discrete model was used to predict the relative abundance of transcripts under repressed and derepressed conditions as a result of RNAP collision (Fig. 3 A and B and Tables S1 and S2). Under the repressed condition, a higher number of transcription events are predicted to give rise to full-length X RNA, compared with the depressed condition, whereas the opposite is predicted for full-length Q RNA. On the other hand, a broader distribution of QAR and XAR RNA is predicted in the repressed state compared with the derepressed state. In the derepressed state, the 8.3-fold stronger PQ not only increases Q transcription but also knocks off most RNAP initiated from PX at loci proximal to the PX promoter. As a result, RNAP collision results in only a moderate decrease in Q transcription, but a rather large decrease in X transcription.
Using the ODE model we next examined the effect of RNAP collision and antisense regulation on the steady-state level of QL in response to the signaling molecule cCF10. The level of QL RNA is an indicator of the state of conjugation, because it encodes genes of conjugation machinery. Steady-state solution of the model shown in Fig. 3C demonstrates a characteristic bistable response of QL to cCF10 concentration. Multiple steady states reside in the S-shaped section of the curve. Two are stable steady states corresponding to on (upper) and off (lower) states, respectively. The unstable steady state in the middle is not observed experimentally. For the system at an off state, as cCF10 concentration increases it moves along the lower stable steady state (QL < 1.4 nM) until cCF10 reaches 3.7 ng/mL, and then the system undergoes a sharp transition to an on state (conjugationally competent) corresponding to a high level of QL (>3.8 nM). Conversely if the system is initially at an on state, it remains at an on state till cCF10 concentration decreases below 3.2 ng/mL where it rapidly changes to an off state. The system is thus marked by well-separated on (cCF10 > 3.7 ng/mL) and off states (cCF10 < 3.2 ng/mL).
We evaluated the relative contribution of RNAP collision and antisense regulation to bistable behavior by eliminating RNAP collision (RC) or antisense regulation (AR) or both from the model (Fig. 3D). AR was eliminated from the model by setting the rate constant for RNA interaction to zero, whereas RC was eliminated by setting the transcription rates of Q and X RNA to RNAP firing rates from PQ and PX, respectively, and XAR and QAR concentration to zero (Table S5). Bistability is observed only when both RC and AR are present (RC/AR). When AR is absent and RC is present (RC/AR−) or both RC and AR (RC−/AR−) are eliminated, no bistability is seen (Table S6). We also examined the case where RC is only partially removed by reducing the RNAP falling-off rate upon RNAP collision. As the falling-off rate decreases to 50%, the steady-state behavior transitions from a bistable to a ramp-like response (Fig. S4B). The results thus hinted that the 104-nt Anti-Q RNA, produced as a result of X RNA processing, alone is insufficient for the bistable behavior. This result is indeed the case for RC−/Anti-Q, where the bistable response to cCF10 is lost and a ramp-like transition is observed (Fig. 3D).
Inverse Expression and Response to Pheromone.
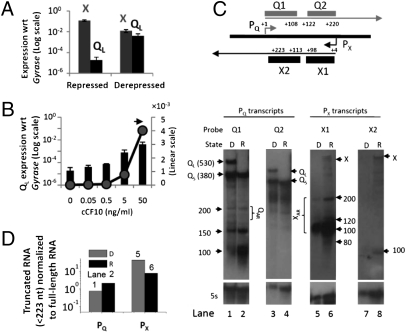
We next measured the levels of transcripts from PX and PQ promoters in strain OG1Sp carrying wild-type (WT) plasmid pCF10. Under repressed conditions the QL transcript level was much lower than the X transcript level even though the strengths of PQ and PX are similar. Upon induction with high levels of cCF10 (50 ng/mL), the QL transcript level increased >200-fold, whereas the X transcript decreased ∼9-fold (Fig. 4A). The opposite trend in their response to derepression is consistent with model predictions (Fig. 3C and Fig. S4C). The level of QL RNA observed was lower than that seen in the lacZ reporter system (Fig. 2). This result could be attributed to a high degradation rate for QL. This notion is consistent with model simulation, where a 10-fold decrease in QL degradation rate abolishes bistable behavior (Fig. S4D).
