Abstract
The long-term persistence of completely asexual species is unexpected. Although asexuality has short-term evolutionary advantages, a lack of genetic recombination leads to the accumulation over time of deleterious mutations. The loss of individual fitness as a result of accumulated deleterious mutations is expected to lead to reduced population fitness and possible lineage extinction. Persistent lineages of asexual, all-female clones (parthenogenetic and gynogenetic species) avoid the negative effects of asexual reproduction through the production of rare males, or otherwise exhibit some degree of genetic recombination. Another form of asexuality, known as androgenesis, results in offspring that are clones of the male parent. Several species of the Asian clam genus Corbicula reproduce via androgenesis. We compared gene trees of mitochondrial and nuclear loci from multiple sexual and androgenetic species across the global distribution of Corbicula to test the hypothesis of long-term clonality of the androgenetic species. Our results indicate that low levels of genetic capture of maternal nuclear DNA from other species occur within otherwise androgenetic lineages of Corbicula. The rare capture of genetic material from other species may allow androgenetic lineages of Corbicula to mitigate the effects of deleterious mutation accumulation and increase potentially adaptive variation. Models comparing the relative advantages and disadvantages of sexual and asexual reproduction should consider the possibility of rare genetic recombination, because such events seem to be nearly ubiquitous among otherwise asexual species.
Keywords: Muller's ratchet, Meselson effect, Hill-Robertson effects, phylogenetics
Although asexual reproduction is widely distributed across eukaryotes, obligate asexuality is comparatively rare, and obligately asexual species tend to be of recent origin (1–4). Because of this observed rarity, there are presumably costs to obligate asexuality that allow sexual lineages to outcompete their asexual relatives, even though sexual reproduction itself carries costs [particularly the “two-fold cost of sex”: the cost of producing males or the cost of meiosis (2, 3)]. The literature on genetic (or mutational) costs to asexuality is extensive, but the main ideas can be broadly summarized as (i) asexuals can only rely on mutation to generate adaptive variation (e.g., refs. 1 and 5); and (ii) deleterious mutations accumulate faster in asexuals than sexuals (e.g., refs. 6–9). When more than one asexual lineage is observed in a closely related group of organisms, some process must either allow for diversification despite the costs, or else repeatedly generate new asexual lineages. Understanding mechanisms that drive asexual diversity can reveal much about the costs and benefits of asexual reproduction.
Androgenesis is a form of asexual reproduction in which offspring are clones of the father (10, 11). Several species of Asian clams in the genus Corbicula produce paternal clones through androgenesis (12–14). Androgenetic Corbicula are hermaphrodites and fertilize both their own eggs and eggs from closely related congeners to make paternal clones that do not incorporate maternal nuclear chromosomes (15, 16). Could the interaction between gametes of closely related Corbicula species allow rare sex between lineages, changing our expectations for the persistence of androgenetic species, and thus the time available to generate asexual diversity? Or is the observed diversity a result of recent, replicate origins [as in ostracods, stick insects, and water fleas (17–19)]?
Many asexual lineages have mechanisms for rare genetic capture (20–24). In parthenogenetic lineages, a male may be produced after several generations of female clonal reproduction, and sex could occur between the parthenogens and the rare male (25). In several pathogenic fungi that seem to be asexual, DNA sequencing revealed the presence of functional genes for mating and meiosis, suggesting that cryptic or unobserved sex could be occurring (26). In one type of asexual reproduction, gynogenesis, females require sperm from males of closely related sexual species to activate embryogenesis, but males do not regularly contribute genes to offspring. For example, even though behaviors associated with sexual reproduction are inherited from a sexual ancestor, Amazon mollies (Poecilia formosa) are considered asexual because daughters are usually genetically identical to their mothers. However, gametic interactions between sexuals and asexuals lead to rare genetic capture: microchromosomes are passed on from host males, and these microchromosomes contain genes with effects on phenotype (27, 28). Some of these microchromosomes are inherited by subsequent generations of female clones (27, 29). Rarely, entire chromosomes of haploid sperm are integrated with the diploid maternal genome, causing triploid, gynogenetic offspring (30), although triploidy is unstable and individuals can end up with both diploid and triploid somatic cells (31). Thus, gynogenetic lineages that gain genetic material from males may persist for longer periods of time than otherwise expected (32).
The ancient asexual bdelloid rotifers are often considered an exception to the rule of persistent asexuality. No males have been observed in either contemporary or fossil populations going back at least 35 million years (33), and genomes of many bdelloids contain highly divergent alleles (34)—some to the point where two alleles may have different functions (35). However, during cycles of desiccation, the genome is fragmented. When the rotifer is rehydrated, DNA from the environment may be incorporated into the genome during chromosome repair, resulting in genetic recombination (36). Between morphologically and genetically distinct species, some nearly identical alleles are shared, even though these species seem to have been reproducing asexually for many millions of years (37).
