Abstract
Background
We investigated the relationship between fatty acid desaturase (FADS) gene polymorphisms and insulin resistance (IR) in association with serum phospholipid polyunsaturated fatty acid (FA) composition in healthy Korean men.
Methods
Healthy men (n = 576, 30 ~ 79 years old) were genotyped for rs174537 near FADS1 (FEN1-10154G>T), FADS2 (rs174575C>G, rs2727270C>T), and FADS3 (rs1000778C>T) SNPs. Dietary intake, serum phospholipid FA composition and HOMA-IR were measured.
Results
Fasting insulin and HOMA-IR were significantly higher in the rs174575G allele carriers than the CC homozygotes, but lower in the rs2727270T allele carriers than the CC homozygotes. The proportion of linoleic acid (18:2ω-6, LA) was higher in the minor allele carriers of FEN1-10154G>T, rs174575C>G and rs2727270C>T than the major homozygotes, respectively. On the other hand, the proportions of dihomo-γ-linolenic acid (20:3ω-6, DGLA) and arachidonic acid (20:4ω-6, AA) in serum phospholipids were significantly lower in the minor allele carriers of FEN1-10154 G>T carriers and rs2727270C>T than the major homozygotes respectively. AA was also significantly lower in the rs1000778T allele carriers than the CC homozygotes. HOMA-IR positively correlated with LA and DGLA and negatively with AA/DGLA in total subjects. Interestingly, rs174575G allele carriers showed remarkably higher HOMA-IR than the CC homozygotes when subjects had higher proportions of DLGA (≥1.412% in total serum phospholipid FA composition) (P for interaction = 0.009) or of AA (≥4.573%) (P for interaction = 0.047).
Conclusion
HOMA-IR is associated with FADS gene cluster as well as with FA composition in serum phospholipids. Additionally, HOMA-IR may be modulated by the interaction between rs174575C>G and the proportion of DGLA or AA in serum phospholipids.
Keywords: FADS gene, homeostasis model assessment-insulin resistance, linoleic acid, dihomo-γ-linolenic acid, arachidonic acid
Backgrounds
It has been reported that fatty acid (FA) composition in serum phospholipids as well as a high fat intake might influence the insulin sensitivity and the progression of obesity [1-3]. People with insulin resistance (IR) or metabolic syndrome (MetS) had high levels of palmitic acid (16:0, PA) and low levels of linoleic acid (18:2ω-6, LA) in serum phospholipids [4,5]. Lopes-Alvarenga et al [1] showed that serum triglyceride (TG) level in MetS subjects was positively associated with saturated FAs (SFA), but negatively with long-chain polyunsaturated fatty acids (PUFA). According to Leeson et al [5], higher proportions of docosahexaenoic acid (22:6ω-3, DHA) in erythrocyte lipids were associated with the improved endothelial function, particularly in young smokers who had some features of the IR.
FA composition in serum lipid esters is known to reflect the dietary FA composition during recent one or two months [6-9]. Serum PUFA compositions are influenced by both the FA metabolisms and the genetic variations [10-12]. The key enzymes involved in PUFA metabolism are the Δ5 desaturase (D5D) and Δ6 desaturase (D6D) which are encoded by the FA desaturase 1 (FADS1) and FADS2 genes located on chromosome 11 (11q12-13.1) [13-16]. This gene cluster also includes a FADS3, another desaturase gene which shares 52 and 62% sequence of the FADS1 and FADS2 genes, respectively [13], but the activity has not been fully identified yet. Among the single nucleotide polymorphisms (SNPs) in FADS gene cluster, rs174537 (FEN1, flapstructure specific endonuclease) near FADS1 shows the most significant association with FAs in a genome-wide association study. Minor T allele homozygotes of the rs174537 (FEN1-10154G>T) are associated with lower levels of plasma arachidonic acid (20:4ω-6, AA) and higher levels of LDL cholesterol and total cholesterol than the major G allele homozygotes [16].
Recently, Dupis et al. [17] and Ingelsson et al [18] reported that FADS1 (rs174550) was strongly associated with fasting glucose, fasting insulin, homeostasis model assessment of IR (HOMA-IR), HOMA-beta cell function and type 2 diabetes in Caucasian population. However, there were no studies on the relationship between FADS gene and IR in association with serum PUFA composition in healthy Koreans. Therefore, this present study aimed to investigate whether FADS gene polymorphisms are associated with IR as well as with serum phospholipid FA composition in healthy Korean men.
