Abstract
The monosaccharide addition of an N-acetylglucosamine to serine and threonine residues of nuclear and cytosolic proteins (O-GlcNAc) is a posttranslational modification emerging as a general regulator of many cellular processes, including signal transduction, cell division, and transcription. The sole mouse O-GlcNAc transferase (OGT) is essential for embryonic development. To understand the role of OGT in mouse development better, we mapped sites of O-GlcNAcylation of nuclear proteins in mouse embryonic stem cells (ESCs). Here, we unambiguously identify over 60 nuclear proteins as O-GlcNAcylated, several of which are crucial for mouse ESC cell maintenance. Furthermore, we extend the connection between OGT and Polycomb group genes from flies to mammals, showing Polycomb repressive complex 2 is necessary to maintain normal levels of OGT and for the correct cellular distribution of O-GlcNAc. Together, these results provide insight into how OGT may regulate transcription in early development, possibly by modifying proteins important to maintain the ESC transcriptional repertoire.
Keywords: EED, Host Cell Factor C1, electron transfer dissociation
The posttranslational modification O-GlcNAc is the enzyme-mediated glycosylation of serine and threonine residues of cytosolic and nuclear proteins by a single N-acetylglucosamine. (1). Unlike membrane-bound and secreted protein glycosylation, such as N-linked glycans, mucin type O-glycosylation, or GPI anchors, O-GlcNAcylation is a reversible and dynamic posttranslational modification (PTM). Although O-GlcNAcylation has been implicated in nearly every cellular process, from nutrient sensing and insulin signaling (2) to synaptic plasticity (3), our knowledge of the exact functions of this widely distributed monosaccharide PTM is still in its infancy.
O-GlcNAcylation is akin to phosphorylation in that it is coupled to and hydrolyzed from specific sites on proteins to modulate their function. Hence, cycling of O-GlcNAc may affect the activity, stability, subcellular localization, and biomolecular interactions of modified proteins. Some of the activities of O-GlcNAcylation may occur as a result of cross-talk between O-GlcNAcylation and other PTMs, a phenomenon documented for protein phosphorylation (4, 5) and ubiquitination (6). The cross-talk with phosphorylation is particularly interesting, as there is only one conserved mammalian O-GlcNAc transferase (OGT), with its extensive tetratricopeptide repeat protein-protein interaction domain, that must cooperate and/or compete with hundreds of protein kinases that recognize their substrates individually (7–9).
OGT is essential for mouse development, as Ogt mutant embryos die shortly after implantation (10). OGT interacts with and may modify OCT4, a transcription factor that is essential for postimplantation development (11–14). Studies in Drosophila melanogaster provide additional mechanistic insight into the developmental role for OGT. Mutants of the fly Ogt homolog, super sex combs (sxc), exhibit phenotypes similar to the loss of Polycomb group (PcG) proteins (15–17). PcG proteins assemble into two Polycomb repressive complexes (PRC1 and PRC2) that are necessary for developmentally regulated transcriptional silencing of many genes crucial for early embryonic patterning (18). Two lines of evidence link fly OGT to PcG proteins. First, there is significant overlap in genomic occupancy between O-GlcNAc and subunits of PRC2 and PRC1. It is likely that O-GlcNAcylation regulates a function other than PRC recruitment to target genes, because PRC binding to the majority of its targets is unaffected in sxc mutants (16). Second, a Drosophila PRC1 protein, Polyhomeotic (Ph), interacts with wheat germ agglutinin, a feature of O-GlcNAcylated proteins (16). O-GlcNAcylation of the mammalian Ph homolog, PHL3, and a second mammalian PcG protein homolog, YY1, extends the relationship between OGT and PcG to mammals (19–21).
Mouse embryonic stem cells (ESCs) provide a mammalian model system for deciphering the role of OGT in early development. ESCs are pluripotent and can differentiate into each cell type in the adult and developing embryo. In addition, they can be cultured in the pluripotent state indefinitely (22). Although OGT mutant ESC lines cannot be derived (10, 23), PRC2 mutant ESCs are viable and exhibit inappropriate expression of genes that are important in later developmental stages (24). Thus, PRC2 mutant ESC lines provide a system to study the interaction between OGT and PcG function.
To explore the role for O-GlcNAc modification in ESCs, we used an unbiased strategy for enrichment of native O-GlcNAcylated nuclear peptides and high-resolution electron transfer dissociation tandem mass spectrometry (ETD MS/MS) to map their sites of modification. Here, we report the unambiguous identification of O-GlcNAc modification sites on over 60 proteins, many of which have documented roles in ESC pluripotency or self-renewal. In addition, we discover that PRC2 is necessary for maintaining normal levels of OGT in ESCs. The decrease in OGT abundance in PRC2 mutant ESCs is accompanied by an alteration in the amounts and distribution of O-GlcNAcylation in the ESC nuclear O-GlcNAcylated proteome. Together, these results suggest that normal OGT activity in ESCs is influenced by PRC2, providing a functional connection between the two pathways.
Results
Identification of O-GlcNAc–Modified Peptides in Mouse ESC Nuclei.
