Abstract
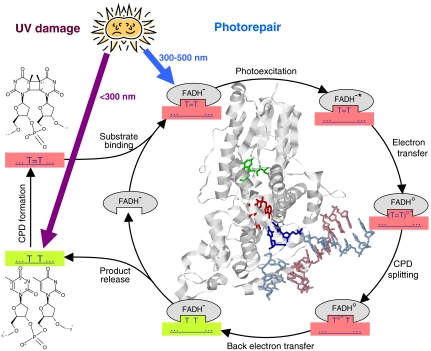
CPD photolyase uses light to repair cyclobutane pyrimidine dimers (CPDs) formed between adjacent pyrimidines in UV-irradiated DNA. The enzyme harbors an FAD cofactor in fully reduced state (FADH-). The CPD repair mechanism involves electron transfer from photoexcited FADH- to the CPD, splitting of its intradimer bonds, and electron return to restore catalytically active FADH-. The two electron transfer processes occur on time scales of 10-10 and 10-9 s, respectively. Until now, CPD splitting itself has only been poorly characterized by experiments. Using a previously unreported transient absorption setup, we succeeded in monitoring cyclobutane thymine dimer repair in the main UV absorption band of intact thymine at 266 nm. Flavin transitions that overlay DNA-based absorption changes at 266 nm were monitored independently in the visible and subtracted to obtain the true repair kinetics. Restoration of intact thymine showed a short lag and a biexponential rise with time constants of 0.2 and 1.5 ns. We assign these two time constants to splitting of the intradimer bonds (creating one intact thymine and one thymine anion radical T∘-) and electron return from T∘- to the FAD cofactor with recovery of the second thymine, respectively. Previous model studies and computer simulations yielded various CPD splitting times between < 1 ps and < 100 ns. Our experimental results should serve as a benchmark for future efforts to model enzymatic photorepair. The technique and methods developed here may be applied to monitor other photoreactions involving DNA.
Keywords: DNA repair, flavin adenine dinucleotide, transient absorption spectroscopy, UV damage
Exposure of living organisms to UV light from the sun induces harmful lesions in DNA. To overcome this threat, specific repair enzymes have evolved, probably the most ancient and widespread one being CPD photolyase (1, 2). It uses light to repair cis-syn cyclobutane pyrimidine dimer (CPD) lesions, formed by a UV-induced [2 + 2] cycloaddition of two adjacent pyrimidines (mostly thymines) in the same strand (Fig. 1). CPD photolyase has been found in organisms from all kingdoms of life, except placental mammals (including humans) that rely on nucleotide excision repair for CPD lesions. Among the various DNA repair mechanisms known, that of CPD photolyase is considered the most “cost-efficient” and least error-prone (3, 4). Note, however, that for CPDs containing cytosine, deamination of the cytosine may occur prior to repair. Uracil containing CPD cleavage by photolyase would then result in harmful cytosine-to-uracil mutations (5).
Fig. 1.
Reaction cycle of CPD repair by photolyase. Light green and light red boxes represent intact and damaged DNA, respectively. T = T denotes a CPD formed from two thymines. (T = T)∘- and T∘- T are intermediates of T = T repair. Gray ellipses represent photolyase with its FAD cofactor in different states as indicated. Inside the repair cycle, a structural overview is shown of photolyase from A. nidulans with a bound double-stranded DNA oligomer that contained a CPD lesion flipped out of the double helix into the enzyme’s binding pocket (1tez.pdb) (10). Red: FAD cofactor (partially hidden). Green: antenna pigment. Blue: DNA strand containing the CPD lesion. Purple: complementary DNA strand. Note that the two intradimer bonds are already split in this structure.
Photolyase is a globular single chain protein of approximately 60 kDa that harbors two buried cofactors: flavin adenine dinucleotide (FAD) in its fully reduced state (FADH-), which is the essential catalytic cofactor, and an antenna pigment, either a folate or a flavin derivative that absorbs blue or near UV light much stronger than FADH- does and efficiently transfers the excitation energy to FADH-.
It is widely accepted (1, 2) that CPD repair by photolyase proceeds by the following steps (see Fig. 1). A CPD recognized by the enzyme is flipped out of the double helix in order to enter the substrate binding pocket and to closely approach the FAD cofactor (see structure in Fig. 1). Once the substrate is bound, light is required to excite FADH-, either directly or via energy transfer from the excited antenna pigment. The photoexcited flavin (FADH-∗) transfers an electron to the CPD; consequently the two intra-CPD bonds split, and then the electron returns to the flavin (transiently present as neutral radical FADH∘) in order to restore catalytically active FADH-.