Subjects and Methods
Study population
Study participants (men) were recruited from the Health Service Center (HSC) in the course of a routine checkup visit or by a newspaper announcement for health examination. Subjects were excluded if they have orthopedic limitations, weight loss/gain over the previous 6 months, or any diagnosis of vascular disease, diabetes mellitus, cancer (clinically or by anamnesis), renal disease, liver disease, thyroid disease, and acute or chronic inflammatory diseases. None of all subjects were taking any medications (antihypertensive, antidyslipidemic, antithrombotic and antidiabetic drugs). Finally, 576 healthy men (aged from 30 to 79 years old) were enrolled in the study. Written informed consent was obtained from all subjects, and the protocol was approved by the Institute of Review Board of Yonsei University.
Anthropometric parameters and blood collection
Body weight and height were measured unclothed and without shoes in the morning. Body mass index (BMI) was calculated as body weight in kilograms divided by height in square meters (kg/m2). Blood pressure (BP) was obtained from the left arm of seated patients with an automatic blood pressure monitor (TM-2654, A&D, Tokyo, Japan) after 20 min of rest. Study subjects were interviewed for their smoking and drinking behavior at their visit. Smoking habit was categorized to 'current smoker' and 'non-smoker', and drinking habit was also categorized to 'current drinker (current alcohol consumption)' and 'non-drinker'.
After overnight fast, venous blood specimens were collected in EDTA-treated and plain tubes. The tubes were immediately placed on ice until they arrived at the analytical laboratory (within 1-3 h). Then, the blood specimens were separated into plasma (i.e. glucose and insulin) or serum (i.e. lipid profiles, apolipoproteins and fatty acid compositions), and stored at -70°C until analysis.
Genomic DNA was extracted from 5 mL whole blood using a commercially available DNA isolation kit (WIZARDR Genomic DNA purification kit, Promega, Madison, WI, USA) according to the manufacturer's protocol.
Genotyping of FADS gene cluster
Genomic DNA was extracted from 5 mL whole blood using a commercially available DNA isolation kit (WIZARDR Genomic DNA purification kit, Promega, Madison, WI, USA) according to the manufacturer's protocol. Based on the previous reports of genetic studies as well as the public databases on the FADS gene cluster [16,19-21] and HapMap project, http://www.hapmap.org), 8 relevant FADS single nucleotide polymorphisms (SNPs) were pre-screened in randomly selected 217 subjects [rs174537 (FEN1-10154G>T near FADS1), rs174575 (FADS2), rs1000778 (FADS3), rs2727270 (FADS2), rs174576 (FADS2), rs174570 (FADS2), rs174583 (FADS2), rs174456 (FADS3)]. Each genotyping reaction was performed with Taqman assay (Applied Biosystems, Foster City, CA, USA).
Serum Lipid profile, Apolipoprotein A1 and B
Fasting serum total cholesterol and TG were measured using commercially available kits on a Hitachi 7150 Autoanalyzer (Hitachi Ltd. Tokyo, Japan). After precipitation of serum chylomicron, low density lipoprotein (LDL) and very low density lipoprotein (VLDL) with dextran sulfate-magnesium, high density lipoprotein cholesterol (HDL-C) left in the supernatant was measured by an enzymatic method. LDL cholesterol (LDL-C) was estimated indirectly using the Friedewald formula for subjects with serum TG concentrations <400 mg/dL (4.52 mol/L). In subjects with serum TG concentrations ≥400 mg/dL (4.52 mol/L), LDL-C was measured by an enzymetic method on a Hitachi 7150 Autoanalyzer directly. Each sample was measured duplicate and their average value was used for one value. If there is big variation between the two values, the samples were remeasured. Serum apolipoprotein (Apo) A1 and B were determined by turbidometry at 340 nm using a specific anti serum (Roche, Basel, Switzerland).
Glucose, Insulin and HOMA-IR
Fasting plasma glucose was measured by a glucose oxidase method using the Glucose Analyzer (Beckman Instruments, Irvine, CA, USA). Plasma Insulin was measured by radioimmuno-assays with commercial kits from Immuno Nucleo Corporation (Stillwater, MN, USA). Insulin resistance (IR) was calculated with the homeostasis model assessment (HOMA) using the following equation: IR = {fasting insulin (μIU/ml) × fasting glucose (mmol/l)}/22.5 [22].
