Abstract
Interaction forces of membrane protein subunits are of importance in their structure, assembly, membrane insertion, and function. In biological membranes, and in the photosynthetic apparatus as a paradigm, membrane proteins fulfill their function by ensemble actions integrating a tight assembly of several proteins. In the bacterial photosynthetic apparatus light-harvesting complexes 2 (LH2) transfer light energy to neighboring tightly associated core complexes, constituted of light-harvesting complexes 1 (LH1) and reaction centers (RC). While the architecture of the photosynthetic unit has been described, the forces and energies assuring the structural and functional integrity of LH2, the assembly of LH2 complexes, and how LH2 interact with the other proteins in the supramolecular architecture are still unknown. Here we investigate the molecular forces of the bacterial LH2 within the native photosynthetic membrane using atomic force microscopy single-molecule imaging and force measurement in combination. The binding between LH2 subunits is fairly weak, of the order of kBT, indicating the importance of LH2 ring architecture. In contrast LH2 subunits are solid with a free energy difference of 90 kBT between folded and unfolded states. Subunit α-helices unfold either in one-step, α- and β-polypeptides unfold together, or sequentially. The unfolding force of transmembrane helices is approximately 150 pN. In the two-step unfolding process, the β-polypeptide is stabilized by the molecular environment in the membrane. Hence, intermolecular forces influence the structural and functional integrity of LH2.
Keywords: atomic force microscopy, photosynthesis, protein unfolding, membrane protein assembly
Membrane protein function is intimately linked and influenced by its structural integrity and molecular environment (1–3). Photosynthesis is a paradigm of such orchestrated molecular action, for which the implicated proteins have evolved to form supramolecular assemblies (4). Knowledge of supramolecular architecture and interaction forces and energies are indispensable for a complete understanding of a membrane protein structure and function.
The atomic force microscope (AFM) (5) can be used as a high-resolution imaging (6) and as a force-measurement tool (7). Most elegantly the two applications were combined allowing the attribution of structural changes to force events (8). More recently, the AFM has proven the unique tool for studying the supramolecular assembly of the bacterial photosynthetic unit (9–12) and has provided a solid basis for the understanding of the ensemble function of the photosynthetic proteins (4, 13–15).
The light-harvesting process in photosynthetic bacteria starts with photon capture at the peripheral light-harvesting complex 2 (LH2) that transfers the absorbed excitation energy to the light-harvesting complex 1 (LH1) associated with the reaction center (RC). The LH2 structure is well described by X-ray crystallography (16, 17) and AFM in native membranes (12, 18–20). Typically, the LH2 complexes appear as circular oligomers of nine αβ subunits, including 18 long-wavelength-absorbing bacteriochlorophyll (BChl) molecules sandwiched between the inner ring of α-polypeptides and the outer ring of β-polypeptides. Another series of nine BChl molecules, closer to the cytoplasmic membrane surface, occupy gaps between the β-polypeptides. The LH2 structures are found to be variable, in terms of the number of units forming the rings, depending on the species and environmental conditions (17, 19, 21, 22). A recent study showed that the synthesis and assembly processes of LH2 complexes are not coupled (23).
What are the forces within the LH2 complexes that assure their structural and functional integrity in the native membrane? In this work we performed for the first time AFM single-molecule force measurements combined with imaging of native photosynthetic membranes from Rhodospirillum (Rsp.) photometricum in order to provide answers to the above question.
Results and Discussion
Compared to a well-controlled system (i.e., unfolding purified proteins) (7, 8), the study of a native system is more complex, as more and different molecules with variable molecular interactions integrate the measurement. However, in compensation, it provides the possibility of more physiologically relevant measurements. The photosynthetic membranes of Rsp. photometricum can be well physi-adsorbed as native chromatophores to freshly cleaved mica (14). The supramolecular protein assembly of such membrane has been described in great detail (11, 14, 19, 24) and shows only minor LH2 size heterogeneity (11, 19) and only a single pair of puc genes (see SI Appendix). These membranes are further studied in the present work. In comparison, the vesicular chromatophores of Rhodobacter (Rb.) sphaeroides and the photosynthetic membranes of Rhodopseudomonas (Rps.) palustris, are much less adequate for force measurements, because of the flexible membrane surface of vesicles and significant LH2 type and size variation, respectively (9, 21).