A combination of lectin weak affinity chromatography (LWAC) and ETD MS/MS (19, 25, 26) was used to identify native O-GlcNAc–modified peptides from mouse ESC nuclear proteins. One hundred forty-two O-GlcNAc sites were detected on 62 proteins (Fig. S1 and Table S1). Although proteins implicated in virtually every nuclear function were found to be O-GlcNAc–modified, the majority are involved in transcriptional regulation. For example, two transcription factors essential for ESC self-renewal, SOX2 (Fig. 1) and ZFP281 (Fig. S1), were modified. BPTF, which is a subunit of the histone remodeling complex NURF and is necessary for pluripotency (27, 28), was also O-GlcNAc–modified. Sin3a, an essential transcriptional regulator in ESCs (29), and several of its interacting proteins were O-GlcNAcylated. For some of these targets, such as SOX2 and the Sin3a interaction protein Host Cell Factor C1 (HCF-1), O-GlcNAcylation was previously reported, although no specific modification sites were determined (30, 31). ETD MS/MS enabled the unambiguous site assignments for these proteins.
Fig. 1.
ETD MS/MS spectra of the two positional isomers of the O-GlcNAcylated SOX2 peptide. Adequate c- and z-series ions allow the unambiguous site assignment of the HexNAc mass modification to serine 248 (Upper) and threonine 258 (Lower). Charged reduced species and common neutral losses (55) are not labeled.
PRC2 Is Required for Normal OGT Stability and O-GlcNAc Distribution.
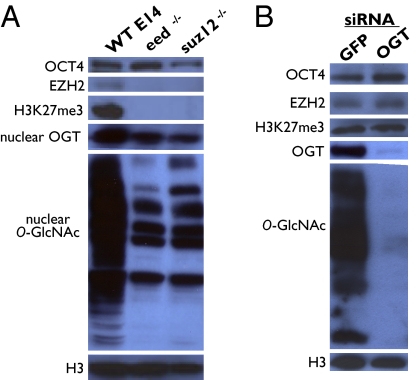
The link between OGT and PcG proteins in D. melanogaster prompted us to investigate whether PRC2-null ESCs exhibited any alterations in O-GlcNAcylation patterns. Mouse ESC lines with mutations in eed or suz12, two core components of PRC2, were analyzed for levels of OCT4, a pluripotency marker, as well as OGT and O-GlcNAcylated proteins. These PRC2 mutant ESCs exhibited levels of OCT4 similar to those in WT ESCs, indicating that they are largely undifferentiated. There was a marked drop in nuclear and whole-cell OGT (Fig. 2 and Fig. S2). Consistent with a decrease in OGT levels, there was a large decrease in the abundance of O-GlcNAcylated nuclear proteins in PRC2 ESCs (Fig. 2A), whereas whole-cell O-GlcNAc patterns were altered compared with WT (Fig. S2).
Fig. 2.
Western blot analysis of the relationship between PRC2 and OGT. (A) Disruption of PRC2 by mutations in either two, eed or suz12, of the three core components abrogates complex formation and methyltransferase activity as seen by EZH2 levels (third core component) and H3K27me3 levels, respectively. However, the pluripotency marker, OCT4, is unaffected. OGT and general O-GlcNAc levels are altered by PRC2 disruption. (B) siRNA knockdown of OGT does not significantly affect levels of OCT4, EZH2, or H3K27me3.
We next asked whether changes in OGT levels affect PRC2 activity in ESCs. After knockdown of OGT in WT ESCs, no disruption of PRC2 activity was seen, as measured by EZH2 and H3K27me3 levels (Fig. 2B).
PRC2 Mutants Exhibit Altered O-GlcNAcylation Patterns.
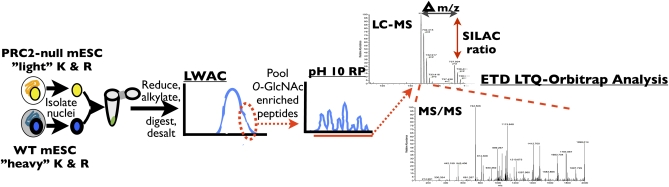
Western blot analysis of PRC2-null ESCs revealed changes in O-GlcNAcylation of nuclear proteins compared with WT cells. To identify specifically what O-GlcNAcylation changes were occurring in the absence of PRC2, we used stable isotopic labeling of amino acids in culture (SILAC) (32) along with LWAC ETD MS/MS to measure the relative abundance of O-GlcNAcylated peptides between WT and eed−/− nuclear proteins (Fig. 3). We found that 56 of the 80 O-GlcNAc–modified peptides exhibited alterations in SILAC ratios greater than 25% between WT and eed−/− mutant ESCs (Table S2). Although proteins implicated in a wide set of biological functions were differentially modified in eed−/− ESCs compared with WT ESCs, only O-GlcNAc–modified peptides from nuclear pore complex (NPC) proteins were consistently less abundant in eed−/− ESCs (Table S2).
Fig. 3.
SILAC LWAC liquid chromatography-MS/MS workflow. Nonquantitative samples were prepared similarly, except a single cell type was used. LC, liquid chromatography; LTQ, linear trap quadrupole.