In androgenetic Corbicula, genetic recombination could occur when gametes interact during reproduction. After fertilization, maternal nuclear chromosomes are extruded from the egg cell; only paternal nuclear chromosomes from the sperm remain in the zygote (12–14). Mitochondria, however, are maternally inherited, so offspring possess the maternal mitochondrial genome and the paternal nuclear genome. Phylogenies built from mitochondrial vs. nuclear markers can be compared to identify between-species genetic recombination (10). Well-supported phylogenetic incongruence between mitochondrial and nuclear gene trees would be a signal of egg-capture (and thus mitochondrial genome-capture) in the genus (10, 15, 16). Nuclear gene-capture by one (paternal) species would be indicated if nuclear alleles found within a single individual were highly divergent and also had different evolutionary histories (as seen after sexual hybridization between species). We used these assumptions to compare a mitochondrial phylogeny of Corbicula (from ref. 16) to two phylogenies built using single-copy nuclear introns. We find evidence that multiple androgenetic lineages have captured mitochondrial and nuclear DNA from other species and suggest that rare sex could generate genetic diversity and allow androgenetic lineages to persist for longer than expected.
Results
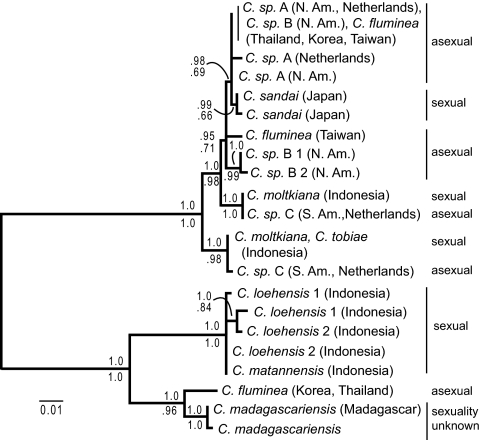
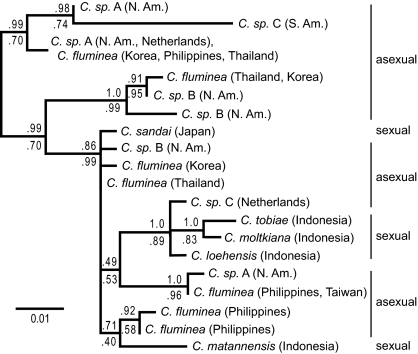
We estimated phylogenies for two single-locus nuclear markers: the third intron of the α-amylase gene (amy; Fig. 1) and a putative intron of the α subunit of adenosine triphosphate synthase (atps-α; Fig. 2). When sexual individuals have more than one allele at a locus, those alleles are closely related to each other. Some asexual individuals, however, contain very divergent alleles at each locus. Hypothesis testing indicates that, in both nuclear trees, alleles within a single androgenetic individual are not always monophyletic, even when other androgenetic alleles are permitted to be nested within that individual's clade. Neither are asexual lineages as a whole monophyletic (Figs. 1 and 2 and Table S1). However, in both gene trees, there is one allele that appears in most of the asexual populations.
Fig. 1.
Maximum likelihood gene tree estimate of the third intron of the nuclear α-amylase gene. Numbers above the branch are Bayesian posterior probabilities, and numbers below are likelihood bootstrap proportions. Species are labeled as putatively sexual or asexual on the basis of morphology and/or lack of population-level genetic variation (Table S2). Two individuals were sequenced for C. sp. B and C. loehensis; these are distinguished by number following the species name.
Fig. 2.
Maximum likelihood gene tree estimate of a putative nuclear intron of the α subunit of adenosine triphosphate synthase. Numbers above the branch are Bayesian posterior probabilities, and numbers below are likelihood bootstrap proportions. Species are labeled as sexual or asexual on the basis of morphology and lack of population-level genetic variation (Table S2).
Three phylogenies, amy, atps-α, and the mitochondrial cytochrome oxidase I (cox-I) data from Hedtke et al. (16), were tested to find whether observed differences in topology were a result of statistically significant differences in phylogenetic signal among the data sets (Table S1). For the six taxa sequenced in all three data sets, we tested support for each bipartition found in any of the three trees by comparing the posterior probabilities between data sets. To test whether there was a single taxon responsible for gene tree incongruence, we deleted each taxon and determined posterior probabilities for each reduced backbone topology. We found no single taxon responsible for gene tree incongruence among the data sets. The clades with posterior probabilities >0.5 in any data set are found in Table S1. These results indicate well-supported incongruence in relationships of taxa among gene trees.