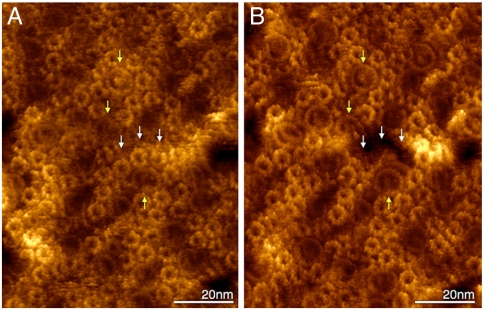
The high-resolution imaging capability of the AFM allowed to circumvent the problem of native sample heterogeneity by topographic analysis of the sample before and after force measurements. As a result, some of the force measurements were unambiguously assigned to specific unfolding events. Individual LH2 were imaged at submolecular resolution prior to force measurements in their complex native assembly with light-harvesting complexes 1 (LH1) and reaction center (RC) core-complexes (Fig. 1A). Imaging the same membrane region after force measurements evidenced the removal of individual LH2 complexes (Fig. 1B).
Fig. 1.
Imaging of the periplasmic surface of a native Rsp. photometricum photosynthetic membrane before (A) and after (B) force measurements. High-resolution topographical information allowed submolecular resolution precise alignment of images (three individual core complexes are outlined by yellow arrows). Force measurements have unfolded and removed individual LH2 complexes from the membrane (white arrows) without disruption of the wider membrane environment.
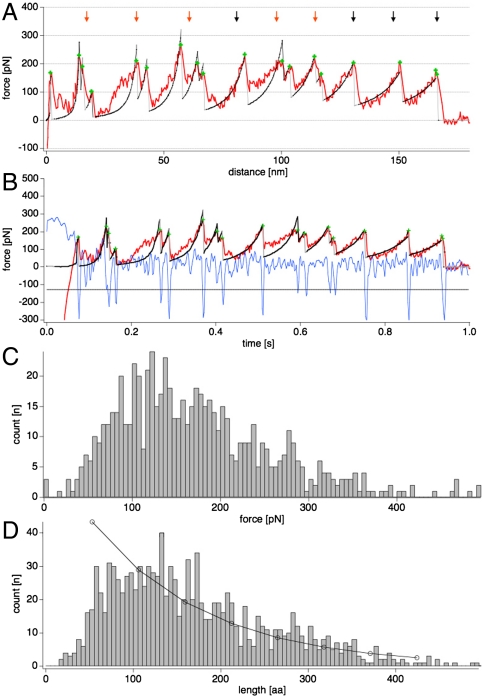
The unfolding of individual LH2 complexes resulted in force-distance curves with approximately periodic unfolding events corresponding to the subunits forming the LH2 ring (Fig. 2A). We have translated the cantilever deflection corrected force-distance curves into force-time curves (Fig. 2B) to aid automated unfolding peak detection (see Methods). The periodic force peaks are interpreted as the force response of the LH2 subunits upon unfolding and extraction from the membrane as documented by imaging (see Fig. 1). While unfolding of some subunits revealed single force events (Fig. 2A, black arrows), other subunits unfolded in a two-step process, with two close sequential force peaks (Fig. 2A, red arrows). The periodicity of the major unfolding events appeared with an average length corresponding to 53 amino acids (Fig. 2B) and an average unfolding rupture force of approximately 150 pN (Fig. 2C), comparable to the forces, between 100 pN and 200 pN, found during unfolding helix pairs of bacteriorhodopsin at the same pulling velocity (8, 25). The total lengths of the unfolding curves are highly variable (Fig. 2D) because the complex is not one single protein chain but a sequence of tightly associated polypeptides. Such a sequence of subunits can break at any subunit interface within the nonameric LH2 ring, leading to a shorter unfolding curve. Accordingly, the probability of a ring to be entirely unfolded corresponds to the addition of the probabilities of subsequent subunits to hold and not to detach. In agreement, the full unfolding length histogram declines with increasing length and can be fitted with an exponential decay (Fig. 2D). It should be mentioned that the minor size heterogeneity of LH2 complexes in Rsp. photometricum photosynthetic membranes (19) cannot be excluded to cause some of the length variability of unfolding curves. From this holding versus breaking probability (Phold ∼ 70%), we calculated the interaction constant K that is approximately 2 and an interaction energy of about kBT (see Methods). Hence LH2 subunits interact by noncovalent self-assembly nature with sufficient strength allowing the complex sometimes to be completely unfolded. Certainly the complex stability in the membrane is assured by the ring formation, in which each subunit has two subunit–subunit interfaces; furthermore, the closed ring prohibits exposed subunit overhang. The relatively weak bonding of subunits within a ring might appear surprising; it is, however, in complete accordance with the finding of subunit mixing in LH2 rings as revealed by single-molecule spectroscopy (26) and combined cross-linking and mass spectrometry (23) analysis, and with the observation of incomplete complexes in the membrane (11, 19).