In contrast to NPC proteins, the glycopeptides identified from other proteins exhibited variable differences between WT and eed−/− ESCs (Table 1). For example, the four different peptides derived from EMSY, two of which derive from the region that interacts with BRCA2 (33), varied greatly in their SILAC ratios. Two O-GlcNAcylated peptides exhibited a decreased abundance in eed−/− mutant ESCs. The I518-K530 peptide O-GlcNAcylated on S520 (gS520) exhibited a decrease of ∼95%, and the gS200-containing T194-K204 peptide showed a decrease of roughly 50%. One glycopeptide, T498-K509 containing either gT499 or gS500, was equivalent between cell types. The fourth glycopeptide, I225-K241, exhibited a 30–40% increase in the relative abundance in eed−/− mutant cells if modified at T228. Thus, the loss of EED affects some sites of modification and not others, suggesting that the O-GlcNAcylation of different regions within the same protein can be regulated independently.
Table 1.
Selected O-GlcNAc–modified peptides and SILAC ratios of discussed proteins
 |
Positional isomers are only reported for verified O-GlcNAcylation sites. All O-GlcNAcylated positional isomers from three biological replicates, as reported by Protein Prospector output, although not confirmed as bona fide sites, were combined for ratios and SDs. SDs are reported if the particular glycopeptide was identified between biological replicates. The total SILAC dataset can be found in Table S2.
In some instances, the same peptide exhibited variable SILAC ratios depending on which sites were modified. For example, the gT1710 containing T1710-K1727 BPTF peptide was equally abundant between cell lines, whereas the same peptide doubly modified at two other residues, gT1713 and gT1716, was 80% less abundant in the eed−/− ESCs. Similarly, a second BPTF glycopeptide, S1750-K1761, showed variable differences depending on which modification sites were used. The gS1750 positional isomer was nearly 50% less abundant in eed−/− mutant cells, whereas the isomer modified at two other sites, gT1755 and gT1760, was ∼80% less abundant. These data suggest that the O-GlcNAc modification of the same peptide can be regulated independently, with different positional isomers having different requirements for EED.
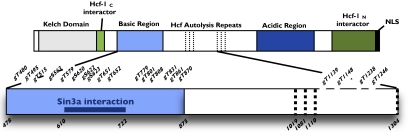
In other instances, a decrease in the abundance of one O-GlcNAcylated positional isomer of a peptide was accompanied by an increase in another, as is illustrated by HCF-1, which contains 20 sites of O-GlcNAcylation (Fig. 4). In eed−/− ESCs, the triply modified T612-K637 glycopeptide containing gS620/gS622/gS623 was almost 70% lower and the doubly modified peptide was ∼25% lower, whereas the singly modified gS623-containing peptide was 75% higher. These data indicate that for this HCF-1 peptide, the extent of glycosylation is hampered when PRC2 function is disrupted.
Fig. 4.
Diagram for landmarks of HCF-1. A zoomed-in view of the basic region and region of alternative proteolysis shows the extent and location of O-GlcNAc sites identified in this study. Numbers indicate landmark residues (Lower) or O-GlcNAc sites (Upper). g, glycosylation; NLS, nuclear localization sequence. Adapted from Wysocka et al. (44).
Discussion
Although the existence of O-GlcNAcylation of nucleocytoplasmic serine and threonine residues has been known for almost 3 decades, the technologies to identify these modification sites precisely have only been recently developed (19, 34), opening up many new lines of investigation into the function of this PTM. Our analysis of ESC nuclear O-GlcNAcylation revealed modifications of more than 60 proteins involved in virtually every nuclear process, although predominantly in transcriptional regulation. Several of these sites of O-GlcNAcylation have been reported in human cell lines and other mouse tissues (4, 19), providing evidence of site-specific conservation (Table S3). Additionally, we find that PRC2 is necessary for normal levels of OGT itself. The decrease in OGT levels in PRC2 mutant ESCs is consistent with an altered distribution of O-GlcNAcylation of nuclear proteins relative to WT ESCs. Together, these results show that proper PRC2 function is necessary for normal OGT abundance and O-GlcNAc distribution.
SOX2 and ZFP281 May Participate in Phosphorylation/O-GlcNAcylation Cross-Talk.
SOX2, OCT4, and Nanog form the core transcription factor network for pluripotency in ESCs (35). These transcription factors interact with themselves and a variety of other transcriptional regulatory proteins to up-regulate genes involved in pluripotency and down-regulate developmental genes. SOX2 and OCT4 cooccupy and coregulate many genes and form one arm of the self-renewal transcription factor network (36). Although there is some overlap with known SOX2/OCT4 targets, Nanog regulates a largely unique set of genes and appears to function in a separate pathway (37). ZFP281 interacts with Nanog and is also necessary for self-renewal (38, 39). Here, we have identified two sites of O-GlcNAcylation, gS248 and gT258, in the transactivation domain of SOX2 and three O-GlcNAcylation sites on ZFP281: gS691, gS889, and gT888. These results suggest that OGT may regulate both the SOX2/OCT4 and Nanog/ZFP281 pathways. Therefore, altered function of these pluripotency transcription factors may contribute to the lack of viability observed for OGT mutant ESCs.