Fatty acid composition in serum phospholipids
Serum phospholipid fatty acid (FA) composition was analyzed using the modified method of Folch et al. [23] and Lepage et al. [24] with gas chromatography (Hewlett Packard 5890A, CA, USA). Individual FAs were calculated as a relative percentage with the elevated FAs set at 100% using Chemstation software.
The assessment of dietary intake/physical activity level
The subjects' usual diet information was obtained using both a 24-hour recall method and a semi-quantitative food frequency questionnaire (SQFFQ) of which the validity had been previously tested [25]. We used the former to carry out analyses and the latter to check if the data collected by 24-hour recall methods was representative of the usual dietary pattern. All subjects were given written and verbal instructions by a registered dietitian on completion of a 3-day (2 week days and 1 weekend) dietary record. Dietary energy values and nutrient content from a 3-day food records were calculated using the Computer Aided Nutritional analysis program (CAN-pro 2.0, Korean Nutrition Society, Seoul, Korea). Total energy expenditure (TEE) (kcal/day) was calculated from activity patterns including basal metabolic rate, physical activity for 24 hours [26], and specific dynamic action of food. Basal metabolic rate for each subject was calculated with the Harris-Benedict equation [27].
Statistical analysis
Statistical analyses were performed with SPSS version 15.0 for Windows (Statistical Package for the Social Science, SPSS Ins., Chicago, IL, USA). Hardy-Weinberg Equilibrium (HWE) and the linkage disequilibrium (LD) tests were examined using the Haploviewer 4.1 (Broad Inst, MA, USA). In order to test the difference in general characteristics and biochemical parameters (ie. lipid profiles, glucose, insulin resistance, fatty acid composition and dietary intakes), student t-test (independent t-test) between the two groups (major allele homozygotes vs minor allele carriers) and one-way analysis of variance (ANOVA) among the genotype groups followed by Bonferroni correction for multiple comparisons to reduce the rate of false positive was used. General linear model analysis (GLM, Post hoc multiple comparison tests) followed by Bonferroni correction for multiple comparisons was also performed to see the differences in IR and FA composition between genotype groups with adjustment for confounders (i.e. age, body mass index, cigarette smoking, alcohol drinking, blood pressure, triglyceride, total energy intake and expenditure etc.). The Pearson correlation analyses were used to evaluate the relationship between insulin resistance (HOMA-IR) and general or biochemical parameters including FA composition. In addition, multiple stepwise regression analysis was performed to see whether FADS SNPs remain significant influent factor on HOMA-IR together with other markers related to HOMA-IR. Frequencies were tested by chi-square test. We determined whether each variable was normally distributed before statistical testing, and logarithmic transformation was performed on skewed variables. For descriptive purposes, mean values are presented using untransformed values. Results are expressed as means ± standard deviation (S.D.) or %. A two-tailed value of P < 0.05 was considered statistically significant.
Results
General characteristics of study population and genotype distribution of pre-screened SNPs
Table 1 presents demographic and metabolic parameters of whole study population. All the 8 SNPs which were pre-screened in 271 subjects satisfied the Hardy Weinberg Equilibrium (HWE) (P > 0.05). From the Linkage Disequilibrium (LD) test, rs174537 (FEN1-10154G>T) and rs174575 (FADS2 rs174575C>G) were found highly linked (D' = 0.93; P < 0.001) even though r2 value was not high (r2 = 0.14). rs174537 and rs2727270 (FADS2 rs2727270C>T) were also found highly linked (D' = 0.99; P < 0.001) and relatively highly correlated (r2 = 0.74). Interestingly, rs174537 was found very highly linked with rs174576, rs174570 and rs174583 (D' = 1.00, r2>0.95 for all). Based on the the result above, we realized that rs174537 is sufficient to cover the genetic information of last three SNPs (rs174576, rs174570 and rs174583) as a tagging SNP. In addition, rs1000778 (FADS3 rs1000778C>T) is very highly linked to rs174456 (FADS3 rs174456T>C) (D' = 1, r2 = 1). Therefore, we finally included 4 SNPs (FEN1-10154 rs174537G>T, FADS2 rs174575C>G, FADS2 rs2727270C>T and FADS3 1000778C>T) for further genotyping in the whole study subjects.