Fig. 4.
Inverse relationship of QL and X expression and presence of truncated RNAs in the prgQ–prgX locus. (A) QL and X RNA levels measured by qPCR using X-specific and QL-specific primers from OG1Sp (pCF10) under repressed and derepressed conditions. (B) Levels of QL RNA in response to different concentrations of cCF10. Data shown in A and B are averages of three independent experiments (error bars are SDs from mean values). (C) (Upper) Map for the single-stranded RNA probes Q1, Q2, X1 and X2 used for Northern blotting experiments. (Lower) Northern blots to detect transcripts from PQ and PX in cells carrying WT pCF10, in either the derepressed (D) or the repressed (R) state. Probes and lanes: Q1, lanes 1 and 2; Q2, lanes 3 and 4; X1, lanes 5 and 6; and X2, lanes 7 and 8. (D) Quantitative estimation of intensity of bands for truncated PQ and PX transcripts in lanes 1 and 2 and 5 and 6 normalized to intensity of QS + QL and X, respectively, using ImageJ software (National Institutes of Health).
A dose/response curve of QL transcript to cCF10 showed that QL expression remains low below 0.5 ng/mL of cCF10, followed by an 8.3-fold increase at 5 ng/mL of cCF10 (Fig. 4B and Fig. S4E), characteristic of a switch from the off to the on state as predicted by the model (Fig. 3C). Flow cytometric data using GFP protein expression as an indicator of on state confirm that the system transitions from an off to an on state between 0.5 and 5 ng/mL (Fig. S4F).
Truncated RNA from PQ and PX.
To verify the existence of the truncated transcripts (XAR and QAR) in pCF10 carrying donor (OG1Sp) cells, we used Northern blots with sets of probes complementary to the 5′ (Q1 and X1) and 3′ segments (Q2 and X2) within the overlapping region of the respective RNAs from each promoter (Fig. 4C, Upper). Shorter (truncated) PQ (Fig. 4C, lanes 1–4) and PX (Fig. 4C, lanes 5–8) transcripts within the overlapping region were indeed observed under both repressed and derepressed conditions. The Q1 probe detected truncated transcripts of ∼100-, 150-, and 200-nt sizes, with higher relative abundance in repressed cells (Fig. 4C, lane 2 vs. lane 1, and Fig. S5C). Similarly, the X1 probe detected truncated RNAs in the size range between 80 and 200 nt (Fig. 4C, lanes 5 and 6). The prominent band of ∼100 nt corresponds to the 104 nt Anti-Q RNA reported previously (17). The levels of truncated transcripts detected by probes Q1 and X1 (Fig. 4C, lanes 1 and 2 and 5 and 6) proximal to the 5′ end are higher than those detected by distal probes Q2 and X2 (Fig. 4C, lanes 3 and 4 and 7 and 8), consistent with the higher collision frequency near the 5′ end. The presence of a short 100-nt band detected by the X2 probe (Fig. 4C, lane 8) indicates an additional mechanism involved in processing of longer PX transcripts. However, this truncated transcript is unlikely to have a regulatory role in the switch behavior, due to the absence of stem loops I and II required for antisense interaction with PQ transcripts.
The high abundance of truncated RNA in the repressed state (Fig. 4C, lane 2 vs. 1 and lane 6 vs. 5, and Fig. S5C) is consistent with the prediction of the RNAP collision model (Fig. 3 A and B). The size of shorter RNAs observed falls within the range of QAR and XAR considered in the model. However, it is possible that these short RNAs arise from degradation of full-length transcripts. In this case, the ratio of truncated RNA to full-length RNA would be similar under derepressed and repressed conditions. Conversely if these RNAs are the products of RNAP collision, we expect this ratio to increase for PX transcripts in the derepressed state as X RNA decreases, whereas the ratio corresponding to PQ transcripts should decrease because of a dramatic increase in Q (sum of QS + QL) transcripts upon induction. Quantification of Northerns in Fig. 4C (lanes 1 and 2 and 5 and 6) indeed shows that the ratio of truncated to full-length RNA increases for PX transcripts and decreases for PQ transcripts in the derepressed state compared with the repressed state (Fig. 4D).