The molecular mechanisms by which O-GlcNAcylation regulates transcription factor function are not fully understood. One well-documented mechanism for control of transcription factor activity is phosphorylation (40, 41), and cross-talk between these two PTMs is emerging as a potential means of regulation (5). Our findings suggest the potential for cross-talk between O-GlcNAcylation and phosphorylation in the modulation of SOX2 and ZFP281 function. In human ESCs, the residue homologous to S248 of SOX2, which is O-GlcNAcylated in mouse ESCs, is phosphorylated (42). Because S248 cannot be simultaneously phosphorylated and O-GlcNAcylated, this may represent an O-GlcNAcylation–phosphorylation switch involved in the regulation of SOX2 activity. In this study, ZFP281 O-GlcNAc modified at gT888 is concurrently phosphorylated at T886, whereas no phosphorylation is detected when ZFP281 is O-GlcNAcylated at S889. This result suggests that at least two populations of modification states for ZFP281 exist within ESCs, suggesting combinatorial control by these two PTMs. Assessment of the stoichiometry of the modification states for the individual proteins as well as mutational analysis of PTM sites will be necessary to understand fully whether O-GlcNAcylation/phosphorylation cross-talk plays a role in the control of pluripotency and self-renewal through SOX2 and ZFP281.
O-GlcNAcylation of Multiple Proteins Associated with Sin3a.
Sin3a is essential for ESC viability (29) and is a core component of several transcriptional corepressor complexes, including the Sin3/HDAC and NCor/Sin3a complexes (43). We identified several proteins implicated in Sin3a-mediated repression as O-GlcNAcylated in ESCs, including RBBP7, SAP130, SAP30bp, SOX2, HCF-1, NCoR1, NCoR2, and Cnot2. There is evidence of a stable interaction between OGT and a subset of these targets, because OGT can be copurified with Sin3a and HCF-1 (44). Moreover, Sin3a has been shown to recruit OGT to promoters to repress transcription (45). Our finding that several other Sin3a partners are also O-GlcNAc–modified suggests that the role for this PTM in the regulation of Sin3a corepressor complexes may be more extensive than previously appreciated. Mutational analysis will be necessary to determine the significance of O-GlcNAcylation of Sin3a and its interacting proteins for Sin3a-mediated transcriptional repression.
Loss of PRC2 Leads to Dysregulation of OGT and O-GlcNAcylation.
We have shown that PRC2 mutant ESCs exhibit a notable decrease in OGT levels, whereas ESCs depleted for OGT exhibit normal PRC2 function. These data indicate that OGT functions downstream of PRC2 in ESCs, as is also observed in Drosophila (16). The decrease in OGT protein levels in PRC2 mutant ESCs is not a consequence of decreased Ogt transcription, because OGT mRNA levels are not significantly altered in eed−/− or suz12 mutant ESCs (46, 47). It is also unlikely that PRC2 directly regulates OGT stability, because neither Suz12 nor EED forms stable complexes with OGT as measured by coimmunoprecipitation (48, 49). However, it is possible that the expression of factors affecting the stability of OGT depends on PRC2.
Western blot analysis of eed mutant ESCs showed the drop in OGT levels coincided with decreased nuclear O-GlcNAcylation levels. The decrease in O-GlcNAcylation is not a consequence of up-regulation of the sole O-GlcNAc hydrolase (50) (Fig. S3). Our SILAC analyses revealed that not all the O-GlcNAc sites were equally affected by this decrease in OGT levels. In fact, there were complex patterns of alterations in OGT target site use in eed−/− mutant ESCs. For the NPC proteins, modified peptides were not as abundant in the eed mutant ESCs as in WT ESCs, consistent with an overall decrease in O-GlcNAcylation. For other proteins, such as EMSY and HCF-1, some modified peptides were more abundant, some were less abundant, and some were unchanged in the mutant ESCs compared with WT. For BPTF, the same peptide exhibited two patterns of O-GlcNAcylation, and loss of eed affected the two positional isomers differently. Finally, a peptide within the Sin3a interaction region of HCF-1 (44) revealed evidence of an alteration in OGT processivity, because a decrease in the abundance of triply and doubly modified glycopeptides was accompanied by an increase in the abundance of the singly modified peptide (Table 1).
Our finding that certain O-GlcNAc sites are influenced by PRC2, whereas others are unaffected, along with previous studies showing the vast majority of the OGT substrates identified here are not directly transcriptionally regulated by PRC2 (24, 51), leads us to believe that the connection between PRC2 and O-GlcNAc distribution is likely indirect. We speculate that the change in O-GlcNAcylation patterns in PRC2-null ESCs is attributable to transcriptional changes in proteins that influence OGT target site selection. PRC2-null ESCs show defects in transcriptional repression of proteins involved in lineage-specific differentiation (24). The aberrant transcription of these genes in PRC2-null ESCs may provide a different set of cofactors that regulate OGT specificity, resulting in inappropriate increases and decreases of O-GlcNAcylation at a subset of target sites. A change in the expression of cofactors that regulate OGT target site selection may also explain the decrease in OGT levels, if these factors also affect OGT stability or turnover. Protein-protein interaction studies aimed at identification of the subproteome that interacts with OGT in ESCs will help to dissect the mechanism by which PRC2 influences OGT and O-GlcNAcylation.