Table 1.
Demographic and metabolic parameters of study population
| Total (n = 567) | Range (Min-Max) | |
|---|---|---|
| Age (year) | 48.7 ± 9.3 | 30.0 - 69.0 |
| Body mass index (kg/m2) | 24.4 ± 2.7 | 18.2 - 35.0 |
| Waist circumference (cm) | 85.5 ± 7.3 | 65.0 - 127.0 |
| Current smokers (%) | 63.0 | |
| Current drinkers (%) | 16.0 | |
| MetS (%) | 17.8 | |
| SBP (mmHg) | 123.1 ± 14.3 | 90.0 - 180.0 |
| DBP (mmHg) | 76.9 ± 11.0 | 48.0 - 120.0 |
| TG (mg/dL) | 134.8 ± 73.8 | 27.0 - 477.0 |
| Total-C (mg/dL) | 192.2 ± 33.3 | 113.0 - 306.0 |
| HDL-C (mg/dL) | 50.4 ± 13.1 | 21.0 - 121.0 |
| LDL-C (mg/dL) | 115.1 ± 31.5 | 37.8 - 227.6 |
| NON-HDL-C (mg/dL) | 141.8 ± 33.9 | 55.0 - 266.0 |
| ApoA1 (mg/dL) | 140.5 ± 24.6 | 92.0 - 238.0 |
| ApoB (mg/dL) | 87.1 ± 23.5 | 45.0 - 162.0 |
| Glucose (mg/dL) | 92.9 ± 10.7 | 60.0 - 122.0 |
| Insulin (μIU/mL) | 8.9 ± 4.3 | 1.6 - 44.7 |
| HOMA-IR | 2.06 ± 1.1 | 0.4 - 11.4 |
| Total energy expenditure (TEE, kcal) | 2337.5 ± 202.5 | 361.0 - 2956.3 |
| Total calorie intake (TIC, kcal) | 2424.7 ± 208.2 | 1631.1 - 3055.6 |
| TEE/TCI | 0.965 ± 0.05 | 0.15 - 1.12 |
| % of carbohydrates | 61.7 ± 1.3 | 57.3 - 71.0 |
| % of protein | 16.9 ± 1.3 | 12.2 - 21.7 |
| % of fat | 21.5 ± 1.4 | 17.1 - 26.6 |
| SFA(g) | 8.3 ± 5.4 | 0.3 - 54.5 |
| MUFA (g) | 11.6 ± 7.0 | 0.6 - 110.3 |
| PUFA (g) | 11.3 ± 5.4 | 0.3 - 33.6 |
| Cholesterol (mg) | 271.2 ± 116.3 | 26.1 - 809.2 |
| Fiber (g) | 10.4 ± 4.7 | 2.5 - 39.7 |
Means ± S.D. or percentage
SBP: systolic blood pressure, DBP: diastolic blood pressure, TG: triglyceride, C: cholesterol, Apo: apolipoprotein, HOMA-IR (homeostatis model assessment-insulin resistance): {fasting insulin (μIU/ml) × fasting glucose (mmol/l)}/22.5, SFA: saturated fatty acid, MUFA: monounsaturated fatty acid, PUFA: polyunsaturated fatty acid
Genotype distribution of four selected SNPs
The selected 4 SNPs which were genotyped in the whole subjects satisfied the HWE (P > 0.05) (Additional file 1). As shown in the prescreening test, rs174537 and rs174575 were found highly linked (D' = 0.95; P < 0.001) even though r2 value was not high (r2 = 0.18). rs174537 and rs2727270 were also found highly linked (D' = 0.98; P < 0.001) and relatively highly correlated (r2 = 0.67). On the other hand, rs1000778 is weakly linked to three other SNPs (rs174537: D' = 0.21, r2 = 0.04; rs174575: D' = 0.46, r2 = 0.04; rs2727270: D' = 0.13, r2 = 0.01). The haplotype distribution of rs174537-rs174575-rs2727270-rs1000778 was like this: GCCC was most highly frequent (0.528), GCCT was second-highly frequent (0.153), TCTC was thirdly frequent (0.144) and TCTT was fourthly frequent (0.087).