Discussion
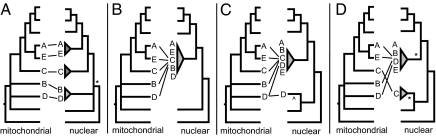
Androgenetic clams in the genus Corbicula are morphologically and genetically distinct (e.g., 11, 13, 37–42). Several hypotheses could explain this observed androgenetic species diversity, given that androgenetic species do not share a most recent common ancestor on a mitochondrial gene tree (15, 16, 41) (Fig. 3). First, androgenesis might have multiple origins as a result of repeated loss-of-function mutations or hybridization events between sexual species. Second, androgenesis might have a single origin; divergent mitochondrial lineages within asexuals might be the result of egg capture by an androgenetic sperm with complete maternal nuclear chromosome extrusion. Third, androgenesis might have a single origin, and egg capture by androgenetic species might have been rarely accompanied not only by capture of maternal mitochondrial, but also nuclear, DNA. Last, combinations of these processes may have acted in this system to form morphologically and genetically distinct androgenetic species.
Fig. 3.
Expected relationships between mitochondrial and nuclear markers in androgenetic Corbicula, given four possible scenarios. Central lines between trees indicate where taxa in the mitochondrial tree are found on the nuclear tree. Androgenetic Corbicula are in red and referred to by letter for ease of comparison. A: C. sp. A (North America, The Netherlands); B: C. sp. B (North America); C: C. sp. C (South America, The Netherlands); D: C. fluminea (Korea, Thailand); E: C. fluminea (Taiwan); F: C. fluminea (Philippines). (A) Multiple origins or single origin at * and reversion; (B) single origin with mitochondrial capture; (C) single origin with mitochondrial and nuclear capture at ^; (D) two origins at * with mitochondrial capture.
Potential Sources of Phylogenetic Incongruence.
Each of the three individual gene trees has statistically supported incongruent relationships between species (Figs. 1 and 2 and Table S1). Gene tree incongruence can be caused by systematic error in the analysis. For example, one gene may be prone to long-branch attraction, whereas another is not because of differences in the rate of evolution (43). Of the three gene trees considered, the only obvious topology for which long-branch attraction might have been a problem was cox-I (16). When the outgroups to Corbicula (Neocorbicula and Polymesoda) and to freshwater Corbicula (Corbicula cf japonica) were removed from analysis, relationships within freshwater Corbicula did not change significantly. This suggests that incongruence between mitochondrial and nuclear gene trees was not due to long-branch attraction between any freshwater taxon and the long-branched outgroups.
There are biological processes that can lead to topological incongruence between gene trees. Given the short branch lengths inferred between taxa, freshwater Corbicula seem to have recently radiated, with rapid morphological evolution (41), making incomplete lineage-sorting between sexual taxa a plausible source of incongruence. The analogous process in asexuals would be retention of ancestral polymorphism (via heterozygosity of the newly asexual clam) and would result in divergent alleles within individuals. However, because some identical alleles are held in common across different species of asexual clam, for ancestral polymorphism to adequately explain the observed topologies, androgenesis would need to have arisen multiple times within an ancestral population, and there would need to be a mechanism that would slow evolution of the shared allele but not of other alleles at that locus. Gene duplication and subsequent loss also cannot be rejected as possible processes that could confuse topological inference, although to explain the observed topologies, duplication would have occurred early in the history of the genus, and different copies independently lost in almost all species.
Topological incongruence between gene trees can be caused by hybridization and capture of mitochondria. Given known mechanisms of androgenesis in Corbicula (12–14), these processes are highly likely to have caused discordance between mitochondrial and nuclear gene trees. These gene tree incongruences, rather than obscuring origins of androgenetic species diversity, can instead reveal processes important to the evolution of androgenetic Corbicula.
Origins of Androgenesis.
There are multiple morphologically distinct species of Corbicula identified as androgenetic through cytological examination of fertilization (12–14) or the presence of genetic invariance, polyploidy, and biflagellate sperm (15, 38–42). Our phylogenetic trees do not support a simple scenario of repeated, multiple origins of androgenesis, which would require congruence between nuclear and mitochondrial gene trees (Fig. 3A). Nuclear alleles across androgenetic taxa are not monophyletic, which counters expectations under a single origin and subsequent diversification of androgenetic taxa (Fig. 3B). Instead, the observed phylogenetic pattern is what we expect under a relatively recent origin of androgenesis, followed by postorigin nuclear hybridization as asexual taxa spread and came into contact with different sexual species (Fig. 3C).
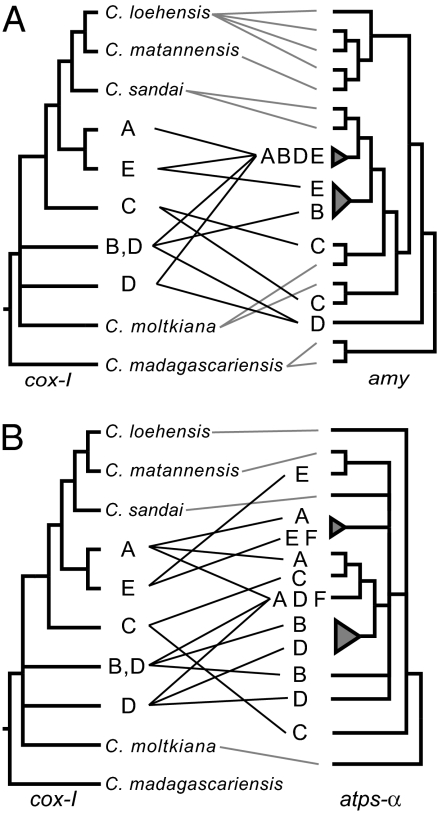
Most androgenetic individuals share a single allele in common (e.g., “ABDE” in Fig. 4A) or have alleles found in a clade of closely related sequences (Fig. 4B). This suggests that at least a portion of the genome of all androgenetic species shares a common history that excludes sexual taxa. Because these alleles are identical or very similar, the shared history seems to be relatively recent; alleles have not had time to diverge since their common ancestor. This suggests a relatively recent origin of androgenesis in the genus, likely from an ancestor of Corbicula sandai, the sexual species found most closely related to androgenetic taxa across phylogenies.