For the CPD formed of two thymines (T = T), the two electron transfer processes occur on time scales of 10-10 and 10-9 s, respectively (6). Finally, the two restored thymines are released from the enzyme’s binding pocket within 50 μs (7). The quantum yield of T = T repair is a matter of debate; while a value close to 100% (upon direct excitation of FADH-) is repeatedly mentioned (1), a recent actinometric study reported a value of approximately 50% (8).
The very process of enzymatic CPD splitting has been characterized only poorly so far. Transient absorption spectroscopy in the absorption band of intact pyrimidines around 265 nm appears to be an obvious choice to monitor CPD repair. However, the only available relevant study (9) did not provide conclusive results. In fact, there are a number of serious difficulties.
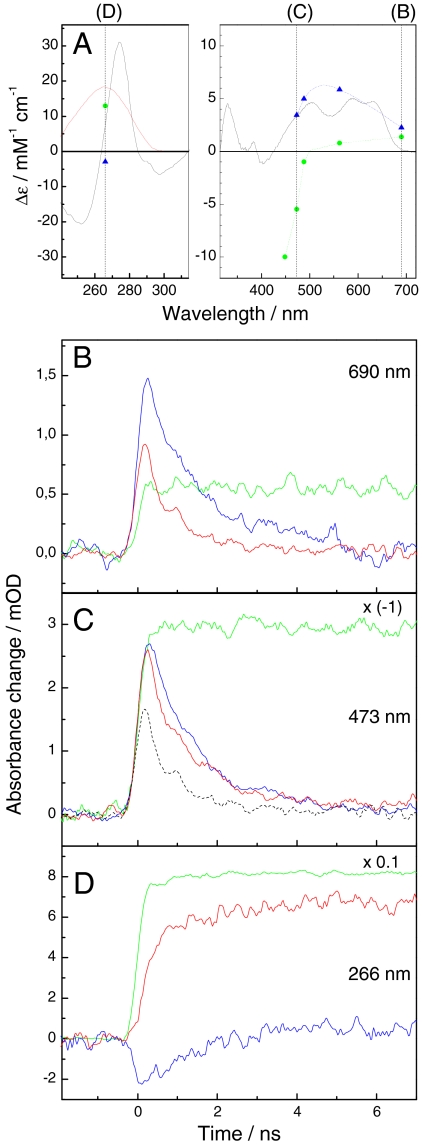
First, in the spectral region around 265 nm, transient absorption spectroscopy is much more difficult to realize than in the visible. Moreover, sample transmission is decreased by background absorption around 265 nm from protein and intact nucleotides flanking the CPD (which are required for proper binding of the substrate to the enzyme) (10). Extensive signal averaging commonly used to improve the signal-to-noise ratio is in conflict with the requirement to replace repaired DNA by damaged DNA after each turnover. Finally, transitions of the FAD cofactor (formation and decay of the excited state FADH-∗, formation of FADH∘ upon electron transfer from FADH-∗ to the CPD, and restoration of FADH- by electron return after CPD splitting) contribute considerably to the absorption changes around 265 nm (Fig. 2A).
Fig. 2.
Time-resolved spectroscopic data on CPD repair by photolyase. (A) Red line: repaired-minus-unrepaired difference spectrum for CPD repair in the present substrate (8). Black line: FADH∘-minus-FADH- difference spectrum obtained here by photoreduction (8) of photolyase and scaled to 5 mM-1 cm-1 at 586 nm (23). Blue triangles: FADH-∗-minus-FADH− differential absorption coefficients obtained in this study (see SI Text). Green circles: [Ru(bpy)3]2+∗-minus-[Ru(bpy)3]2+ differential absorption coefficients obtained here (see SI Text). Dotted curves serve to guide the eye. Vertical lines indicate the wavelengths of transient absorption measurements. (B–D) Transient absorption data at 690, 473, and 266 nm, respectively. Blue traces: photolyase without substrate. Red traces: photolyase with substrate. Green traces: aqueous solution of Ru(bpy)3Cl2 (21 μM). Photolyase concentration: 37, 37, and 3.7 μM, respectively. Excitation energy: 0.6, 0.9, and 11.6 mJ/cm2, respectively, at 355 nm. Substrate concentration was 2.5 to 3 times the photolyase concentration. The red trace in D is the average of 361 single shot experiments. All other solid traces are averages of 100 to 200 shots. Black dotted trace in C: calculated contribution of FADH-∗ to the transient absorption signal at 473 nm in the presence of substrate (see SI Text).