Since haplotype analysis did not provide information beyond that revealed by each SNP (data not shown), we presented only the results of individual SNPs. Particularly, we presented the results especially by the form of the homozygotes for a given allele and the carrier of the alternative allele because their general and biochemical characteristics (i.e. insulin level, HOMA-IR and FA composition) were shown similarly with those among 3 genotype groups, furthermore the subject number of the minor homozygotes of rs rs174575 was so small (n = 7).
Clinical and biochemical characteristics according to FEN1 -10154G>T, FADS2 rs174575C>G, FADS2 rs2727270C>T and FADS3 rs1000778C>T
Table 2 shows clinical and biochemical characteristics of study subjects according to the genotypes of the FEN1 -10154G>T, FADS2 rs174575C>G, FADS2 rs2727270C>T and FADS3 rs1000778C>T, respectively. No significant genotype-associated differences were observed for age, BMI, systolic blood pressure (SBP), diastolic blood pressure (DBP), serum concentrations of TG, total cholesterol (Total-C), HDL-C, LDL-C, ApoA1, ApoB (Table 2), total energy intake, proportions of energy intake derived from carbohydrates and fat in each of 4 FADS SNPs (Additional file 2). Interestingly, fasting insulin and HOMA-IR were significantly higher in subjects having the minor G allele of FADS2 rs174575C>G than those having the major CC homozygotes. On the other hand, the levels of these two markers were significantly lower in the subjects carrying the minor T allele of FADS2 rs2727270C>T than those having major CC homozygote (Table 3). These significant associations still maintained after adjustment for confounders (age, BMI, cigarette smoking, alcohol consumption, blood pressure, triglyceride, total energy expenditure and total calorie intake) which were followed by Bonferroni correction for multiple comparisons (Table 3).
Table 2.
Demographic and metabolic parameters of study population according to FEN1 -10154G>T, FADS2 rs174575, FADS2 rs2727270 and FADS3 rs1000778C>T
| FEN1 -10154G>T | FADS2 rs174575 | FADS2 rs2727270 | FEN1 -10154G>T | |||||
|---|---|---|---|---|---|---|---|---|
| GG (n = 259) | GT+TT (n = 308) | CC (n = 471) | CG+GG (n = 96) | CC (n = 323) | CT+TT (n = 244) | GG (n = 259) | GT+TT (n = 308) | |
| Age (year) | 48.6 ± 9.8 | 48.7 ± 9.0 | 48.9 ± 9.4 | 47.7 ± 9.0 | 48.4 ± 9.6 | 49.0 ± 9.0 | 49.1 ± 9.6 | 48.3 ± 9.1 |
| BMI (kg/m2) | 24.4 ± 2.9 | 24.4 ± 2.5 | 24.3 ± 2.7 | 24.8 ± 2.6 | 24.4 ± 2.8 | 24.4 ± 2.5 | 24.3 ± 2.6 | 24.5 ± 2.8 |