Fig. 4.
Observed relationships between mitochondrial and nuclear markers in androgenetic Corbicula. Central lines between trees indicate where taxa in the mitochondrial tree are found on the nuclear tree. Androgenetic Corbicula are in red and referred to by letter for ease of comparison. A: C. sp. A (North America, The Netherlands); B: C. sp. B (North America); C: C. sp. C (South America, The Netherlands); D: C. fluminea (Korea, Thailand); E: C. fluminea (Taiwan); F: C. fluminea (Philippines). (A) Comparison between the mitochondrial cox-I tree from Hedtke et al. (15) and the nuclear amy tree. (B) Comparison between the mitochondrial cox-I tree and the nuclear atps-α tree.
Some individual asexuals contain a highly divergent allele (e.g., “D” in Fig. 4A or “E” in Fig. 4B). The “Meselson effect,” in which alleles within an asexual lineage diverge as they are retained and accumulate mutations over time (4, 44), is not a convincing explanation for the within-individual diversity observed. In such a scenario, all alleles of a particular gene would be expected to accumulate differences. However, in asexual Corbicula, one common (or similar) allele is shared among most species, whereas a second, highly divergent allele may be unique to a particular species. This pattern suggests that the divergent alleles within each species are not derived directly from the common allele by substitutions within species, but rather are the result of genetic capture of nuclear DNA from other, more distantly related species. Because the androgenetic species that share an allele have only one allele in common, there is no genetic signature that suggests androgenesis arose through a single hybridization event between two sexual species.
Multiple hybrid origins of androgenesis could potentially explain the observed distribution of alleles in the gene trees among androgenetic species, if a single sexual species hybridized with a number of other sexual species, and genetic interactions between incompatible genomes led to independent origins of asexuality. For this scenario to be true, the common sexual species in these multiple hybridization events would have been widely distributed and cooccurred with several other sexual species. No sexual species is known to be currently distributed throughout the ranges of the various androgenetic species. Because androgenetic Corbicula, which do have a widespread geographic distribution, seem to be able to parasitize eggs of other species (15, 16), multiple hybrid events involving a single sexual species seems less likely than genetic capture by an androgenetic lineage after the evolution of androgenesis.
Although most species of androgenetic Corbicula seem to have a common origin (on the basis of the common allele shared among most species), there is evidence of a possible second origin of androgenesis (Fig. 3D). We did not sequence the common amy allele found in most androgenetic lineages (“ABDE” of Fig. 4A) in Corbicula sp. C. This species groups with Corbicula moltkiana across phylogenies and could have a separate independent origin from a C. moltkiana-like ancestor. However, this conclusion is highly dependent on whether all alleles were successfully amplified and sequenced in this species. In the phylogeny based on atps-α, an allele from C. sp. C is found in a common clade of androgenetic individuals (Fig. 4B). If the shared amy allele was simply not sampled in C. sp. C, then the divergent sequence found in that gene would be another instance of genetic capture, rather than evidence for a separate origin.
Mechanistically, genetic capture by androgenetic clams could have happened as a polyploidization event. Meiosis of the maternal genome is completed after sperm fertilization of the egg in Corbicula (12, 14). In sexual species, the meiotic axis rotates to become perpendicular to the cell cortex, and the product of the meiotic division closest to the cell cortex is extruded as a polar body, leaving the haploid maternal genome to join with the paternal genome to form a zygote. In eggs fertilized by androgenetic Corbicula, this rotation of the meiotic axis does not occur, and so the entire maternal genome typically is extruded as two polar bodies during meiosis (12, 14), leaving only the paternal nuclear genome to form the zygote. However, within-species polyploidization has been observed in the laboratory in some androgenetic lineages, presumably because normal spindle fiber orientation allowed half of the maternal genome to be added to the unreduced paternal genome from the sperm (45). Corbicula seems tolerant of polyploidy [diploid, triploid, and tetraploid Corbicula have been found (46–49)]. Alternatively, only a portion of the maternal genome could be retained through recombination between paternal and maternal chromosomes, before the formation of polar bodies. The incorporation of maternal DNA from different species seems to be relatively rare, however, because androgenetic species of Corbicula in sympatry remain genetically and morphologically distinct (16).