Fig. 2.
LH2 unfolding. (A) Force-distance curve and (B) force-time curve of an individual LH2 complex unfolding. In both graphs, the corresponding WLC fits of unfolding events are shown in black on the red raw-data force curve. Black and red arrows indicate one-step and two-step unfolding events, respectively, during the subunit unfolding. In B the blue trace is the derivative of the measured force-time curve and the horizontal black line depicts five times the standard deviation (5σ) of the noise of the force curve. Automated unfolding events detection was defined by cross-over of the force derivative plot with the 5σ-trace, indicated by green crosses. (C) Rupture force, and (D) force curve length histograms of unfolding curves analysis and an exponential fit to the decay.
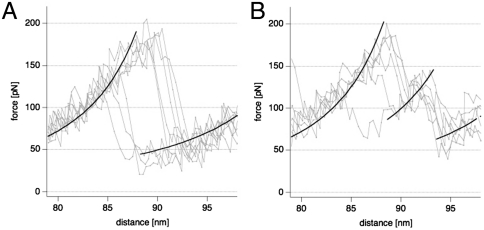
As mentioned above, due to the variable length of the force curves and the fact that individual subunits may either unfold in a one-step or a two-step process at variable positions during LH2 ring unfolding, entire unfolding curves could not be overlaid and averaged. Nonetheless, the individual subunit unfolding pathways can be overlaid and averaged (Fig. 3). The one-step unfolding force-distance graphs were best fitted with a worm-like chain (WLC) fit of 53 amino acids length (from the last unfolding event) at an unfolding force of approximately 150 pN (Fig. 3A). The two-step subunit unfolding curves were best fitted with two WLC fits, one with the 53 amino acids periodicity and rupture force of approximately 150 pN, and an intermediate peak at 23 amino acids length and a rupture force slightly above 100 pN (Fig. 3B).
Fig. 3.
Overlay of forces curves (n = 7) of the two different LH2 subunit-unfolding pathways. (A) One-step subunit unfolding with periodicity of 53 amino acids length. (B) Two-step subunit unfolding with an intermediate unfolding barrier at 23 amino acids length and the next at 30 amino acids.
Given the remarkable theoretical achievement of Jarzynski (27) providing basis to derive equilibrium free energy differences from many nonequilibrium measurements, proven applicable for single-molecule experiments (28) and notably AFM protein unfolding (29), we calculated (Supplementary Information 1 in SI Appendix) the free energy difference ΔG for LH2 subunit unfolding of 90 kBT or 53 kcal·mol-1 was derived, similar to helix-hairpins in bacteriorhodopsin (29). Certainly this solidity constitutes a prerequisite for holding the pigments precisely in space for light-harvesting function.