| Waist (cm) | 85.1 ± 6.9 | 85.9 ± 7.5 | 85.3 ± 7.3 | 86.8 ± 7.2 | 85.1 ± 7.0 | 86.0 ± 7.5 | 85.4 ± 7.3 | 85.6 ± 7.3 |
| Current smokers (%) | 34.7 | 39.0 | 38.0 | 32.3 | 35.3 | 39.3 | 35.7 | 38.3 |
| Current drinkers (%) | 83.4 | 84.4 | 83.4 | 86.5 | 84.8 | 82.8 | 79.6 | 88.2 |
| Sysptolic BP (mmHg) | 123.0 ± 14.7 | 123.2 ± 14.0 | 122.9 ± 14.3 | 124.0 ± 14.3 | 123.1 ± 14.7 | 123.2 ± 13.8 | 123.1 ± 13.7 | 123.1 ± 14.8 |
| Diastolic BP (mmHg) | 76.4 ± 11.1 | 77.3 ± 10.9 | 76.8 ± 10.9 | 77.4 ± 11.6 | 76.3 ± 11.1 | 77.6 ± 10.9 | 76.9 ± 10.5 | 76.9 ± 11.4 |
| TG (mg/dL)§ | 137.9 ± 76.2 | 132.1 ± 71.6 | 133.1 ± 73.7 | 143.1 ± 73.8 | 136.7 ± 75.2 | 132.2 ± 71.9 | 135.5 ± 80.7 | 134.1 ± 66.5 |
| Total-C (mg/dL)§ | 194.5 ± 32.5 | 190.3 ± 33.8 | 192.4 ± 33.3 | 191.4 ± 33.2 | 194.4 ± 33.4 | 189.3 ± 33.0 | 192.5 ± 32.3 | 192.0 ± 34.2 |
| HDL-C (mg/dL)§ | 50.0 ± 12.3 | 50.7 ± 13.7 | 50.4 ± 13.3 | 50.2 ± 12.1 | 50.5 ± 12.3 | 50.3 ± 14.0 | 49.6 ± 12.9 | 51.1 ± 13.2 |
| LDL-C (mg/dL) | 117.2 ± 31.4 | 113.4 ± 31.6 | 115.6 ± 31.8 | 112.6 ± 30.3 | 116.8 ± 32.1 | 112.9 ± 30.8 | 116.2 ± 31.3 | 114.0 ± 31.8 |
| non-HDL-C (mg/dL) | 144.5 ± 33.1 | 139.6 ± 34.5 | 142.0 ± 34.2 | 141.2 ± 32.6 | 143.9 ± 33.8 | 139.1 ± 34.0 | 142.8 ± 33.7 | 140.8 ± 34.2 |
| ApoA1 (mg/dL) | 139.9 ± 23.0 | 141.0 ± 26.1 | 140.4 ± 24.0 | 140.9 ± 28.6 | 141.9 ± 24.5 | 138.9 ± 24.8 | 140.8 ± 26.7 | 140.2 ± 22.7 |
| ApoB (mg/dL)§ | 88.6 ± 22.3 | 85.9 ± 24.6 | 87.0 ± 24.0 | 87.8 ± 21.3 | 88.6 ± 22.9 | 85.4 ± 24.3 | 85.4 ± 22.9 | 88.7 ± 24.1 |
| Glucose (mg/dL) | 93.3 ± 11.1 | 92.5 ± 10.4 | 92.7 ± 10.9 | 93.9 ± 9.8 | 93.4 ± 10.7 | 92.2 ± 10.8 | 92.3 ± 10.6 | 93.4 ± 10.9 |
Means ± S.D. or %, §Tested by log-transformed, Performed by independent t-test or qui-square test, *P < 0.05, **P < 0.01 and ***P < 0.001 compared with major allele for each genotype.
BMI: body mass index, BP: blood pressure, TG: triglyceride, C: cholesterol, Apo: apolipoprotein, HOMA-IR(homeostatis model assessment-insulin resistance): {fasting insulin (μIU/ml) × fasting glucose (mmol/l)}/22.5.
Table 3.
Insulin, HOMA-IR and Fatty acid composition (%) in serum phospholipids according to FEN1 -10154G>T, FADS2 rs174575, FADS2 rs2727270 and rsFADS3 1000778C>T
| FEN1 -10154G>T | FADS2 rs174575 | FADS2 rs2727270 | FEN1 -10154G>T | |||||
|---|---|---|---|---|---|---|---|---|
| GG (n = 259) | T carrier (n = 308) | CC (n = 471) | G carrier (n = 96) | CC (n = 323) | T carrier (n = 244) | GG (n = 259) | GT+TT (n = 308) | |
| Insulin (μIU/mL)§ | 9.1 ± 4.2 | 8.8 ± 4.3 | 8.7 ± 3.8 | 9.7 ± 5.9* | 9.3 ± 4.8 | 8.4 ± 3.3* | 8.6 ± 3.6 | 9.2 ± 4.8 |