This study unambiguously identifies 142 O-GlcNAc modification sites on 62 proteins. We show that OGT modifies transcription factors and components of transcriptional regulatory complexes that are essential to maintain the ESC-specific expression profile, and thus are central to stem cell maintenance. In addition, this work extends the relationship between OGT and Polycomb from Drosophila to mammalian systems. These data provide evidence toward the nature of the relationship between PRC2 and OGT, wherein OGT levels and O-GlcNAcylation patterns are influenced by PRC2 activity. Together, these results indicate that O-GlcNAc is directly involved in the transcriptional regulation during early embryonic development.
Methods
Sample Preparation.
WT ESCs were grown without feeders under standard conditions. For SILAC experiments, WT ESCs were grown in 13C6 arginine and 13C6, 15N2 lysine supplemented with 200 mM proline (Invitrogen) to avoid arginine-to-proline conversion. The suz12−/− (52) and eed−/− (53) ESC lines were grown on γ-irradiated mouse embryonic fibroblast feeder cells until the final passage, where the feeders were depleted twice by preferential adherence to gelatinized culture dishes.
Cells were harvested, washed twice with cold PBS, and combined at a 1:1 ratio after counting with a hemocytomer. Combined cells were resuspended in nuclear preparation buffer I [320 mM sucrose, 10 mM Tris (pH 8.0), 3 mM CaCl2, 2 mM Mg(OAc)2, 0.1 mM EDTA, 0.1% Triton X-100, 20 μM Thiamet G (Caymen Chemicals), protease inhibitors (Roche), and 1× phosphatase inhibitors (Invitrogen)] and dounce-homogenized on ice until >95% of nuclei stained by Trypan blue. Two volumes of nuclear preparation buffer II [2.0 M sucrose, Tris (pH 8.0), 5 mM Mg(OAc)2, 5 mM DTT, 20 μM Thiamet G, protease inhibitors (Roche), and phosphatase inhibitors (Invitrogen)] were added to the nuclei suspension. Nuclei were pelleted by ultracentrifugation at 130,000 × g at 4 °C for 45 min. Pelleted nuclei were washed with cold PBS and stored at −80 °C.
Nuclei, containing ∼3 mg of protein were resuspended in 6 M guanidine hydrochloride, 25 mM ammonium bicarbonate, 20 μM Thiamet G, protease inhibitors (Roche), and phosphatase inhibitors (Invitrogen); sonicated; and cleared by centrifugation. Soluble proteins were reduced using 2 mM Tris(2-carboxyethyl)phosphine (Thermo) for 1 h at 55 °C and alkylated using 10 mM iodoacetamide (Sigma) before digestion for 18 h using modified trypsin at 37 °C (part no. 9PIV5113; Promega). After digestion, the sample was acidified with formic acid and desalted using a C18 Sep-Pak (Waters), before drying down by vacuum centrifugation.
O-GlcNAcylated Peptide Chromatography-LWAC and High-pH RP.
O-GlcNAc–modified peptides were enriched as previously described (19). The O-GlcNAc–enriched fractions of the chromatograph were pooled, desalted, and dried. The O-GlcNAc–enriched pool was resuspended in 2 mM NH4OH (pH 10) and 5%(vol/vol) acetonitrile and separated on a Phenomenex Gemini 3-μm C18 reverse-phase column. The gradient of buffer A (2 mM NH4OH and 5% acetonitrile) to buffer B (2 mM NH4OH and 50%(vol/vol) acetonitrile) increased from 0–100% over 6 mL at 50 μL/min. Twenty-four 250-μL fractions were dried by vacuum centrifugation and resuspended in 0.1% formic acid for subsequent MS analysis.
LC-MS/MS.
LC-MS/MS of O-GlcNAcylated peptides was performed as previously described (19).
Data Analysis.
Fragment mass spectra were converted into peaklists using the in-house software PAVA. Collision activated dissociation (CAD) and electron transfer dissociation (ETD) data were searched separately using Protein Prospector version 5.6.0 against the UniProt database (accessed June 15, 2010) with a concatenated database. Only mouse and human genomes were used for the database searching. Precursor tolerance was set to 20 ppm, whereas fragment mass error tolerance was set to 0.6 Da. Cysteine residues were assumed to be carbamidomethylated, whereas N terminus acetylation, methionine oxidation, loss of N-terminal methionine, and glutamate conversion to pyroglutamate were variable modifications. For ETD data, N-acetylhexosamine (HexNAc) modifications to serine and threonine residues and phosphorylation to serine/threonine/tyrosine were allowed as variable mass modifications. The same modifications were allowed for CAD data, except the neutral loss of 203 Da (HexNAc) or 80 Da (phosphate) was considered. For SILAC experiments, the data were searched with arginine (+6 Da) and lysine (+8 Da) either as constant modifications or constantly absent. SILAC ratios were determined at a resolution of 30,000. All SILAC ratios and spectra of modified peptides were examined manually to assess spectral quality and site-assignment accuracy.
siRNA Knockdown.
siRNAs were created by in vitro cleavage of Ogt double-stranded RNA (54). A PCR product for in vitro transcription of Ogt RNA was generated using the primers OgtF GGGCGGGTAAAGAGAAGGGCAGTGTTGC and OgtR GGGCGGGTCTTGTGAATGGAGGCCAGAT.