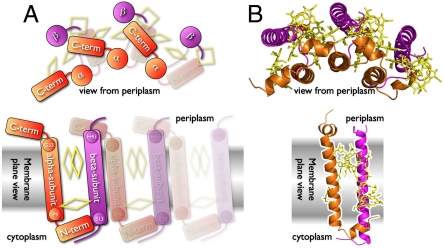
The atomic structures of the LH2 complexes from Rhodopseudomonas (Rps.) acidophila (30, 31) and Phaeospirillum (Ph.) molischianum (17) have been solved by X-ray crystallography. The LH2 complex of Rps. acidophila is a nonamer (α9β9), whereas that of Ph. molischianum is an octamer (α8β8). For structural interpretation of our results, we compare with the atomic structure of the Rps. acidophila LH2 (16, 30), as the large majority of LH2 observed in Rsp. photometricum are nonameric (11, 19, 32). The sequence identities of the α- and β-polypeptides of Rsp. photometricum and Rps. acidophila are 29% and 26%, respectively (Supplementary Information 2 in SI Appendix). Each subunit consists of an α-polypeptide and a β-polypeptide that sandwich the pigment molecules (Fig. 4). Most of the interactions between α-polypeptide and β-polypeptide in LH2 occur close to the membrane-water interfaces, between the C and N termini lying on the membrane, while the pigments (both BChl and carotenoid molecules) mediate most of the contacts between α and β in the hydrophobic phase (33, 34). The AFM unfolding experiments were performed on LH2 complexes viewed from the periplasmic photosynthetic membrane surface (see Fig. 1). The tip attached to the C-terminal helix of the α-polypeptide lies exposed on the periplasmic surface and protrudes out further than the β-polypeptide (Fig. 4 A and B). First, tip sample separation led to unfolding the α-helix of the α-polypeptide unwinding the 24 transmembrane helix amino acids from αG33 to αP9 (αS35 to αP11 in 1KUZ). This is in good agreement with the unfolding length of the intermediate peak in the two-step unfolding curves corresponding to 23 amino acids length (see Fig. 3B). The α- and the β-polypeptides are tightly associated (Supplementary Information 3 in SI Appendix) by an antiparallel N-terminal stretch over αN8 and βS9 (αN10 and βA1 in 1KUZ) (Fig. 4 A and B). Second, the AFM tip pulling unfolded 30 amino acids within the β-polypeptide transmembrane helix from βE13 to βH43 (βA5 to βS35 in 1KUZ). The length of this helix is in perfect agreement with the second unfolding peak in the two-step subunit unfolding process with length of 30 amino acids (see Fig. 3B). The C-terminal interfacial periplasmic helix of the next subunit α-polypeptide bridges to the C terminus of the unfolded β-polypeptide, allowing sequential LH2 subunit unfolding, though this interaction is weak (see Fig. 2D). Nevertheless, the importance of the C termini for intersubunit interaction has been shown by molecular dynamics simulations (35) and by mutation screens (36). In total the LH2 subunit comprises 54 transmembrane helix amino acids, in good agreement with the one-step unfolding length of 53 amino acids (see Fig. 3A) and the combined length of 23 plus 30 amino acids measured by the two-step unfolding process (see Fig. 3B).
Fig. 4.
Structural interpretation of LH2 unfolding measurements. (A) schematic and (B) experimental structure (PDB ID code 1KZU) representation. Side views (in membrane plane): The LH2 subunit protrudes on the periplasmic surface by a membrane-parallel C-terminal helix of the α-polypeptide. The pigments are sandwiched in between the α- (orange) and the β-polypeptide (magenta). The transmembrane helices have lengths of 24 (αG33 to αP9) and 30 (βE13 to βH43) amino acids.
Obviously, the β-polypeptide helix can be unfolded alone or together with the α-polypeptide. Typically, the β-polypeptide remained stable in subunits that are unfolded early and unfolded together with the α-polypeptide toward the end of long force curves unfolding several subunits. As example, LH2-ring unfolding shown in Fig. 2 revealed stabilized β-polypeptide helices in subunits 1, 2, 3, 5, and 6 (red arrows), while it was unzipped together with the α-polypeptides in subunits 4, 7, 8, and 9 (black arrows). Therefore we concluded that the stabilization of the β-polypeptide during the unfolding of the first subunits of the complex is due to the firmness of the molecular environment, while subunits unfolded toward the end are more loosely within the empty membrane space left by the unfolding of the first subunits. The stabilization of the β-polypeptide helix by the molecular environment is furthermore strong evidence that a peripheral helix in one LH2 ring interacts with comparable strength to the neighboring protein complex as to the helix within the ring.
In conclusion, combined AFM imaging and single-molecule force spectroscopy provide deep qualitative and quantitative insight into LH2 structural integrity: Weakly bound subunits self-assemble to form rings. The ring-shape itself guaranties the stability of the complex. Individual subunits are solid and their stability is further supported and depends on their specific molecular environment within the native membrane. The acquired information will facilitate the comprehension of how molecular interactions drive photosynthetic protein assembly.
Methods
Sample Preparation.