| HOMA-IR§ | 2.11 ± 1.04 | 2.03 ± 1.12 | 2.00 ± 0.96 | 2.28 ± 1.53* | 2.17 ± 1.23 | 1.93 ± 0.83* | 1.98 ± 0.91 | 2.15 ± 1.23 |
| Total SFA | 54.724 ± 0.340 | 53.694 ± 0.339* | 54.374 ± 0.265 | 53.139 ± 0.583 | 54.487 ± 0.305 | 53.739 ± 0.389 | 54.039 ± 5.359 | 54.288 ± 6.117 |
| 12:0§ | 0.367 ± 0.014 | 0.411 ± 0.016 | 0.392 ± 0.012 | 0.386 ± 0.026 | 0.377 ± 0.013 | 0.410 ± 0.018 | 0.394 ± 0.264 | 0.388 ± 0.247 |
| 14:0§ | 0.595 ± 0.017 | 0.683 ± 0.056 | 0.650 ± 0.037 | 0.604 ± 0.031 | 0.599 ± 0.016 | 0.700 ± 0.070 | 0.599 ± 0.254 | 0.685 ± 1.019 |
| 16:0 | 32.736 ± 0.325 | 32.206 ± 0.298 | 32.509 ± 0.247 | 32.152 ± 0.474 | 32.666 ± 0.283 | 32.161 ± 0.348 | 32.403 ± 4.945 | 32.493 ± 5.514 |
| 18:0§ | 19.067 ± 0.204 | 18.487 ± 0.198* | 18.886 ± 0.156 | 18.092 ± 0.348** | 18.892 ± 0.186 | 18.566 ± 0.223 | 18.676 ± 3.181 | 18.826 ± 3.604 |
| Total MUFA | 11.353 ± 0.134 | 11.514 ± 0.146 | 11.352 ± 0.104 | 11.877 ± 0.297* | 11.483 ± 0.134 | 11.385 ± 0.151 | 11.574 ± 2.491 | 11.311 ± 2.282 |
| 16:1§ | 0.628 ± 0.028 | 0.733 ± 0.048 | 0.650 ± 0.025 | 0.859 ± 0.117 | 0.656 ± 0.032 | 0.724 ± 0.052 | 0.681 ± 0.721 | 0.689 ± 0.655 |
| 18:1 ω-9 | 6.917 ± 0.101 | 6.996 ± 0.108 | 6.903 ± 0.077 | 7.234 ± 0.225 | 7.002 ± 0.103 | 6.903 ± 0.108 | 7.015 ± 1.869 | 6.905 ± 1.678 |
| 18:1 ω-7 | 1.729 ± 0.025 | 1.712 ± 0.027 | 1.715 ± 0.020 | 1.744 ± 0.050 | 1.739 ± 0.024 | 1.694 ± 0.029 | 1.755 ± 0.434 | 1.685 ± 0.445 |
| Total PUFA | 23.978 ± 0.377 | 24.046 ± 0.349 | 23.863 ± 0.278 | 24.761 ± 0.648 | 24.073 ± 0.343 | 23.939 ± 0.385 | 24.135 ± 5.709 | 23.897 ± 6.455 |
| Total ω-6 FA | 19.237 ± 0.297 | 19.229 ± 0.274 | 19.082 ± 0.217 | 19.973 ± 0.528 | 19.304 ± 0.272 | 19.138 ± 0.298 | 19.286 ± 4.365 | 19.180 ± 5.178 |
| 18:2 ω-6 | 12.062 ± 0.186 | 12.853 ± 0.181** | 12.303 ± 0.139 | 13.417 ± 0.352** | 12.241 ± 0.172 | 12.824 ± 0.200* | 12.394 ± 2.794 | 12.587 ± 3.410 |
| 18:3 ω-6§ | 0.218 ± 0.014 | 0.233 ± 0.014 | 0.227 ± 0.012 | 0.225 ± 0.021 | 0.223 ± 0.013 | 0.231 ± 0.017 | 0.228 ± 0.258 | 0.225 ± 0.228 |
| 20:2 ω-6§ | 0.856 ± 0.104 | 0.880 ± 0.102 | 0.864 ± 0.082 | 0.898 ± 0.161 | 0.855 ± 0.094 | 0.888 ± 0.115 | 0.855 ± 1.790 | 0.883 ± 1.686 |
| 20:3 ω-6 | 1.504 ± 0.034 | 1.405 ± 0.031* | 1.440 ± 0.024 | 1.498 ± 0.063* | 1.492 ± 0.032 | 1.394 ± 0.033* | 1.458 ± 0.513 | 1.442 ± 0.580 |
| 20:4 ω-6 | 5.016 ± 0.112 | 4.323 ± 0.099*** | 4.690 ± 0.084 | 4.391 ± 0.162 | 4.907 ± 0.100 | 4.285 ± 0.111*** | 4.806 ± 1.834 | 4.478 ± 1.748* |
| 22:4 ω-6§ | 0.247 ± 0.020 | 0.217 ± 0.012 | 0.230 ± 0.012 | 0.232 ± 0.024 | 0.243 ± 0.017 | 0.214 ± 0.012 | 0.219 ± 0.191 | 0.242 ± 0.314 |