In the absence of reliable experimental data on the kinetics of enzymatic CPD splitting, predictions by computer simulations (reviewed in ref. 2) are of particular interest. According to the only quantum mechanical/molecular mechanical dynamics simulation that included the entire enzyme (11), cleavage of the T = T occurs within 400 fs upon uptake of an electron.
We have now managed to monitor enzymatic splitting of T = T and restoration of intact thymines experimentally by transient absorption spectroscopy. We applied an extension to 266 nm of a real time transient absorption setup developed recently in our lab (12) that allowed obtaining the kinetics by averaging relatively few single shot experiments (approximately 102 vs. approximately 106 in ref. 9; see also Discussion). A recently introduced substrate (8) in which a T = T is flanked by 5,6-dihydrothymidines that do not absorb around 265 nm, allowed to work at large substrate excess. Flavin transitions overlaying the repair-associated absorption changes at 266 nm were monitored independently in the visible and subtracted to obtain true T = T repair kinetics. In order to avoid interference by excitation energy transfer, we used a photolyase from the cyanobacterium Anacystis nidulans that was devoid of the antenna pigment due to heterologous expression in Escherichia coli.
Results
In order to resolve the reaction sequence and kinetics of T = T repair by photolyase, we monitored flash-induced absorbance changes at three principal wavelengths, 690, 473, and 266 nm (Fig. 2 B–D, respectively), where major contributions from FADH-∗, FADH∘, and restoration of intact thymines, respectively, are expected (see Fig. 2A). Photolyase in the presence of T = T containing oligonucleotides (red traces) was compared with photolyase in the absence of substrate (blue traces) to distinguish between reactions related to T = T repair and to the intrinsic behavior of FADH-∗ in photolyase. Absorbance changes from the well characterized, photostable ruthenium complex [Ru(bpy)3]2+ (green traces) were recorded as a reference for proper scaling of the photolyase data (see SI Text).
FADH-∗ can be monitored directly at 690 nm, because no other species in the photolyase-substrate system is expected to contribute significantly at this wavelength. The decay of FADH-∗ was strongly accelerated in the presence of substrate (Fig. 2B) as observed previously (6, 13) and attributed to electron transfer to T = T (see SI Text for details).
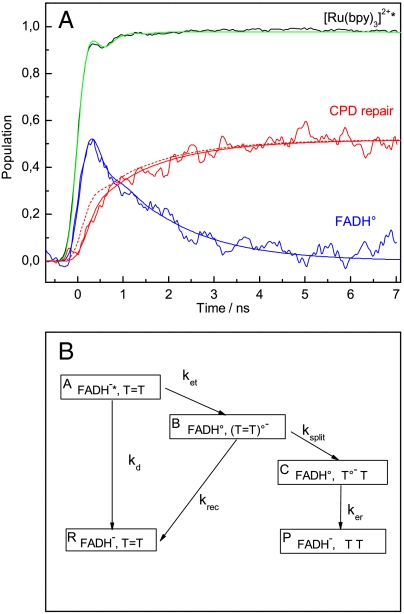
Monitoring the kinetics of FADH∘ is complicated by the transient absorption of FADH-∗ present throughout the visible absorption bands of FADH∘ (Fig. 2A). The true kinetics of FADH∘ (noisy blue trace in Fig. 3A) was obtained from the transient absorption data at 473 and 690 nm by correcting the signal observed at 473 nm in the presence of substrate (Fig. 2C, red trace) for the contribution of FADH-∗ (Fig. 2C, black dotted trace) as deduced from the data at 690 nm (see SI Text). The amplitude was converted from absorbance change to population (FADH∘ per originally excited photolyase) using the [Ru(bpy)3]2+ signal at 473 nm as a reference (see SI Text). The FADH∘ population rose to a maximal value of 0.5 at approximately 300 ps after excitation and decayed on a time scale of a few ns with apparently biphasic kinetics. Similar data were obtained from measurements at 488 and at 562 nm (see SI Text). The kinetics of FADH∘ is analyzed in more detail below.
Fig. 3.
Modeling of the experimental population kinetics of FADH∘ and of T = T repair. (A) Noisy blue and red traces: population kinetics of FADH∘ and of T = T repair (restoration of thymine pairs) deduced from experimental data as outlined in the text and in the SI Text. Black trace: transient absorption signal for Ru(bpy)3Cl2 from Fig. 2D scaled to unity amplitude. The four smooth traces are convolutions (see SI Text) of the instrument response function (SI Text) with the following model kinetics: Blue and red solid traces: population kinetics of FADH∘ and of CPD repair, respectively, according to the reaction scheme presented in B with kd = (1.25 ns)-1, ket = (0.1 ns)-1, krec = (0.35 ns)-1, ksplit = (0.26 ns)-1, ker = (1.55 ns)-1. Red dashed trace: same as red solid trace, except that ksplit = (1 ps)-1, and that the amplitude was scaled down by a factor of 0.58. Green trace: step function with unity amplitude. (B) Kinetic scheme used to model the experimental results. Analytical expressions for the population kinetics according to this scheme are presented in SI Text.