Decoupling RNAP Collision and Antisense Regulation.
The relative contribution of RNAP collision and antisense regulation to switch response was evaluated by decoupling these effects using single or convergent promoter constructs in host strains OG1Sp and 100-5 for derepressed and repressed conditions, respectively (Fig. 5A). Three cases were analyzed: convergent transcription (RC/AR), no RNAP collision in absence of antisense RNA (RC−/AR−), and presence of antisense RNA supplied in trans (RC−/AR) using plasmid constructs shown in Fig. 5A.
Fig. 5.
Decoupling the effects of RNAP collision (RC) and antisense regulation (AR) in the prgQ–prgX locus. (A) Plasmid constructs containing either PQ (red) or PX (green) or both promoters used to decouple RC and AR effects (lanes indicated). Derepressed vs. repressed states are compared using strains OG1Sp (derepressed) vs. 100-5 (repressed). Antisense interactions in the absence of RC were assessed by supplying complementary RNAs in trans using the plasmids diagrammed on the Right. (B) (Upper) Transcripts from PX probed with X1. (Lower) Transcripts from PQ probed with Q1. Lanes and corresponding constructs/host strains used are as indicated in A.
Transcription from PX in the presence of convergent transcription (RC/AR) showed abundant truncated RNA from a repressed PQ compared with a derepressed PQ (Fig. 5B, lane 2 vs. lane 1). The longer (>700 nt), likely X transcript, was more abundant in the repressed condition, consistent with our model prediction (Fig. 3B and Fig. S4C). The distribution of transcripts was somewhat different from that in Fig. 4C because of different combinations of host cells and plasmid constructs.
The absence of convergent transcription (RC−/AR−) resulted in increased expression of both Anti-Q RNA (∼100-nt band) and longer X transcripts (Fig. 5B, lane 3), compared with the case with convergent transcription (lane 1), consistent with the negative effects of RNAP collision and antisense regulation on PX expression. A 200-nt band observed in the RC−/AR− case was not observed in the RC/AR case. This result could be caused by the presence of intrinsic pause sites, which were identified during in vitro transcription assays using conditions of RC−/AR− (Fig. S5B). It is possible that in the RC/AR case, converging RNAP from PQ knocks off elongating RNAP from PX before reaching this locus.
The antisense effect (RC−/AR) was restored (Fig. 5B, lane 4) by providing excess amounts of shorter 253-nt RNA from PQ in trans from a high copy number plasmid to the construct used in lane 3. The 253-nt RNA species contained the sequence of the overlapping region and was used to represent QAR species in the mathematical model. Compared with the RC−/AR− case (Fig. 5B, lane 3), providing excess antisense QAR RNA reduced the expression of X and Anti-Q transcripts significantly. However, the extent of suppression of X (>700 nt) was lower than that observed in the RC/AR case, as the X transcript was still present even when large amounts of QAR RNA were supplied (Fig. S5D). Comparison of the intensity of X transcripts normalized to 5s RNA shown in lanes 3 (∼0.9) and 4 (∼0.3) (Fig. S5D) indicates that RNAP collision is responsible for two-thirds of the decrease of X transcription in the derepressed state, exerting a stronger effect on X transcription than antisense regulation. These results suggest that both RNAP collision and antisense regulation are required to achieve high degree of suppression of the X transcript upon derepression, although RNAP collision plays a more dominating role as predicted by the model (Fig. 3B). The presence of shorter RNA (100–223 nt) in lane 4 compared with lane 3 in Fig. 5B suggests that these are products of endonucleolytic cleavage within paired regions formed by the “kissing” interactions between the loop structures of QAR and PX transcripts (Fig. S3) (32).