Western Blot Analysis.
Antibodies were purchased from Abcam (OCT4 ab19857, EZH2 ab3748, and H3 ab1791), Millipore (H3K27me3 07-499), ProteinTech (OGA 14711-1-AP), Sigma (OGT SAB2101676 and O-GlcNAc CTD 110.6), and BioRad (secondary antibodies 172-1019 and 172-1011) and used at recommended concentrations.
Supplementary Material
Acknowledgments
We thank J. Trinidad, R. Chalkley, and M. Ali for technical assistance; R. Chalkley, R. Bradshaw, and T. Quan for critical review of the manuscript; and B. Hamilton and J. Fistorino for discussion. This work was supported by National Institutes of Health Grant R01 GM085186 and National Center for Research Resources (NCRR) Grant P41RR001614. S.A.M. is supported by a National Institutes of Health National Institute of General Medical Sciences T32 training grant and the Genentech Predoctoral Fellowship Program.
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1019289108/-/DCSupplemental.
References
- 1.Torres CR, Hart GW. Topography and polypeptide distribution of terminal N-acetylglucosamine residues on the surfaces of intact lymphocytes. Evidence forO-linked GlcNAc. J Biol Chem. 1984;259:3308–3317. [PubMed] [Google Scholar]
- 2.Yang X, et al. Phosphoinositide signalling links O-GlcNAc transferase to insulin resistance. Nature. 2008;451:964–969. doi: 10.1038/nature06668. [DOI] [PubMed] [Google Scholar]
- 3.Tallent MK, et al. In vivo modulation of O-GlcNAc levels regulates hippocampal synaptic plasticity through interplay with phosphorylation. J Biol Chem. 2009;284:174–181. doi: 10.1074/jbc.M807431200. [DOI] [PubMed] [Google Scholar]
- 4.Wang Z, et al. Extensive crosstalk between O-GlcNAcylation and phosphorylation regulates cytokinesis. Sci Signal. 2010 doi: 10.1126/scisignal.2000526. 10.1126/scisignal.2000526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hu P, Shimoji S, Hart GW. Site-specific interplay between O-GlcNAcylation and phosphorylation in cellular regulation. FEBS Lett. 2010;584:2526–2538. doi: 10.1016/j.febslet.2010.04.044. [DOI] [PubMed] [Google Scholar]
- 6.Guinez C, et al. Protein ubiquitination is modulated by O-GlcNAc glycosylation. FASEB J. 2008;22:2901–2911. doi: 10.1096/fj.07-102509. [DOI] [PubMed] [Google Scholar]
- 7.Butkinaree C, Park K, Hart GW. O-linked beta-N-acetylglucosamine (O-GlcNAc): Extensive crosstalk with phosphorylation to regulate signaling and transcription in response to nutrients and stress. Biochim Biophys Acta. 2010;1800:96–106. doi: 10.1016/j.bbagen.2009.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jínek M, et al. The superhelical TPR-repeat domain of O-linked GlcNAc transferase exhibits structural similarities to importin alpha. Nat Struct Mol Biol. 2004;11:1001–1007. doi: 10.1038/nsmb833. [DOI] [PubMed] [Google Scholar]
- 9.Clarke AJ, et al. Structural insights into mechanism and specificity of O-GlcNAc transferase. EMBO J. 2008;27:2780–2788. doi: 10.1038/emboj.2008.186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shafi R, et al. The O-GlcNAc transferase gene resides on the X chromosome and is essential for embryonic stem cell viability and mouse ontogeny. Proc Natl Acad Sci USA. 2000;97:5735–5739. doi: 10.1073/pnas.100471497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pardo M, et al. An expanded Oct4 interaction network: Implications for stem cell biology, development, and disease. Cell Stem Cell. 2010;6:382–395. doi: 10.1016/j.stem.2010.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.van den Berg DL, et al. An Oct4-centered protein interaction network in embryonic stem cells. Cell Stem Cell. 2010;6:369–381. doi: 10.1016/j.stem.2010.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Webster DM, et al. O-GlcNAc modifications regulate cell survival and epiboly during zebrafish development. BMC Dev Biol. 2009;9:28. doi: 10.1186/1471-213X-9-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lemischka IR. Hooking up with Oct4. Cell Stem Cell. 2010;6:291–292. doi: 10.1016/j.stem.2010.03.011. [DOI] [PubMed] [Google Scholar]
- 15.Ingham PW. A gene that regulates the bithorax complex differentially in larval and adult cells of Drosophila. Cell. 1984;37:815–823. doi: 10.1016/0092-8674(84)90416-1. [DOI] [PubMed] [Google Scholar]
- 16.Gambetta MC, Oktaba K, Müller J. Essential role of the glycosyltransferase sxc/Ogt in polycomb repression. Science. 2009;325:93–96. doi: 10.1126/science.1169727. [DOI] [PubMed] [Google Scholar]