Formation of intact thymines as the result of T = T repair is accompanied by a rather strong absorption increase at 266 nm (Fig. 2A, red line). In fact, flash excitation of photolyase in the presence of substrate induced a persistent absorption increase at 266 nm (Fig. 2D, red trace). Its rise was somewhat slower than the quasi-instantaneous rise of the [Ru(bpy)3]2+ reference signal (green trace). For photolyase in the absence of substrate (blue trace), a small negative signal due to formation and decay of FADH-∗ was observed. FADH-∗ should contribute with similar amplitude, but accelerated decay kinetics, to the 266 nm signal in the presence of substrate. In addition, a significant positive contribution from transient FADH∘ is expected at 266 nm (see Fig. 2A). In order to recover the true kinetics of thymine restoration/ T = T repair, the observed 266 nm kinetics (Fig. 2D, red trace) had to be corrected for the flavin-based contributions (see SI Text). The resulting population kinetics of T = T repair (restored thymine pairs per originally excited photolyase) is shown in Fig. 3A, red trace. Formation of intact thymine turned out to be clearly delayed and slower than the formation of FADH∘, and appears to be biphasic with a first phase faster than 1 ns and a second phase with a time constant of approximately 1.5 ns. The second phase mirrors the slower phase of the decay of FADH∘. This coincidence was expected, as restoration of the second intact thymine should occur simultaneously with the reduction of FADH∘ by electron return from T∘- (see Fig. 1). The asymptotic population of restored thymine pairs (quantum yield of T = T repair) was 52 ± 5%.
Our observations deviate in two important points from what was expected on the grounds of previous findings and reasonings (1, 2):
Formation of intact thymine is significantly delayed with respect to formation of FADH∘ (i.e., with respect to electron transfer from FADH-∗ to the CPD).
The quantum yield of T = T repair is clearly below 100%.
Computer simulations of T = T splitting by photolyase (11) predicted virtually barrierless, ultrafast (< 1 ps) splitting of the two intradimer bonds upon electron addition to the CPD. If this were true, there should be no significant delay between formation of FADH∘ and restoration of the first thymine of the CPD. In addition, the quantum yield of T = T splitting should be close to 100%, because the only significant loss process would be the decay of the excited state FADH-∗ to the ground state (1.25 ns, as observed in the absence of substrate), which competes only poorly with electron transfer from FADH-∗ to the CPD (approximately 0.2 ns) (6).
The simplest reaction scheme that could account for our experimental observations is shown in Fig. 3B. It includes an additional loss process, recombination (i.e., electron back transfer) between the dimer anion radical (T = T)∘- and FADH∘ before splitting of the intradimer bonds (rate constant krec). The rate constants for the three electron transfer processes between FAD and T = T, viz. ket, krec, and ker , may be of the same order of magnitude ( i.e., approximately 109 s-1) as the electron transfer distances are similar (if not identical) for the three processes; because of different driving forces, the three rates are, however, not expected to be identical. Were the rate of dimer splitting (ksplit) comparable to krec, this would qualitatively account for both a repair quantum yield in the order of 50% and for the relatively slow (compared to formation of FADH∘) rise of intact thymine restoration.
For quantitative analysis, we convoluted the kinetics predicted by the scheme in Fig. 3B with the instrument response of our setup (see SI Text) and adjusted the rate constants iteratively to obtain simultaneously optimal descriptions of the observed population kinetics of both T = T repair and FADH∘ (Fig. 3A, smooth solid traces). The rate constants of this fit are: kd = (1.25 ns)-1, ket = (0.1 ns)-1, krec = (0.35 ns)-1, ksplit = (0.26 ns)-1, ker = (1.55 ns)-1. As expected, the splitting rate constant is comparable to recombination prior to splitting and much slower than predicted (11) by computer simulations (ksplit > (1 ps)-1).
We also checked how much a simulation assuming ultrafast splitting deviates from the observed repair kinetics, ignoring the discrepancy in absolute repair quantum yield between our experimental data (52%) and a model assuming ultrafast splitting (close to 100%). For this aim, ksplit was fixed to (1 ps)-1 and all the other rate constants were left unchanged. In order to fit the observed asymptotic population of restored thymine pairs, the simulated population kinetics had to be scaled down artificially by a factor of 0.58. The resulting simulation (Fig. 3A, broken red line) rises significantly faster than the experimental curve, so that ultrafast T = T splitting would be inconsistent with our experimental results even if we had strongly underestimated the absolute repair quantum yield.