Similarly, during convergent transcription QL was expressed at higher levels in the depressed state than in the repressed state (Fig. 5B, lane 5 vs. lane 6), whereas higher abundance of truncated RNA was seen under the repressed state (Fig. 5B, lane 6). In the absence of RNAP collision and antisense regulation, longer QL and QS RNAs were expressed from PQ, even under repressed state (Fig. 5B, lane 7, RC−/AR−). The lack of truncated RNA in this case in the absence of RNAP collision (Fig. 5B, lane 7) is consistent with our model that these RNAs are mainly a result of convergent transcription. When we provided excess amounts of antisense 104 nt Anti-Q RNA, representing XAR species in the model, in trans from a high copy number plasmid (Fig. 5B, lane 8), QL expression decreased (Fig. 5B, lane 7 vs. lane 8); however, it was visible at longer exposures. Importantly, QL RNA was expressed in RC−/AR− and RC−/AR cases even under repressed conditions. Intensity of bands corresponding to both X (Fig. 5B, lanes 1–4) and QL RNA in (Fig. 5B, lanes 5–8) normalized to 5s RNA shows that the high degree of suppression of X in the derepressed state (Fig. 5B, lane 1) and QL in the repressed state (Fig. 5B, lane 6) occurs only during convergent transcription (Fig. S5D). These results are consistent with model prediction.
Discussion
Our results demonstrate that a head-to-head convergent promoter system can accomplish two levels of regulation: (i) enhancing the degree of repression through RNAP collision and (ii) suppressing transcript levels through interactions with cis-encoded antisense RNA produced from the overlapping region. With RNAP collision alone, an 8.3-fold increase in PQ strength is predicted to give rise to a 12-fold increase in successful transcription from PQ (Fig. 3A) relative to the repressed state. Experimentally a much greater difference in QL transcript level was observed using both a reporter system (Fig. 2) and direct measurement of QL transcripts (Fig. 4A). This enhanced difference is accomplished by a greater degree of suppression of QL transcript level under the repressed conditions through the products of RNAP collision, namely truncated RNAs that have an antisense regulatory effect on transcripts from both PQ and PX. Using a combination of host strains and constructs to mimic conditions of RC and AR, we show that QL transcript level that was suppressed under repressed conditions with RC/AR became elevated when RC or both RC and AR were eliminated (Fig. 5B). Providing constant levels of Anti-Q RNA did not restore switch behavior, as demonstrated both experimentally (Fig. 5B, lane 8) and theoretically (Fig. 3D). The sharp difference in the level of Anti-Q/XAR and Q species between the repressed and the derepressed state due to RC and AR contributes to the switch behavior.
Both mathematical modeling (Fig. 3 A and B) and experimental observations (Fig. 4C) indicate the presence of certain prominent sizes of truncated RNA. However, the stochasticity of RNAP firing from promoters is expected to blur the discrete nature of truncated RNAs, pointing toward the presence of potential RNAP pause sites within overlapping DNA, thus increasing the probability of collision at those sites. Such an effect will further strengthen the bistability due to enhanced effects of RNAP collision and antisense regulation (10).
Many biological switches have been shown to exhibit bistability in steady-state analysis of their system models (34, 35). The pivotal role of RC and AR in affording a bistable switch behavior is predicted by our model (Fig. 3 C and D). Bistability allows for a binary behavior. Because conjugation is an energy-intensive process, the donor cells must turn their conjugation system on only in response to true signal. A switch type of regulatory mechanism allows for a tighter control of conjugation.
Transcription interference has been reported to exert a suppressive effect on transcription from the weaker promoter in pR-pL and PR-PRE promoter pairs of bacteriophages 186 and λ, respectively (10, 11). In these studies, RNAP collision was the major contributor to the observed interference whereas antisense effects were concluded to be insignificant. Convergent transcription in the IME4 locus of Saccharomyces cerevisiae was shown to result in antisense and sense transcripts that were inversely related between diploid and haploid cell types (36). Again, in this case, the inverse relationship was ascribed to transcriptional interference but not to antisense regulation. Other convergent transcription systems have also been reported to have inverse transcript levels under different conditions, including the ecsit gene involved in mouse stem cell development (37) and the mogR locus of L. monocytogenes controlling cell motility (6). In a study of transcription of the Pot2 locus in rice blast fungus, truncated transcripts were observed that led to the identification of an antisense promoter (38). In silico analysis on the mouse genome and a follow-up study using an oligo-microarray revealed nearly 2,000 sense–antisense transcripts (39). The presence of multiple-sized transcripts without poly(A) appeared to hint at the presence of truncated transcripts.