Rsp. photometricum (DSM 122) was grown anaerobically and photoheterotrophically on modified Hutners media under medium-light (30 W·m-2) conditions and harvested in late-log phase. Cells were harvested and washed twice with 10 mM Tris-HCl, pH 7.0, then broken in 20 mM Tris pH 8.0, 0.5 mM EDTA, DNAase 25 μg·mL-1 by a single passage through a French pressure cell. Lysates were loaded directly onto 5–60% sucrose gradients and centrifuged for 1.5 h. The membranes corresponding to the major pigmented band sedimented to about 40% sucrose and contained the different proteins of the photosynthetic apparatus. The membranes were then washed with 10 mM Tris-HCl, pH 8.0, in a centrifugal concentrator (220,000 × g, 1.5 h) and kept at 4 °C for AFM analysis. Absorption spectra were recorded at room temperature by a Lambda 800 spectrophotometer with a bandwidth of 2 nm (PerkinElmer) using a 1 cm pathway cuvette.
Gene Sequencing.
Attempts to sequence genomic DNA after cloning or inverse PCR were unsuccessful, because of the high GC content of the DNA stretches in the puc operon regions. To obtain the sequences of all the operons genomic DNA was sequenced using 454 technology and the resulting reads automatically assembled (20× coverage). Partial sequences corresponding to a single potential puc operon were identified by homology to the genes of the pucBA operons of Ph. molischianum (DSM119), and partial N-terminal sequence data from purified LH2 polypeptides. The partial sequences identified were then completed by genome walking and sequencing on direct DNA amplification. The genomic DNA sequences corresponding to the operon encoding the α- and β-polypeptides shown in Fig. S2 in SI Appendix have been deposited in the EMBL nucleotide sequence database (accession numbers FR848366 and FR848367).
Atomic Force Microscopy (AFM).
First, native Rsp. photometricum chromatophores were adsorbed to the AFM support immersed in 50 μl adsorption buffer (10 mM Tris-HCl, pH 7.2, 150 mM KCl, 25 mM MgCl2). After approximately 1 hr the sample was rinsed with recording buffer (10 mM Tris-HCl, pH 7.2, 150 mM KCl) (14). A Nanoscope-E AFM (5) (Veeco), equipped with a 160-μm scanner (J-scanner) and oxide-sharpened Si3N4 cantilevers (length = 100 μm, k = 0.09 N·m-1, Olympus Ltd.) was operated in contact mode for imaging and force curve mode for unfolding experiments, at ambient temperature and pressure. For imaging minimal loading forces of approximately 100 pN were applied, at scan frequencies of 4–7 Hz using optimized feedback parameters. Force measurements were performed at 298 K repeating tip approach and retraction cycles with a z-ramp size of 200 nm, and at a tip velocity of 200 nm·s-1 on the photosynthetic membranes. We allowed the tip to adhere the protein by a controlled loading force ≤ 1 nN during ≤ 1 s. Force peak events were observed in about 10% of all force curves. Many of the force curves had to be rejected because the force rupture events were incompatible with a polymer-unfolding model (37). Finally, 650 curves entered the final analysis.
Data Analysis
All data analysis was performed using self-written routines (38) for IGOR PRO.
Force-Distance Curves and Worm-Like Chain (WLC) Fitting.
Initially, the raw data cantilever-deflection versus piezo-drive curves were converted into force-distance curves that inform at any point about the real distance between the surface and the tip, correcting for the cantilever deflection and knowing the cantilever sensitivity and spring constant (red curve in Fig. 2A). The unfolding events in the force-distance curves were fitted using the worm-like chain (WLC) model (7, 37), following
where F(x) is the force at distance x, kB is the Boltzmann constant, b = 4 Å is the persistence length, L the contour length of the unfolded polypeptide chain, and T is the temperature = 298 K.
Peak Selection Criteria.
To validate force peak assignment to LH2 protein unfolding the χ2 value was calculated for all automatically detected unfolding peaks, a measure for the quality of the WLC fit to the raw data force curve (normalized square deviation between curve fit and data). χ2 = 1 documents an optimal fit; here we accepted χ2 values less than 2.
Force-Time Curves and Force-Time Derivative for Peak Detection.