In our simulations, it is assumed that the absorption of (T = T)∘- at 266 nm is negligible, and that the species (T∘- T) in the substrate binding pocket absorbs as one isolated thymine at 266 nm. MacFarlane and Stanley (9) suggested that (T = T)∘- might have an absorption coefficient of 1.6 mM-1 cm-1 ( i.e., approximately 10% of two intact thymines). According to a theoretical calculation (14), the cumulated oscillator strength in the 260- to 290-nm range of (T = T)∘- is 28% of that of (T∘- T). Possibly, the slightly positive amplitude of our experimental T = T repair curve (noisy red trace in Fig. 3A) at early times is in fact due to the absorption of (T = T)∘- formed simultaneously with FADH∘.
Discussion
Using a previously unreported real time transient absorption setup with monitoring light from a low noise CW laser at 266 nm, we succeeded in monitoring the kinetics of T = T splitting and restoration of intact thymines during T = T repair by photolyase. Flavin transitions occurring on the same time scale and contributing at 266 nm were monitored independently in the visible spectral region. For relative scaling of measurements at different wavelengths and absolute quantum yield determinations, we developed a method using the long-lived absorption changes of a ruthenium complex as a standard. Apart from applications in photorepair of DNA [including repair of pyrimidine (6-4) pyrimidone photoproducts] (15), the setup and methods presented here could be used for time-resolved studies of electron and hole transfer within DNA and from type I photosensitizers to DNA (16).
Our results reveal that restoration of the first thymine of the dimer (i.e., cleavage of both intradimer bonds) is significantly delayed with respect to formation of FADH∘ (i.e., electron transfer from FADH-∗ to the dimer), and that the total amount of restored thymine corresponds to only 0.5 dimers repaired per excited flavin (Fig. 3A). These two observations could be explained in a kinetic scheme (Fig. 3B) that assumes competition between dimer splitting and a recombination reaction (electron back transfer from (T = T)∘- to FADH∘ prior to dimer splitting) that have comparable rate constants of approximately (0.3 ns)-1.
Our data on the kinetics of FADH∘ are in accordance with observations by Kao and coworkers (6). These authors also reported that the decay of FADH∘ deviated from monoexponential kinetics and described it either by a stretched exponential or by a double-exponential decay (with lifetimes of 0.46 and 1.24 ns). It was suggested that the non-single-exponential decay reflected strong coupling of electron return (from split T = T to FADH∘) with slow solvation at the active site (6). In contrast, according to our simulation, the faster phase reflects the decay of FADH∘ by recombination prior to T = T splitting, and only the slower phase reflects electron return after splitting. We do not have to postulate coupling with slow solvation to explain the non-monoexponential decay of FADH∘.
MacFarlane and Stanley (9) attempted to monitor T = T repair by ultrafast pump-probe transient absorption at 265 nm, using photolyase from Anacystis nidulans and a pentameric thymine oligonucleotide containing arbitrarily distributed CPDs as substrate. For each sample, signals were averaged for approximately 1 h at a repetition rate of 1 kHz. A spinning cell was used for replacing the excited volume between shots. The pump pulses (at 398 nm) had an energy of 4–5 μJ and a beam diameter of 150–200 μm; this yields 13–28 mJ/cm2, compared to approximately 10 mJ/cm2 (at 355 nm) in our UV experiments. The amount of T = T repair per excited FADH- was apparently four times lower than in the present study (judged from the ratio of absorbance increase at 3 ns after excitation in the presence of substrate compared to the initial absorbance decrease in the absence of substrate). In the previous work (9), it could not be corrected for the contributions of FADH-∗ and FADH∘, because these two species were not monitored independently. The uncorrected rise kinetics at 265 nm was described by a minor phase with a lifetime τ = 32 ± 20 ps and a major phase with τ = 0.6 ± 0.3 ns (9). A 32-ps phase would have escaped detection in our setup because of its lower time resolution. The 0.6 ns rise in ref. (9) is likely to reflect a superposition of several processes, including decay of FADH-∗, formation and decay of FADH∘, and restoration of intact thymines. Surprisingly, the 265-nm absorbance change reported in ref. 9 lost amplitude beyond 1.5 ns and until the end of the time window at 3 ns, which was described by a decay time constant of 6.5 ns and was tentatively attributed to electron escape from T∘- to an unidentified acceptor (instead of electron return to FADH∘). We did not observe any such decay and would hence exclude the electron escape suggested in ref. 9.