The fact that convergent transcription is ubiquitous and has persisted in evolution (40) is perhaps an indication that such gene organizations confer fundamental mechanisms of gene regulation. We show that using a single repressor, convergent transcription allows for simultaneous regulation of two genes, giving rise to a complex, tightly regulated switch behavior. Critical to conferring the sophisticated control is the combined use of RNAP collision and antisense regulation. The collision frequency and the level of transcription suppression are highly dependent on many factors, including relative promoter strength, the length of overlap, and the exact sequence that causes RNAP pausing and interaction. With such a wide range of possible outcomes, using subtle structural tuning, convergent transcription may be highly adaptable to become a robust controller for many complex cellular events.
Materials and Methods
Details for strains, reporter constructs, medium, and growth conditions are provided in SI Text. Details for Northern blotting and qPCR are provided in SI Text. The numerical solution to stiff differential equations for steady-state analysis was obtained using the MATLAB function solve. For scanning parameter space, the MATLAB function polyxpoly was used for checking multiple steady states.
Supplementary Material
Acknowledgments
We thank Dawn Manias for conducting the Western blot for PrgX, Laura C. C. Cook for flow cytometric data, and Sarika Mehra and Salim Charaniya for useful discussions. This work was supported by National Institutes of Health Grants GM081888 (to W.-S.H.) and GM049530 (to G.M.D.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1101569108/-/DCSupplemental.
References
- 1.Katayama S, et al. RIKEN Genome Exploration Research Group; Genome Science Group (Genome Network Project Core Group); FANTOM Consortium. Antisense transcription in the mammalian transcriptome. Science. 2005;309:1564–1566. doi: 10.1126/science.1112009. [DOI] [PubMed] [Google Scholar]
- 2.Yelin R, et al. Widespread occurrence of antisense transcription in the human genome. Nat Biotechnol. 2003;21:379–386. doi: 10.1038/nbt808. [DOI] [PubMed] [Google Scholar]
- 3.Misra S, et al. Annotation of the Drosophila melanogaster euchromatic genome: A systematic review. Genome Biol. 2002;3(12):research0083.1–0083.22. doi: 10.1186/gb-2002-3-12-research0083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yamada K, et al. Empirical analysis of transcriptional activity in the Arabidopsis genome. Science. 2003;302:842–846. doi: 10.1126/science.1088305. [DOI] [PubMed] [Google Scholar]
- 5.Toledo-Arana A, et al. The Listeria transcriptional landscape from saprophytism to virulence. Nature. 2009;459:950–956. doi: 10.1038/nature08080. [DOI] [PubMed] [Google Scholar]
- 6.Güell M, et al. Transcriptome complexity in a genome-reduced bacterium. Science. 2009;326:1268–1271. doi: 10.1126/science.1176951. [DOI] [PubMed] [Google Scholar]
- 7.Dornenburg JE. Widespread antisense transcription in. Escherichia coli. MBio. 2010;1(1):e00024–e000210. doi: 10.1128/mBio.00024-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brantl S. Regulatory mechanisms employed by cis-encoded antisense RNAs. Curr Opin Microbiol. 2007;10:102–109. doi: 10.1016/j.mib.2007.03.012. [DOI] [PubMed] [Google Scholar]
- 9.Shearwin KE, Callen BP, Egan JB. Transcriptional interference—a crash course. Trends Genet. 2005;21:339–345. doi: 10.1016/j.tig.2005.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Palmer AC, Ahlgren-Berg A, Egan JB, Dodd IB, Shearwin KE. Potent transcriptional interference by pausing of RNA polymerases over a downstream promoter. Mol Cell. 2009;34:545–555. doi: 10.1016/j.molcel.2009.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Callen BP, Shearwin KE, Egan JB. Transcriptional interference between convergent promoters caused by elongation over the promoter. Mol Cell. 2004;14:647–656. doi: 10.1016/j.molcel.2004.05.010. [DOI] [PubMed] [Google Scholar]