Evolutionary Consequences of Androgenesis.
When obligate androgenesis arises, under most conditions androgenetic individuals are expected to have a reproductive advantage over sexual members of the same population, and obligate androgenesis will spread to fixation (50). In addition to the direct reproductive benefits of egg capture, androgenesis could benefit from infrequent chromosomal rescue. If harmful mutations are recessive, they could be masked by nuclear gene capture or polyploidization. Alternatively, recombination with the maternal genome, gene conversion, or postcapture gene loss could replace alleles in the paternal genome. This would allow usually clonal androgenetic species to slow the rate of accumulation of deleterious mutations and potentially introduce beneficial allelic variants.
Persistent asexual lineages are expected to become extinct over evolutionary time, as they accumulate deleterious mutations, cannot free beneficial alleles from a poor genetic background, and must rely on mutation (not recombination) to introduce new beneficial alleles. Therefore, the persistence of asexual lineages is made more probable through rare genetic capture from divergent lineages. Even a limited amount of recombination could ameliorate the negative effects associated with asexual reproduction (e.g., refs. 24 and 51). Thus, any gene capture that occurs during androgenetic reproduction in Corbicula reduces the extinction risk of these otherwise asexual species.
Primarily asexual lineages have increasingly been shown to have mechanisms for rare acquisition of genetic material from other lineages (e.g., refs. 20–27 and 36). Capture of complete or partial maternal genomes in androgenetic Corbicula seems to be an additional mechanism of rare genetic recombination in otherwise asexual lineages. Therefore, modeling asexual reproduction as the complete lack of genetic recombination no longer seems appropriate in considering the evolution and persistence of asexual lineages. To understand the distribution and diversity of sexual vs. asexual species, we should turn to models that explore not whether sex is advantageous under a given set of conditions, but rather, how much sex is sufficient to counteract the deleterious effects of mutation accumulation (e.g., refs. 3 and 52–54).
Materials and Methods
Sequencing.
DNA was extracted from frozen tissue specimens of Corbicula from North and South America and the Netherlands using the Viogene DNA Blood and Tissue Genomic extraction kits (Viogene Biotek). DNA was extracted from ethanol-preserved samples of Corbicula australis, Corbicula cf elongata, Corbicula fluminea, Corbicula fluminalis, Corbicula loehensis, Corbicula matannensis, C. moltkiana, Corbicula tobiae, Corbicula leana, and Corbicula madagascariensis using a standard phenol-chloroform protocol modified by suggestions in ref. 55. A total of 19 specimens were used (Table S2). Primers for PCR amplification were developed using an α-amylase gene sequence from Corbicula (GenBank accession no. AF468016). Primers for atps-α were designed from sequence obtained using universal primers designed to amplify specific introns of the atps-α gene (56), but we could not confirm the identify of sequences amplified by these primers, because there are no existing sequences for these introns on GenBank for any Corbicula or their close relatives. SI Materials and Methods provides details on primer development, sequences, and amplification conditions.
Binning of Alleles.
Cloning and sequencing introduce noise into a data set because Taq polymerase makes errors in replication (57, 58), and these errors can be sampled by cloning. Multiple sequences from one individual may not represent genetic alleles but rather PCR error. To reduce noise arising from PCR error, clones were binned such that a separate allele was called if there were more than three base pair differences between that sequence and another group of sequences. The consensus sequence for each bin was the most common base pair sampled across sequences in the bin, or coded using International Union of Pure and Applied Chemistry ambiguity rules if two base pairs were equally represented between sequences at a given site. The number of alleles inferred for each marker was consistent with the ploidy expected (triploid for most androgenetic clams, diploid for sexual species; Table S2).
Alignment.
Alignments of sequence data were constructed using MacClade (59). Each alignment was trimmed at each end to exclude regions of missing data. For each aligned data set, we ran analyses in which all indels were included or excluded (amy: 560 bp with indels included, 379 bp with indels excluded; atps-α: 347 bp with indels included, 263 bp with indels excluded). Excluding indels did not make a significant difference in phylogenetic inference, and we present results for analyses in which indels were excluded. We did not find evidence for recombinant sequences (detailed in SI Materials and Methods).
Phylogenetic Analyses.
We performed phylogenetic analyses on each data set using maximum likelihood as implemented in GARLI v0.96 (60) (SI Materials and Methods) and Bayesian phylogenetic analysis using MrBayes v.3.1.2 (61) (SI Materials and Methods). The outgroup to freshwater Corbicula, Corbicula cf japonica, did not amplify or was too divergent from the freshwater ingroup for successful gene alignment. Topologies are therefore unrooted for these markers (although midpoint rooting is used for the convenience of visualization). Details of hypothesis testing are presented in the SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank R. Linder and the members of the D.M.H., Bull, and Cannatella laboratory groups at the University of Texas at Austin for advice on project development and constructive criticism of the manuscript; S. Otto, B. Normark, and D. Mark Welch for providing additional helpful comments; D. ÓFoighil, C. Ituarte, and A. de Vaate for contributing tissue samples used in this study; R. Heineman, N. Wilson, and B. Ramesh for assisting with laboratory work; and Thomas von Rintelen for help during field work, which was supported by Deutsche Forschungsgemeinschaft Grant GL 297/7 (to M.G.). This work was funded by Doctoral Dissertation Improvement Grant DEB-0513426 from the National Science Foundation (to S.M.H.). S.M.H. was also supported by a National Science Foundation Graduate Fellowship.