Our observed T = T repair kinetics can adequately be described by the minimal scheme presented in Fig. 3B. This scheme ignores that T = T cleavage probably proceeds by stepwise splitting of the C5-C5′ and C6-C6′ bonds (11, 14). The suggested reaction intermediate after the first bond splitting (T- - T∘) is unlikely to absorb strongly at 266 nm and would hence not have been detected in our study. However, recombination from this intermediate would restore T = T and FADH- (17). This reaction would be another loss channel (in addition to kd and krec) and might have contributed to the observed FADH∘ population decay.
Interestingly, CPDs composed of cytosines (C = C) are split with a much lower quantum yield than T = T (5% versus 90% according to earlier literature) (18). As pointed out in ref. 19, the lower quantum yield for C = C splitting may reflect less favorable competition between bond splitting and back electron transfer from (C = C)∘- to FADH∘. It would hence be interesting to study the repair of cytosine containing CPDs (C = C, C = T, and T = C) by the methods developed in this contribution [however, care would have to be taken to minimize deamination of cytosine in the CPDs (5) prior to photorepair]. An earlier suggestion (20) that the strain imposed on the cyclobutane ring configuration by the C(5) methyl groups of the thymines favors rapid splitting of T = T may be addressed by time-resolved studies of the repair of CPDs containing uracil (which lacks the methyl group).
Comparison of the observed kinetics of T = T splitting by photolyase to model studies in the absence of enzyme is expected to provide insight into the role the enzyme plays in lowering the activation energy of splitting. Unfortunately, the large variation of reported splitting rates for model systems [< 35 ns (14) to several hundred ns (21)] precludes definitive conclusions.
On the other hand, quantum mechanical/molecular mechanical dynamics simulations that included the entire enzyme predicted ultrafast (< 1 ps) barrierless splitting of T = T CPDs (11). The considerably slower splitting observed in our study now suggests that there is a significant activation barrier to enzymatic T = T splitting. With the transient absorption technique developed in the present study, it should be possible to monitor CPD splitting and electron transfer during enzymatic photorepair as a function of temperature and establish their respective activation energies experimentally.
Materials and Methods
Sample Preparation.
A. nidulans photolyase devoid of the antenna pigment was expressed and purified as described (8). The substrate was a modified thymidine 10-mer with a central T = T and all other bases, except the one at the 3′ end, replaced by 5,6-dihydrothymine (5S∶5R stereoisomer ratio 90∶10) (8). The binding constant of our photolyase to this substrate is at least in the order of 106 M-1 (see SI Text). For comparison, a value of 3 × 108 M-1 was reported for T = T in an unmodified thymidine 20-mer (22). Photolyase samples were made up in 10 mM phosphate buffer, pH 7.0, and 100 mM NaCl, placed in an anaerobic 10 × 2 × 8 mm (length×width×height) inner volume quartz cell with self-masking solid black walls and four clear windows (Starna) and kept on ice all the time, except for optical measurements (10 °C).
Samples for photorepair experiments were prepared anaerobically essentially as described (8). However, not only FADH∘ but also the fraction (approximately 25%) of fully oxidized flavin FADox was photoreduced to FADH- by replacing the 515 nm cutoff filter of the Xe-lamp by 2 mm GG435 (Schott) and 3 mm B390 (Hoya). Complete photoreduction was verified by measuring absorption spectra of the samples (see SI Text). The photolyase concentration was calculated from the absorbance differences between prior to and after the photoreduction using differential absorption coefficients of 5.0 and 2.0 mM-1 cm-1 for FADH∘ minus FADH- at 580 nm and 450 nm, respectively (Fig. 2A), and 0 and 8.5 mM-1 cm-1 for FADox minus FADH- at 580 nm and 450 nm, respectively (23). In presentations of signals measured without and with substrate (Fig. 2), amplitudes were rescaled to compensate for the dilution by substrate addition (< 25%).
Absorption spectra of the samples were measured with an Uvikon XS spectrophotometer (Secomam) along either the 10 mm or the 2 mm path of the cell such that the optical density was as high as possible but not exceeding 1.5.
Transient Absorption Measurements.