In order to precisely detect the force peak of unfolding events the force-distance curves were converted into force-time curves (red curve in Fig. 2B). In the force-time curve the fast snap back of the cantilever upon unfolding was detected definitely in the derivative (blue curve in Fig. 2B). When the derivative was larger than five times the standard deviation (5σ, black horizontal line in Fig. 2B) the routine automatically identified this as a force rupture event (green crosses on the peaks in Fig. 2 A and B). Note the crossing of the derivative with the 5σ-line directly detected one-step and two-step unfolding events (indicated by black and red arrows in Fig. 2A).
Force-Distance Curve Rupture Length Histogram.
The force-distance curve rupture length histogram (Fig. 2D) reports directly about the subunit interaction. It falls off exponentially with the number n of subunits and the probability P that the subunits hold or break when being subsequently pulled out of the native membrane.
When the LH2 complex is pulled out of the membrane, the interaction between the subunits, holding versus breaking, is expressed by the equilibrium constant K.
with n(hold) and n(break) being the average number of interactions per LH2 complex in the associated and dissociated state, respectively.
Thus, when pulling a subunit out of the membrane, the probability P for the next subunit to be attached and also being pulled out of the membrane is given by:
Obviously, an interaction that does not hold leads to an abort of the chain and thus to the end of the measured unfolding force curve.
The probability that an unfolding force curve ends after the nth unfolding peak, from the association and dissociation probabilities P and 1 - P, respectively, is:
With this, the histogram of the measured chain lengths at the last unfolding event can be described as:
where N0 corresponds to the number of force curves taken.
For fitting the histogram as a function of the force curves lengths, which are measured in number of unfolded amino acids, the number of subunits n can be replaced by the average distance between the measured unfolding peaks (i.e., 53 transmembrane amino acids per subunit).
Structure Representation.
The LH2 structure (PDB ID code 1KZU) (16, 31) representation (Fig. 4B) was generated using PyMol (39).
Supplementary Material
Acknowledgments.
This study was supported by the Institut Curie, the Institut National de la Santé et Recherche Médicale, the Center National de la Recherche Scientifique, the Agence Nationale de la Recherche, and the City of Paris.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The sequence reported in this paper for Rhodospirillum photometricum pucA and pucB genes for light-harvesting protein alpha and beta, strain DSM122, has been deposited in the European Nucleotide Archive (accession no. FR848366 and FR848367).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1004205108/-/DCSupplemental.
References
- 1.Bowie JU. Solving the membrane protein folding problem. Nature. 2005;438:581–589. doi: 10.1038/nature04395. [DOI] [PubMed] [Google Scholar]
- 2.Engelman DM. Membranes are more mosaic than fluid. Nature. 2005;438:578–580. doi: 10.1038/nature04394. [DOI] [PubMed] [Google Scholar]
- 3.Sachs JN, Engelman DM. Introduction to the membrane protein reviews: The interplay of structure, dynamics, and environment in membrane protein function. Annu Rev Biochem. 2006;75:707–712. doi: 10.1146/annurev.biochem.75.110105.142336. [DOI] [PubMed] [Google Scholar]
- 4.Scheuring S, Sturgis JN. Atomic force microscopy of the bacterial photosynthetic apparatus: Plain pictures of an elaborate machinery. Photosynth Res. 2009;102:197–211. doi: 10.1007/s11120-009-9413-7. [DOI] [PubMed] [Google Scholar]
- 5.Binnig G, Quate CF, Gerber C. Atomic force microscope. Phys Rev Lett. 1986;56:930–933. doi: 10.1103/PhysRevLett.56.930. [DOI] [PubMed] [Google Scholar]