- 17.Sinclair DA, et al. Drosophila O-GlcNAc transferase (OGT) is encoded by the Polycomb group (PcG) gene, super sex combs (sxc) Proc Natl Acad Sci USA. 2009;106:13427–13432. doi: 10.1073/pnas.0904638106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kerppola TK. Polycomb group complexes—Many combinations, many functions. Trends Cell Biol. 2009;19:692–704. doi: 10.1016/j.tcb.2009.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chalkley RJ, Thalhammer A, Schoepfer R, Burlingame AL. Identification of protein O-GlcNAcylation sites using electron transfer dissociation mass spectrometry on native peptides. Proc Natl Acad Sci USA. 2009;106:8894–8899. doi: 10.1073/pnas.0900288106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hiromura M, et al. YY1 is regulated by O-linked N-acetylglucosaminylation (O-glcNAcylation) J Biol Chem. 2003;278:14046–14052. doi: 10.1074/jbc.M300789200. [DOI] [PubMed] [Google Scholar]
- 21.Love DC, Krause MW, Hanover JA. O-GlcNAc cycling: Emerging roles in development and epigenetics. Semin Cell Dev Biol. 2010;21:646–654. doi: 10.1016/j.semcdb.2010.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wray J, Kalkan T, Smith AG. The ground state of pluripotency. Biochem Soc Trans. 2010;38:1027–1032. doi: 10.1042/BST0381027. [DOI] [PubMed] [Google Scholar]
- 23.O'Donnell N, Zachara NE, Hart GW, Marth JD. Ogt-dependent X-chromosome-linked protein glycosylation is a requisite modification in somatic cell function and embryo viability. Mol Cell Biol. 2004;24:1680–1690. doi: 10.1128/MCB.24.4.1680-1690.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Boyer LA, et al. Polycomb complexes repress developmental regulators in murine embryonic stem cells. Nature. 2006;441:349–353. doi: 10.1038/nature04733. [DOI] [PubMed] [Google Scholar]
- 25.Vosseller K, et al. O-linked N-acetylglucosamine proteomics of postsynaptic density preparations using lectin weak affinity chromatography and mass spectrometry. Mol Cell Proteomics. 2006;5:923–934. doi: 10.1074/mcp.T500040-MCP200. [DOI] [PubMed] [Google Scholar]
- 26.Syka JE, Coon JJ, Schroeder MJ, Shabanowitz J, Hunt DF. Peptide and protein sequence analysis by electron transfer dissociation mass spectrometry. Proc Natl Acad Sci USA. 2004;101:9528–9533. doi: 10.1073/pnas.0402700101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wysocka J, et al. A PHD finger of NURF couples histone H3 lysine 4 trimethylation with chromatin remodelling. Nature. 2006;442:86–90. doi: 10.1038/nature04815. [DOI] [PubMed] [Google Scholar]
- 28.Landry J, et al. Essential role of chromatin remodeling protein Bptf in early mouse embryos and embryonic stem cells. PLoS Genet. 2008;4:e1000241. doi: 10.1371/journal.pgen.1000241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fazzio TG, Huff JT, Panning B. An RNAi screen of chromatin proteins identifies Tip60-p400 as a regulator of embryonic stem cell identity. Cell. 2008;134:162–174. doi: 10.1016/j.cell.2008.05.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Khidekel N, Ficarro SB, Peters EC, Hsieh-Wilson LC. Exploring the O-GlcNAc proteome: Direct identification of O-GlcNAc-modified proteins from the brain. Proc Natl Acad Sci USA. 2004;101:13132–13137. doi: 10.1073/pnas.0403471101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mazars R, et al. The THAP-zinc finger protein THAP1 associates with coactivator HCF-1 and O-GlcNAc transferase: A link between DYT6 and DYT3 dystonias. J Biol Chem. 2010;285:13364–13371. doi: 10.1074/jbc.M109.072579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ong SE, et al. Stable isotope labeling by amino acids in cell culture, SILAC, as a simple and accurate approach to expression proteomics. Mol Cell Proteomics. 2002;1:376–386. doi: 10.1074/mcp.m200025-mcp200. [DOI] [PubMed] [Google Scholar]
- 33.Hughes-Davies L, et al. EMSY links the BRCA2 pathway to sporadic breast and ovarian cancer. Cell. 2003;115:523–535. doi: 10.1016/s0092-8674(03)00930-9. [DOI] [PubMed] [Google Scholar]
- 34.Wang Z, et al. Enrichment and site mapping of O-linked N-acetylglucosamine by a combination of chemical/enzymatic tagging, photochemical cleavage, and electron transfer dissociation mass spectrometry. Mol Cell Proteomics. 2010;9:153–160. doi: 10.1074/mcp.M900268-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Boyer LA, et al. Core transcriptional regulatory circuitry in human embryonic stem cells. Cell. 2005;122:947–956. doi: 10.1016/j.cell.2005.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rizzino A. Sox2 and Oct-3/4: A versatile pair of master regulators that orchestrate the self-renewal and pluripotency of embryonic stem cells. Wiley Interdiscip Rev Syst Biol Med. 2009;1:228–236. doi: 10.1002/wsbm.12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Silva J, et al. Nanog is the gateway to the pluripotent ground state. Cell. 2009;138:722–737. doi: 10.1016/j.cell.2009.07.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wang J, et al. A protein interaction network for pluripotency of embryonic stem cells. Nature. 2006;444:364–368. doi: 10.1038/nature05284. [DOI] [PubMed] [Google Scholar]