Transient absorption changes in the visible were measured using a slightly modified version of a setup described previously (12). In brief, the sample was excited along the 2-mm path of the cell by vertically polarized flashes of 100-ps duration at 355 nm from an Nd/YAG laser (Leopard SS-10 from Continuum). Monochromatic monitoring light at several fixed wavelengths was provided by the CW lasers described previously (12) and a newly acquired laser emitting 50 mW at 473 nm (473L-50-COL-PP from Oxxius). The monitoring light passed the cell along the 10-mm path, was polarized at magic angle (54.7°) with respect to the polarization of the excitation flashes, and was mechanically chopped to provide rectangular pulses of 140-μs duration. The excitation flash was synchronized such that it occurred at approximately 30 μs after the rise of the monitoring light pulse. The detection system for the monitoring light consisted of a photodiode (UPD-200 from Alphalas, rise time 200 ps), 1.5 m coaxial cable, a bias T (5550B from Picosecond Pulse Labs, bandwidth 100 kHz–18 GHz), an electronic amplifier (IV72A from Hahn-Meitner Institut, bandwidth 100 Hz–1.7 GHz, gain 30 dB), and a digital oscilloscope (Infiniium 81004B from Agilent, bandwidth DC—10 GHz, sampling rate 40 Gsa/s).
The excitation beam was shaped to a rectangular area of 10 × 1 mm (width × height) at the position of the cell. The monitoring beams had diameters slightly less than 1 mm and passed centrally through the excited volume. The energy of excitation was measured for each flash by an energy meter (OE25SP-H-MB from Gentec) placed in the excitation beam leaving the cell. Signals presented in the same panel in Fig. 2 were, if necessary, rescaled to compensate for slight differences in the excitation energy.
For transient absorption measurements at 266 nm, a separate setup was constructed. It used a CW laser emitting approximately 20 mW at 266 nm (UW-1020A from Sony) and optical elements optimized for 266 nm. Otherwise, it was identical to the set-up described above.
In order to achieve that the probed volume contained mostly enzymes that were bound to substrate (rather than to repaired DNA or to no DNA at all), the cell was moved vertically by 1 mm between the individual excitation flashes (or between series of 16 flashes in experiments at flash energies < 1 mJ/cm2). Thus, seven experiments could be carried out by moving the cell from the lowest to the highest position, so that every excitation flash (or series of flashes) hit a fresh sample spot. Afterwards, an absorption spectrum was recorded to check the amount of repaired T = T (from the UV spectrum) and the reduction state of the FAD cofactor (from the visible spectrum). The transient absorption experiments were continued as long as less than 25% of the CPDs were repaired and less than 10% of FADH- was reoxidized (see SI Text).
Before, after, and several times during a series of experiments on photolyase, the transient absorption of a 21 μM aqueous solution of Ru(bpy)3Cl2 (from Aldrich) in an identical cell was recorded as a reference. These data were used to define time zero and for proper scaling of experiments on photolyase (see SI Text).
Supplementary Material
Acknowledgments.
This work was supported by Agence Nationale de la Recherche (Grant ANR-05-BLAN-0304-01). We thank Martin Gass from Sony for his dedication and efficiency with the acquisition and installation of the 266 nm laser.
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1101026108/-/DCSupplemental.
References
- 1.Sancar A. Structure and function of DNA photolyase and cryptochrome blue-light photoreceptors. Chem Rev. 2003;103:2203–2237. doi: 10.1021/cr0204348. [DOI] [PubMed] [Google Scholar]
- 2.Brettel K, Byrdin M. Reaction mechanisms of DNA photolyase. Curr Opin Struct Biol. 2010;20:693–701. doi: 10.1016/j.sbi.2010.07.003. [DOI] [PubMed] [Google Scholar]
- 3.Dalhus B, Laerdahl JK, Backe PH, Bjørås M. DNA base repair—recognition and initiation of catalysis. FEMS Microbiol Rev. 2009;33:1044–1078. doi: 10.1111/j.1574-6976.2009.00188.x. [DOI] [PubMed] [Google Scholar]
- 4.Eker APM, Quayle C, Chaves I, van der Horst GTJ. Direct DNA damage reversal: elegant solutions for nasty problems. Cell Mol Life Sci. 2009;66:968–980. doi: 10.1007/s00018-009-8735-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Peng W, Shaw BR. Accelerated deamination of cytosine residues in UV-induced cyclobutane pyrimidine dimers leads to CC → TT transitions. Biochemistry. 1996;35:10172–10181. doi: 10.1021/bi960001x. [DOI] [PubMed] [Google Scholar]