- 12.Napoli S, Pastori C, Magistri M, Carbone GM, Catapano CV. Promoter-specific transcriptional interference and c-myc gene silencing by siRNAs in human cells. EMBO J. 2009;28:1708–1719. doi: 10.1038/emboj.2009.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Greger IH, Aranda A, Proudfoot N. Balancing transcriptional interference and initiation on the GAL7 promoter of Saccharomyces cerevisiae. Proc Natl Acad Sci USA. 2000;97:8415–8420. doi: 10.1073/pnas.140217697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Waters LS, Storz G. Regulatory RNAs in bacteria. Cell. 2009;136:615–628. doi: 10.1016/j.cell.2009.01.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dunny GM. The peptide pheromone-inducible conjugation system of Enterococcus faecalis plasmid pCF10: Cell–cell signalling, gene transfer, complexity and evolution. Philos Trans R Soc B. 2007;362:11185–11193. doi: 10.1098/rstb.2007.2043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nakayama J, Ruhfel RE, Dunny GM, Isogai A, Suzuki A. The prgQ gene of the Enterococcus faecalis tetracycline resistance plasmid pCF10 encodes a peptide inhibitor, iCF10. J Bacteriol. 1994;176:7405–7408. doi: 10.1128/jb.176.23.7405-7408.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bae T, Kozlowicz B, Dunny GM. Two targets in pCF10 DNA for PrgX binding: Their role in production of Qa and prgX mRNA and in regulation of pheromone-inducible conjugation. J Mol Biol. 2002;315:995–1007. doi: 10.1006/jmbi.2001.5294. [DOI] [PubMed] [Google Scholar]
- 18.Johnson CM, et al. Direct evidence for control of the pheromone-inducible prgQ operon of Enterococcus faecalis plasmid pCF10 by a countertranscript-driven attenuation mechanism. J Bacteriol. 2010;192:1634–1642. doi: 10.1128/JB.01525-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Buttaro BA, Antiporta MH, Dunny GM. Cell-associated pheromone peptide (cCF10) production and pheromone inhibition in Enterococcus faecalis. J Bacteriol. 2000;182:4926–4933. doi: 10.1128/jb.182.17.4926-4933.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chandler JR, Flynn AR, Bryan EM, Dunny GM. Specific control of endogenous cCF10 pheromone by a conserved domain of the pCF10-encoded regulatory protein PrgY in Enterococcus faecalis. J Bacteriol. 2005;187:4830–4843. doi: 10.1128/JB.187.14.4830-4843.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shi K, et al. Structure of peptide sex pheromone receptor PrgX and PrgX/pheromone complexes and regulation of conjugation in Enterococcus faecalis. Proc Natl Acad Sci USA. 2005;102:18596–18601. doi: 10.1073/pnas.0506163102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dunny GM, Brown BL, Clewell DB. Induced cell aggregation and mating in Streptococcus faecalis: Evidence for a bacterial sex pheromone. Proc Natl Acad Sci USA. 1978;75:3479–3483. doi: 10.1073/pnas.75.7.3479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Leonard BAB, Podbielski A, Hedberg PJ, Dunny GM. Enterococcus faecalis pheromone binding protein, PrgZ, recruits a chromosomal oligopeptide permease system to import sex pheromone cCF10 for induction of conjugation. Proc Natl Acad Sci USA. 1996;93:260–264. doi: 10.1073/pnas.93.1.260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bensing BA, Manias DA, Dunny GM. Pheromone cCF10 and plasmid pCF10-encoded regulatory molecules act post-transcriptionally to activate expression of downstream conjugation functions. Mol Microbiol. 1997;24:285–294. doi: 10.1046/j.1365-2958.1997.3301710.x. [DOI] [PubMed] [Google Scholar]