- 39.Wang ZX, et al. The transcription factor Zfp281 controls embryonic stem cell pluripotency by direct activation and repression of target genes. Stem Cells. 2008;26:2791–2799. doi: 10.1634/stemcells.2008-0443. [DOI] [PubMed] [Google Scholar]
- 40.Seth A, Alvarez E, Gupta S, Davis RJ. A phosphorylation site located in the NH2-terminal domain of c-Myc increases transactivation of gene expression. J Biol Chem. 1991;266:23521–23524. [PubMed] [Google Scholar]
- 41.Bode AM, Dong Z. Post-translational modification of p53 in tumorigenesis. Nat Rev Cancer. 2004;4:793–805. doi: 10.1038/nrc1455. [DOI] [PubMed] [Google Scholar]
- 42.Swaney DL, Wenger CD, Thomson JA, Coon JJ. Human embryonic stem cell phosphoproteome revealed by electron transfer dissociation tandem mass spectrometry. Proc Natl Acad Sci USA. 2009;106:995–1000. doi: 10.1073/pnas.0811964106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.McDonel P, Costello I, Hendrich B. Keeping things quiet: Roles of NuRD and Sin3 co-repressor complexes during mammalian development. Int J Biochem Cell Biol. 2009;41:108–116. doi: 10.1016/j.biocel.2008.07.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wysocka J, Myers MP, Laherty CD, Eisenman RN, Herr W. Human Sin3 deacetylase and trithorax-related Set1/Ash2 histone H3-K4 methyltransferase are tethered together selectively by the cell-proliferation factor HCF-1. Genes Dev. 2003;17:896–911. doi: 10.1101/gad.252103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yang X, Zhang F, Kudlow JE. Recruitment of O-GlcNAc transferase to promoters by corepressor mSin3A: Coupling protein O-GlcNAcylation to transcriptional repression. Cell. 2002;110:69–80. doi: 10.1016/s0092-8674(02)00810-3. [DOI] [PubMed] [Google Scholar]
- 46.Pasini D, Bracken AP, Hansen JB, Capillo M, Helin K. The polycomb group protein Suz12 is required for embryonic stem cell differentiation. Mol Cell Biol. 2007;27:3769–3779. doi: 10.1128/MCB.01432-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Schoeftner S, et al. Recruitment of PRC1 function at the initiation of X inactivation independent of PRC2 and silencing. EMBO J. 2006;25:3110–3122. doi: 10.1038/sj.emboj.7601187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Peng JC, et al. Jarid2/Jumonji coordinates control of PRC2 enzymatic activity and target gene occupancy in pluripotent cells. Cell. 2009;139:1290–1302. doi: 10.1016/j.cell.2009.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pasini D, et al. JARID2 regulates binding of the Polycomb repressive complex 2 to target genes in ES cells. Nature. 2010;464:306–310. doi: 10.1038/nature08788. [DOI] [PubMed] [Google Scholar]
- 50.Love DC, Hanover JA. The hexosamine signaling pathway: Deciphering the “O-GlcNAc code”. Sci STKE. 2005;2005:re13. doi: 10.1126/stke.3122005re13. [DOI] [PubMed] [Google Scholar]
- 51.Lee TI, et al. Control of developmental regulators by Polycomb in human embryonic stem cells. Cell. 2006;125:301–313. doi: 10.1016/j.cell.2006.02.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Fujimura Y, et al. Distinct roles of Polycomb group gene products in transcriptionally repressed and active domains of Hoxb8. Development. 2006;133:2371–2381. doi: 10.1242/dev.02405. [DOI] [PubMed] [Google Scholar]
- 53.Montgomery ND, et al. The murine polycomb group protein Eed is required for global histone H3 lysine-27 methylation. Curr Biol. 2005;15:942–947. doi: 10.1016/j.cub.2005.04.051. [DOI] [PubMed] [Google Scholar]
- 54.Kittler R, et al. Genome-wide resources of endoribonuclease-prepared short interfering RNAs for specific loss-of-function studies. Nat Methods. 2007;4:337–344. doi: 10.1038/nmeth1025. [DOI] [PubMed] [Google Scholar]
- 55.Fälth M, et al. Analytical utility of small neutral losses from reduced species in electron capture dissociation studied using SwedECD database. Anal Chem. 2008;80:8089–8094. doi: 10.1021/ac800944u. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.