- 6.Kao YT, Saxena C, Wang L, Sancar A, Zhong D. Direct observation of thymine dimer repair in DNA by photolyase. Proc Natl Acad Sci USA. 2005;102:16128–16132. doi: 10.1073/pnas.0506586102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Espagne A, Byrdin M, Eker APM, Brettel K. Very fast product release and catalytic turnover of DNA photolyase. Chembiochem. 2009;10:1777–1780. doi: 10.1002/cbic.200900328. [DOI] [PubMed] [Google Scholar]
- 8.Thiagarajan V, et al. DNA repair by photolyase: A novel substrate with low background absorption around 265 nm for transient absorption studies in the UV. Biochemistry. 2010;49:297–303. doi: 10.1021/bi901562a. [DOI] [PubMed] [Google Scholar]
- 9.MacFarlane AW, Stanley RJ. Cis-syn thymidine dimer repair by DNA photolyase in real time. Biochemistry. 2003;42:8558–8568. doi: 10.1021/bi034015w. [DOI] [PubMed] [Google Scholar]
- 10.Mees A, et al. Crystal structure of a photolyase bound to a CPD-like DNA lesion after in situ repair. Science. 2004;306:1789–1793. doi: 10.1126/science.1101598. [DOI] [PubMed] [Google Scholar]
- 11.Masson F, Laino T, Rothlisberger U, Hutter JA. QM/MM investigation of thymine dimer radical anion splitting catalyzed by DNA photolyase. Chemphyschem. 2009;10:400–410. doi: 10.1002/cphc.200800624. [DOI] [PubMed] [Google Scholar]
- 12.Byrdin M, Thiagarajan V, Villette S, Espagne A, Brettel K. Use of ruthenium dyes for subnanosecond detector fidelity testing in real time transient absorption. Rev Sci Instrum. 2009;80:043102. doi: 10.1063/1.3117208. [DOI] [PubMed] [Google Scholar]
- 13.Okamura T, et al. Picosecond laser photolysis studies on the photorepair of pyrimidine dimers by DNA photolyase. 1. Laser photolysis of photolyase-2-deoxyuridine dinucleotide photodimer complex. J Am Chem Soc. 1991;113:3143–3145. [Google Scholar]
- 14.Chatgilialoglu C, et al. Ring opening of the cyclobutane in a thymine dimer radical anion. Chem Eur J. 2007;13:8979–8984. doi: 10.1002/chem.200700807. [DOI] [PubMed] [Google Scholar]
- 15.Li J, et al. Dynamics and mechanism of repair of ultraviolet-induced (6-4) photoproduct by photolyase. Nature. 2010;466:887–890. doi: 10.1038/nature09192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kumar A, Sevilla MD. Proton-coupled electron transfer in DNA on formation of radiation-produced ion radicals. Chem Rev. 2010;110:7002–7023. doi: 10.1021/cr100023g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Boussicault F, Krüger O, Robert M, Wille U. Dissociative electron transfer to and from pyrimidine cyclobutane dimers: An electrochemical study. Org Biomol Chem. 2004;2:2742–2750. doi: 10.1039/B406923D. [DOI] [PubMed] [Google Scholar]
- 18.Kim ST, Sancar A. Effect of base, pentose, and phosphodiester backbone structures on binding and repair of pyrimidine dimers by Escherichia coli DNA photolyase. Biochemistry. 1991;30:8623–30. doi: 10.1021/bi00099a019. [DOI] [PubMed] [Google Scholar]
- 19.Murphy AK, Tammaro M, Cortazar F, Gindt YM, Schelvis JPM. Effect of the cyclobutane cytidine dimer on the properties of Escherichia coli DNA photolyase. J Phys Chem B. 2008;112:15217–15226. doi: 10.1021/jp806526y. [DOI] [PubMed] [Google Scholar]
- 20.Pac C, Kubo J, Majima T, Sakurai H. Structure-reactivity relationships in redox-photosensitized splitting of pyrimidine dimers and unusual enhancing effect of molecular oxygen. Photochem Photobiol. 1982;36:273–282. [Google Scholar]
- 21.Yeh SR, Falvey DE. Model studies of DNA photorepair: Energetic requirements for the radical anion mechanism determined by fluorescence quenching. J Am Chem Soc. 1992;114:7313–7314. [Google Scholar]
- 22.Malhotra K, Kim S-T, Walsh C, Sancar A. Roles of FAD and 8-hydroxy-5-deazaflavin chromophores in photoreactivation by Anacystis nidulans DNA photolyase. J Biol Chem. 1992;267:15406–154011. [PubMed] [Google Scholar]
- 23.Jorns MS, Wang BY, Jordan SP, Chanderkar LP. Chromophore function and interaction in Escherichia coli DNA photolyase: Reconstitution of the apoenzyme with pterin and/or flavin derivatives. Biochemistry. 1990;29:552–561. doi: 10.1021/bi00454a032. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.