Footnotes
The authors declare no conflict of interest.
Data deposition: The sequences reported in this paper have been deposited in the GenBank database (accession nos. JF899599–JF899769).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1106742108/-/DCSupplemental.
References
- 1.Muller HJ. Some genetic aspects of sex. Am Nat. 1932;66:118–138. [Google Scholar]
- 2.Maynard Smith J. The Evolution of Sex. Cambridge, UK: Cambridge Univ Press; 1978. [Google Scholar]
- 3.Maynard Smith J. Evolutionary Genetics. Oxford, UK: Oxford Univ Press; 1989. [Google Scholar]
- 4.Judson OP, Normark BB. Ancient asexual scandals. Trends Ecol Evol. 1996;11:41–46. doi: 10.1016/0169-5347(96)81040-8. [DOI] [PubMed] [Google Scholar]
- 5.Fisher RA. The Genetical Theory of Natural Selection. Oxford: Clarendon Press; 1930. [Google Scholar]
- 6.Muller HJ. The relation of recombination to mutational advance. Mutat Res. 1964;106:2–9. doi: 10.1016/0027-5107(64)90047-8. [DOI] [PubMed] [Google Scholar]
- 7.Felsenstein J. The evolutionary advantage of recombination. Genetics. 1974;78:737–756. doi: 10.1093/genetics/78.2.737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Charlesworth B, Charlesworth D. Rapid fixation of deleterious alleles can be caused by Muller's ratchet. Genet Res. 1997;70:63–73. doi: 10.1017/s0016672397002899. [DOI] [PubMed] [Google Scholar]
- 9.Keightley PD, Otto SP. Interference among deleterious mutations favours sex and recombination in finite populations. Nature. 2006;443:89–92. doi: 10.1038/nature05049. [DOI] [PubMed] [Google Scholar]
- 10.Hedtke SM, Hillis DM. The potential role of androgenesis in cytoplasmic-nuclear phylogenetic discordance. Syst Biol. 2011;60:87–96. doi: 10.1093/sysbio/syq070. [DOI] [PubMed] [Google Scholar]
- 11.Normark BB. Unusual gametic and genetic systems. In: Hosken DJ, Birkhead T, Pitnick S, editors. Sperm Biology: An Evolutionary Perspective. Amsterdam: Academic Press; 2009. pp. 507–538. [Google Scholar]
- 12.Komaru A, Kawagishi T, Konishi K. Cytological evidence of spontaneous androgenesis in the freshwater clam Corbicula leana Prime. Dev Genes Evol. 1998;208:46–50. doi: 10.1007/s004270050152. [DOI] [PubMed] [Google Scholar]
- 13.Komaru A, Ookubo K, Kiyomoto M. All meiotic chromosomes and both centrosomes at spindle pole in the zygotes discarded as two polar bodies in clam Corbicula leana: Unusual polar body formation observed by antitubulin immunofluorescence. Dev Genes Evol. 2000;210:263–269. doi: 10.1007/s004270050313. [DOI] [PubMed] [Google Scholar]
- 14.Ishibashi R, et al. Androgenetic reproduction in a freshwater diploid clam Corbicula fluminea (Bivalvia: Corbiculidae) Zoolog Sci. 2003;20:727–732. doi: 10.2108/zsj.20.727. [DOI] [PubMed] [Google Scholar]
- 15.Lee T, Siripattrawan S, Ituarte CF, ÓFoighil D. Invasion of the clonal clams: Corbicula lineages in the New World. Am Malacol Bull. 2005;20:113–122. [Google Scholar]
- 16.Hedtke SM, Stanger-Hall K, Baker RJ, Hillis DM. All-male asexuality: Origin and maintenance of androgenesis in the Asian clam Corbicula. Evolution. 2008;62:1119–1136. doi: 10.1111/j.1558-5646.2008.00344.x. [DOI] [PubMed] [Google Scholar]
- 17.Bode SNS, et al. Exceptional cryptic diversity and multiple origins of parthenogenesis in a freshwater ostracod. Mol Phylogenet Evol. 2010;54:542–552. doi: 10.1016/j.ympev.2009.08.022. [DOI] [PubMed] [Google Scholar]
- 18.Morgan-Richards M, Trewick SA, Stringer IAN. Geographic parthenogenesis and the common tea-tree stick insect of New Zealand. Mol Ecol. 2010;19:1227–1238. doi: 10.1111/j.1365-294X.2010.04542.x. [DOI] [PubMed] [Google Scholar]
- 19.Paland S, Colbourne JK, Lynch M. Evolutionary history of contagious asexuality in Daphnia pulex. Evolution. 2005;59:800–813. [PubMed] [Google Scholar]
- 20.Hurst LD, Peck JR. Recent advances in understanding of the evolution and maintenance of sex. Trends Ecol Evol. 1996;11:46–52. doi: 10.1016/0169-5347(96)81041-x. [DOI] [PubMed] [Google Scholar]