- 25.Maamar H, Dubnau D. Bistability in the Bacillus subtilis K-state (competence) system requires a positive feedback loop. Mol Microbiol. 2005;56:615–624. doi: 10.1111/j.1365-2958.2005.04592.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tian T, Burrage K. Bistability and switching in the lysis/lysogeny genetic regulatory network of bacteriophage lambda. J Theor Biol. 2004;227:229–237. doi: 10.1016/j.jtbi.2003.11.003. [DOI] [PubMed] [Google Scholar]
- 27.Pomerening JR, Sontag ED, Ferrell JE., Jr Building a cell cycle oscillator: Hysteresis and bistability in the activation of Cdc2. Nat Cell Biol. 2003;5:346–351. doi: 10.1038/ncb954. [DOI] [PubMed] [Google Scholar]
- 28.Tang GQ, Roy R, Bandwar RP, Ha T, Patel SS. Real-time observation of the transition from transcription initiation to elongation of the RNA polymerase. Proc Natl Acad Sci USA. 2009;106:22175–22180. doi: 10.1073/pnas.0906979106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Neuman KC, Abbondanzieri EA, Landick R, Gelles J, Block SM. Ubiquitous transcriptional pausing is independent of RNA polymerase backtracking. Cell. 2003;115:437–447. doi: 10.1016/s0092-8674(03)00845-6. [DOI] [PubMed] [Google Scholar]
- 30.Liang ST, et al. Activities of constitutive promoters in Escherichia coli. J Mol Biol. 1999;292:19–37. doi: 10.1006/jmbi.1999.3056. [DOI] [PubMed] [Google Scholar]
- 31.Crampton N, Bonass WA, Kirkham J, Rivetti C, Thomson NH. Collision events between RNA polymerases in convergent transcription studied by atomic force microscopy. Nucleic Acids Res. 2006;34:5416–5425. doi: 10.1093/nar/gkl668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shokeen S, et al. Structural analysis of the Anti-Q-Qs interaction: RNA-mediated regulation of E. faecalis plasmid pCF10 conjugation. Plasmid. 2010;64:26–35. doi: 10.1016/j.plasmid.2010.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ding Y, Lawrence CE. A statistical sampling algorithm for RNA secondary structure prediction. Nucleic Acids Res. 2003;31:7280–7301. doi: 10.1093/nar/gkg938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chatterjee A, Kaznessis YN, Hu W-S. Tweaking biological switches through a better understanding of bistability behavior. Curr Opin Biotechnol. 2008;19:475–481. doi: 10.1016/j.copbio.2008.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kobayashi H, et al. Programmable cells: Interfacing natural and engineered gene networks. Proc Natl Acad Sci USA. 2004;101:8414–8419. doi: 10.1073/pnas.0402940101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hongay CF, Grisafi PL, Galitski T, Fink GR. Antisense transcription controls cell fate in Saccharomyces cerevisiae. Cell. 2006;127:735–745. doi: 10.1016/j.cell.2006.09.038. [DOI] [PubMed] [Google Scholar]
- 37.Dinger ME, et al. Long noncoding RNAs in mouse embryonic stem cell pluripotency and differentiation. Genome Res. 2008;18:1433–1445. doi: 10.1101/gr.078378.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kimura M, Yamaguchi I. Convergent transcription units and their promoters at both ends of pot2, an inverted repeat transposon from the rice blast fungus. J Biochem. 1998;124:268–273. doi: 10.1093/oxfordjournals.jbchem.a022106. [DOI] [PubMed] [Google Scholar]
- 39.Kiyosawa H, Mise N, Iwase S, Hayashizaki Y, Abe K. Disclosing hidden transcripts: Mouse natural sense-antisense transcripts tend to be poly(A) negative and nuclear localized. Genome Res. 2005;15:463–474. doi: 10.1101/gr.3155905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chen JJ, Sun M, Hurst LD, Carmichael GG, Rowley JD. Genome-wide analysis of coordinate expression and evolution of human cis-encoded sense-antisense transcripts. Trends Genet. 2005;21:326–329. doi: 10.1016/j.tig.2005.04.006. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.