- 6.Scheuring S, Reiss-Husson F, Engel A, Rigaud JL, Ranck JL. High-resolution AFM topographs of Rubrivivax gelatinosus light-harvesting complex LH2. EMBO J. 2001;20:3029–3035. doi: 10.1093/emboj/20.12.3029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rief M, Gautel M, Oesterhelt F, Fernandez JM, Gaub HE. Reversible unfolding of individual titin immunoglobulin domains by AFM. Science. 1997;276:1109–1112. doi: 10.1126/science.276.5315.1109. [DOI] [PubMed] [Google Scholar]
- 8.Oesterhelt F, et al. Unfolding pathways of individual bacteriorhodopsins. Science. 2000;288:143–146. doi: 10.1126/science.288.5463.143. [DOI] [PubMed] [Google Scholar]
- 9.Bahatyrova S, et al. The native architecture of a photosynthetic membrane. Nature. 2004;430:1058–1062. doi: 10.1038/nature02823. [DOI] [PubMed] [Google Scholar]
- 10.Scheuring S, et al. Nanodissection and high-resolution imaging of the Rhodopseudomonas viridis photosynthetic core complex in native membranes by AFM. Proc Natl Acad Sci USA. 2003;100:1690–1693. doi: 10.1073/pnas.0437992100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Scheuring S, Sturgis JN. Chromatic adaptation of photosynthetic membranes. Science. 2005;309:484–487. doi: 10.1126/science.1110879. [DOI] [PubMed] [Google Scholar]
- 12.Liu LN, Sturgis JN, Scheuring S. Native architecture of the photosynthetic membrane from Rhodobacter veldkampii. J Struct Biol. 2011;173:138–145. doi: 10.1016/j.jsb.2010.08.010. [DOI] [PubMed] [Google Scholar]
- 13.Cogdell RJ, Gall A, Köhler J. The architecture and function of the light-harvesting apparatus of purple bacteria: from single molecules to in vivo membranes. Q Rev Biophys. 2006;39:227–324. doi: 10.1017/S0033583506004434. [DOI] [PubMed] [Google Scholar]
- 14.Liu LN, Duquesne K, Sturgis JN, Scheuring S. Quinone pathways in entire photosynthetic chromatophores of Rhodospirillum photometricum. J Mol Biol. 2009;393:27–35. doi: 10.1016/j.jmb.2009.07.044. [DOI] [PubMed] [Google Scholar]
- 15.Sturgis JN, Niederman RA. Atomic force microscopy reveals multiple patterns of antenna organization in purple bacteria: Implications for energy transduction mechanisms and membrane modeling. Photosynth Res. 2008;95:269–278. doi: 10.1007/s11120-007-9239-0. [DOI] [PubMed] [Google Scholar]
- 16.McDermott G, et al. Crystal structure of an integral membrane light-harvesting complex from photosynthetic bacteria. Nature. 1995;374:517–521. [Google Scholar]
- 17.Koepke J, Hu X, Muenke C, Schulten K, Michel H. The crystal structure of the light-harvesting complex II (B800-850) from Rhodospirillum molischianum. Structure. 1996;4:581–597. doi: 10.1016/s0969-2126(96)00063-9. [DOI] [PubMed] [Google Scholar]
- 18.Scheuring S, et al. AFM characterization of tilt and intrinsic flexibility of Rhodobacter sphaeroides light harvesting complex 2 (LH2) J Mol Biol. 2003;325:569–580. doi: 10.1016/s0022-2836(02)01241-x. [DOI] [PubMed] [Google Scholar]
- 19.Scheuring S, Rigaud JL, Sturgis JN. Variable LH2 stoichiometry and core clustering in native membranes of Rhodospirillum photometricum. EMBO J. 2004;23:4127–4133. doi: 10.1038/sj.emboj.7600429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gonçalves RP, Busselez J, Lévy D, Seguin J, Scheuring S. Membrane insertion of Rhodopseudomonas acidophila light harvesting complex 2 investigated by high resolution AFM. J Struct Biol. 2005;149:79–86. doi: 10.1016/j.jsb.2004.09.001. [DOI] [PubMed] [Google Scholar]
- 21.Scheuring S, Gonçalves RP, Prima V, Sturgis JN. The photosynthetic apparatus of Rhodopseudomonas palustris: Structures and organization. J Mol Biol. 2006;358:83–96. doi: 10.1016/j.jmb.2006.01.085. [DOI] [PubMed] [Google Scholar]
- 22.Kereïche S, et al. The peripheral light-harvesting complexes from purple sulfur bacteria have different ‘ring’ sizes. FEBS Lett. 2008;582:3650–3656. doi: 10.1016/j.febslet.2008.09.050. [DOI] [PubMed] [Google Scholar]