- 21.Little TJ, Hebert PD. Ancient asexuals: Scandal or artifact? Trends Ecol Evol. 1996;11:296. doi: 10.1016/s0169-5347(96)91640-7. [DOI] [PubMed] [Google Scholar]
- 22.Butlin R, Schön I, Martens K. Asexual reproduction in nonmarine ostracods. Heredity. 1998;81:473–480. [Google Scholar]
- 23.Normark BB. Evolution in a putatively ancient asexual aphid lineage: Recombination and rapid karyotypic change. Evolution. 1999;53:1458–1469. doi: 10.1111/j.1558-5646.1999.tb05410.x. [DOI] [PubMed] [Google Scholar]
- 24.D'Souza TG, Michiels NK. The costs and benefits of occasional sex: Theoretical predictions and a case study. J Hered. 2010;101(Suppl 1):S34–S41. doi: 10.1093/jhered/esq005. [DOI] [PubMed] [Google Scholar]
- 25.Schurko AM, Neiman M, Logsdon JM., Jr Signs of sex: What we know and how we know it. Trends Ecol Evol. 2009;24:208–217. doi: 10.1016/j.tree.2008.11.010. [DOI] [PubMed] [Google Scholar]
- 26.Butler G. Fungal sex and pathogenesis. Clin Microbiol Rev. 2010;23:140–159. doi: 10.1128/CMR.00053-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schartl M, et al. Incorporation of subgenomic amounts of DNA as compensation for mutational load in a gynogenetic fish. Nature. 1995;373:68–71. [Google Scholar]
- 28.Lamatsch DK, et al. Distribution and stability of supernumerary microchromosomes in natural populations of the Amazon molly, Poecilia formosa. Cytogenet Genome Res. 2004;106:189–194. doi: 10.1159/000079286. [DOI] [PubMed] [Google Scholar]
- 29.Nanda I, et al. Stable inheritance of host species-derived microchromosomes in the gynogenetic fish Poecilia formosa. Genetics. 2007;177:917–926. doi: 10.1534/genetics.107.076893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schlupp I, et al. Dispensable and indispensable genes in an ameiotic fish, the Amazon molly Poecilia formosa. Cytogenet Cell Genet. 1998;80:193–198. doi: 10.1159/000014979. [DOI] [PubMed] [Google Scholar]
- 31.Lamatch DK, Schmid M, Schartl M. A somatic mosaic of the gynogenetic Amazon molly. J Fish Biol. 2002;60:1417–1422. [Google Scholar]
- 32.Loewe L, Lamatsch DK. Quantifying the threat of extinction from Muller's ratchet in the diploid Amazon molly (Poecilia formosa) BMC Evol Biol. 2008;8:88. doi: 10.1186/1471-2148-8-88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Waggoner BM, Poinar GO., Jr Fossil habrotochid rotifers in Dominican amber. Experientia. 1993;49:354–357. [Google Scholar]
- 34.Mark Welch D, Meselson M. Evidence for the evolution of bdelloid rotifers without sexual reproduction or genetic exchange. Science. 2000;288:1211–1215. doi: 10.1126/science.288.5469.1211. [DOI] [PubMed] [Google Scholar]
- 35.Pouchkina-Stantcheva NN, et al. Functional divergence of former alleles in an ancient asexual invertebrate. Science. 2007;318:268–271. doi: 10.1126/science.1144363. [DOI] [PubMed] [Google Scholar]
- 36.Gladyshev EA, Meselson M, Arkhipova IR. Massive horizontal gene transfer in bdelloid rotifers. Science. 2008;320:1210–1213. doi: 10.1126/science.1156407. [DOI] [PubMed] [Google Scholar]
- 37.Mark Welch DB, Cummings MP, Hillis DM, Meselson M. Divergent gene copies in the asexual class Bdelloidea (Rotifera) separated before the bdelloid radiation or within bdelloid families. Proc Natl Acad Sci USA. 2004;101:1622–1625. doi: 10.1073/pnas.2136686100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hillis DM, Patton JC. Morphological and electrophoretic evidence for two species of Corbicula (Bivalvia: Corbiculidae) in North America. Am Midl Nat. 1982;108:74–80. [Google Scholar]
- 39.McLeod MJ. Electrophoretic variation in North American Corbicula. Am Malacol Bull. 1986;Spec Ed 2:125–132. [Google Scholar]
- 40.Byrne M, et al. Reproduction and development of the freshwater clam Corbicula australis in southeast Australia. Hydrobiologia. 2000;418:185–197. [Google Scholar]
- 41.Glaubrecht M, Rintelen TV, Korniushin AV. Toward a systematic revision of brooding freshwater Corbiculidae in southeast Asia (Bivalvia, Veneroida): On shell morphology, anatomy and molecular phylogenetics of endemic taxa from islands in Indonesia. Malacologia. 2003;45:1–40. [Google Scholar]