- 23.Mascle-Allemand C, Duquesne K, Lebrun R, Scheuring S, Sturgis JN. Antenna mixing in photosynthetic membranes from Phaeospirillum molischianum. Proc Natl Acad Sci USA. 2010;107:5357–5362. doi: 10.1073/pnas.0914854107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Scheuring S, et al. Watching the photosynthetic apparatus in native membranes. Proc Natl Acad Sci USA. 2004;101:11293–11297. doi: 10.1073/pnas.0404350101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Janovjak H, Kessler M, Oesterhelt D, Gaub H, Müller DJ. Unfolding pathways of native bacteriorhodopsin depend on temperature. EMBO J. 2003;22:5220–5229. doi: 10.1093/emboj/cdg509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Brotosudarmo THP, et al. Single-molecule spectroscopy reveals that individual low-light LH2 complexes from Rhodopseudomonas palustris 2.1.6. have a heterogeneous polypeptide composition. Biophys J. 2009;97:1491–1500. doi: 10.1016/j.bpj.2009.06.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jarzynski C. Nonequilibrium equality for free energy differences. Phys Rev Lett. 1997;78:2690–2693. [Google Scholar]
- 28.Liphardt J, Dumont S, Smith SB, Tinoco I, Jr, Bustamante C. Equilibrium information from nonequilibrium measurements in an experimental test of Jarzynski’s equality. Science. 2002;296:1832–1835. doi: 10.1126/science.1071152. [DOI] [PubMed] [Google Scholar]
- 29.Preiner J, et al. Free energy of membrane protein unfolding derived from single-molecule force measurements. Biophys J. 2007;93:930–937. doi: 10.1529/biophysj.106.096982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Papiz MZ, Prince SM, Howard T, Cogdell RJ, Isaacs NW. The structure and thermal motion of the B800-850 LH2 complex from Rps. acidophila at 2.0 Å resolution and 100 K: New structural features and functionally relevant motions. J Mol Biol. 2003;326:1523–1538. doi: 10.1016/s0022-2836(03)00024-x. [DOI] [PubMed] [Google Scholar]
- 31.Prince SM, et al. Apoprotein structure in the LH2 complex from Rhodopseudomonas acidophila strain 10050: Modular assembly and protein pigment interactions. J Mol Biol. 1997;268:412–423. doi: 10.1006/jmbi.1997.0966. [DOI] [PubMed] [Google Scholar]
- 32.Gonçalves RP, Bernadac A, Sturgis JN, Scheuring S. Architecture of the native photosynthetic apparatus of Phaeospirillum molischianum. J Struct Biol. 2005;152:221–228. doi: 10.1016/j.jsb.2005.10.002. [DOI] [PubMed] [Google Scholar]
- 33.Braun P, Gebhardt R, Kwa L, Doster W. High pressure near infrared study of the mutated light-harvesting complex LH2. Braz J Med Biol Res. 2005;38:1273–1278. doi: 10.1590/s0100-879x2005000800017. [DOI] [PubMed] [Google Scholar]
- 34.Papiz MZ, et al. A model for the photosynthetic apparatus of purple bacteria. Trends Plant Sci. 1996;1:198–206. [Google Scholar]
- 35.Janosi L, Keer H, Kosztin I, Ritz T. Influence of subunit structure on the oligomerization state of light-harvesting complexes: A free energy calculation study. Chem Phys. 2006;323:117–128. [Google Scholar]
- 36.Braun P, Olsen JD, Strohmann B, Hunter CN, Scheer H. Assembly of light-harvesting bacteriochlorophyll in a model transmembrane helix in its natural environment. J Mol Biol. 2002;318:1085–1095. doi: 10.1016/S0022-2836(02)00192-4. [DOI] [PubMed] [Google Scholar]
- 37.Bustamante C, Marko JF, Siggia ED, Smith S. Entropic elasticity of lambda-phage DNA. Science. 1994;265:1599–1600. doi: 10.1126/science.8079175. [DOI] [PubMed] [Google Scholar]
- 38.Kessler M, Gaub HE. Unfolding barriers in bacteriorhodopsin probed from the cytoplasmic and the extracellular side by AFM. Structure. 2006;14:521–527. doi: 10.1016/j.str.2005.11.023. [DOI] [PubMed] [Google Scholar]
- 39.DeLano WL. The PyMOL Molecular Graphics System. PaloAlto, CA: DeLano Scientific; 2002. http://www.pymol.org. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.