- 42.Korniushin AV. A revision of some Asian and African freshwater clams assigned to Corbicula fluminalis (Müller, 1774) (Mollusca: Bivalvia: Corbiculidae), with a review of anatomical characters and reproductive features based on museum collections. Hydrobiologia. 2004;529:251–270. [Google Scholar]
- 43.Hedtke SM, Townsend TM, Hillis DM. Resolution of phylogenetic conflict in large data sets by increased taxon sampling. Syst Biol. 2006;55:522–529. doi: 10.1080/10635150600697358. [DOI] [PubMed] [Google Scholar]
- 44.Birky CW., Jr Heterozygosity, heteromorphy, and phylogenetic trees in asexual eukaryotes. Genetics. 1996;144:427–437. doi: 10.1093/genetics/144.1.427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Komaru A, et al. A hypothesis of ploidy elevation by formation of a female pronucleus in the androgenetic clam Corbicula fluminea in the Tone River estuary, Japan. Zoolog Sci. 2006;23:529–532. doi: 10.2108/zsj.23.529. [DOI] [PubMed] [Google Scholar]
- 46.Okamoto A, Arimoto B. Chromosomes of Corbicula japonica, C. sandai and C. (Corbiculina) leana (Bivalvia: Corbiculidae) Venus Jpn J Malacol. 1986;45:194–202. [Google Scholar]
- 47.Komaru A, et al. Hermaphroditic freshwater clams in the genus Corbicula produce non-reductional spermatozoa with somatic DNA content. Biol Bull. 1997;193:320–323. doi: 10.2307/1542934. [DOI] [PubMed] [Google Scholar]
- 48.Komaru A, Konishi K. Non-reductional spermatozoa in three shell color types of the freshwater clam Corbicula fluminea in Taiwan. Zoolog Sci. 1999;16:105–108. [Google Scholar]
- 49.Qiu A, Shi A, Komaru A. Yellow and brown shell color morphs of Corbicula fluminea (Bivalvia: Corbiculidae) from Sichuan Province, China, are triploids and tetraploids. J Shellfish Res. 2001;20:323–328. [Google Scholar]
- 50.McKone MJ, Halpern SL. The evolution of androgenesis. Am Nat. 2003;161:641–656. doi: 10.1086/368291. [DOI] [PubMed] [Google Scholar]
- 51.Pamilo P, Nei M, Li W-H. Accumulation of mutations in sexual and asexual populations. Genet Res. 1987;49:135–146. doi: 10.1017/s0016672300026938. [DOI] [PubMed] [Google Scholar]
- 52.Otto SP, Barton NH. The evolution of recombination: Removing the limits to natural selection. Genetics. 1997;147:879–906. doi: 10.1093/genetics/147.2.879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Barton NH, Charlesworth B. Why sex and recombination? Science. 1998;281:1986–1990. [PubMed] [Google Scholar]
- 54.Barton NH, Otto SP. Evolution of recombination due to random drift. Genetics. 2005;169:2353–2370. doi: 10.1534/genetics.104.032821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Watt EM, Watt VM. DNA fingerprints from minimal blood volumes. Mol Ecol. 1992;1:131–132. doi: 10.1111/j.1365-294x.1992.tb00165.x. [DOI] [PubMed] [Google Scholar]
- 56.Jarman SN, Ward RD, Elliott NG. Oligonucleotide primers for PCR amplification of coelomate introns. Mar Biotechnol (NY) 2002;4:347–355. doi: 10.1007/s10126-002-0029-6. [DOI] [PubMed] [Google Scholar]
- 57.Tindall KR, Kunkel TA. Fidelity of DNA synthesis by the Thermus aquaticus DNA polymerase. Biochemistry. 1988;27:6008–6013. doi: 10.1021/bi00416a027. [DOI] [PubMed] [Google Scholar]
- 58.Kobayashi N, Tamura K, Aotsuka T. PCR error and molecular population genetics. Biochem Genet. 1999;37:317–321. doi: 10.1023/a:1018759210666. [DOI] [PubMed] [Google Scholar]
- 59.Maddison DR, Maddison WP. MacClade 4. Sunderland, MA: Sinauer Associates; 2000. [Google Scholar]
- 60.Zwickl DJ. TX: University of Texas at Austin; 2006. Genetic algorithm approaches for the phylogenetic analysis of large biological sequence data sets under the maximum likelihood criterion. Ph.D. dissertation. [Google Scholar]
- 61.Ronquist F, Huelsenbeck JP. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics. 2003;19:1572–1574. doi: 10.1093/bioinformatics/btg180. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.