| 22:5 ω-6§ | 0.190 ± 0.013 | 0.198 ± 0.012 | 0.191 ± 0.010 | 0.209 ± 0.021 | 0.198 ± 0.012 | 0.190 ± 0.014 | 0.182 ± 0.194 | 0.206 ± 0.227 |
| Total ω-3 FA§ | 4.741 ± 0.123 | 4.818 ± 0.123 | 4.781 ± 0.097 | 4.789 ± 0.199 | 4.769 ± 0.110 | 4.801 ± 0.141 | 4.849 ± 2.125 | 4.717 ± 2.031 |
| 18:3 ω-3§ | 0.135 ± 0.006 | 0.162 ± 0.025 | 0.153 ± 0.017 | 0.134 ± 0.009 | 0.134 ± 0.005 | 0.170 ± 0.032 | 0.135 ± 0.096 | 0.163 ± 0.460 |
| 20:5 ω-3§ | 1.158 ± 0.038 | 1.166 ± 0.042 | 1.178 ± 0.032 | 1.085 ± 0.062 | 1.161 ± 0.034 | 1.165 ± 0.048 | 1.197 ± 0.750 | 1.129 ± 0.602 |
| 22:5 ω-3§ | 0.515 ± 0.017 | 0.546 ± 0.021 | 0.526 ± 0.015 | 0.558 ± 0.036 | 0.523 ± 0.016 | 0.544 ± 0.024 | 0.539 ± 0.333 | 0.525 ± 0.312 |
| 22:6 ω-3§ | 2.833 ± 0.088 | 2.868 ± 0.083 | 2.837 ± 0.066 | 2.925 ± 0.146 | 2.852 ± 0.079 | 2.851 ± 0.093 | 2.897 ± 1.458 | 2.807 ± 1.404 |
| 18:3 ω-6/18:2 ω-6§ | 0.019 ± 0.001 | 0.020 ± 0.002 | 0.019 ± 0.001 | 0.018 ± 0.002 | 0.019 ± 0.001 | 0.020 ± 0.002 | 0.019 ± 0.020 | 0.020 ± 0.028 |
| 20:3 ω-6/18:2 ω-6 | 0.126 ± 0.003 | 0.112 ± 0.003** | 0.120 ± 0.002 | 0.111 ± 0.004 | 0.123 ± 0.002 | 0.113 ± 0.004* | 0.119 ± 0.041 | 0.118 ± 0.059 |
| 20:3 ω-6/18:3 ω-6§ | 13.066 ± 0.679 | 11.460 ± 0.541 | 12.206 ± 0.469 | 12.134 ± 1.055 | 12.527 ± 0.585 | 11.752 ± 0.626 | 12.487 ± 10.074 | 11.907 ± 10.327 |
| 20:4 ω-6/18:2 ω-6 | 0.419 ± 0.010 | 0.339 ± 0.008*** | 0.385 ± 0.007 | 0.327 ± 0.010*** | 0.404 ± 0.008 | 0.338 ± 0.010*** | 0.392 ± 0.157 | 0.359 ± 0.147** |
| 20:4 ω-6/20:3 ω-6§ | 3.509 ± 0.102 | 3.192 ± 0.060** | 3.383 ± 0.065 | 3.112 ± 0.099 | 3.462 ± 0.085 | 3.172 ± 0.069** | 3.387 ± 1.020 | 3.288 ± 1.618* |
Mean ± S.D.
§Tested by log-transformed, Performed by general linear model analysis (GLM, Post hoc multiple comparison tests) with adjustment for age, body mass index, cigarette smoking, alcohol drinking, blood pressure, triglyceride, total energy intake and expenditure, followed by Bonferroni correction for multiple comparison.
Adjusted P-value: *P < 0.05, **P < 0.01 and ***P < 0.001 compared with major allele for each genotype
Fatty acid composition in serum phospholipids according to FEN1 -10154G>T, FADS2 rs174575C>G, FADS2 rs2727270C>T and FADS3 rs1000778C>T
Table 3 shows the FA composition in serum phospholipids according to FEN1 -10154G>T, FADS2 rs174575C>G, FADS2 rs2727270C>T and FADS3 rs1000778C>T, respectively after the adjustment for confounders (age, BMI, cigarette smoking, alcohol consumption, blood pressure, triglyceride, total energy expenditure and total calorie intake) which were followed by Bonferroni correction for multiple comparisons. The minor T allele carriers of FEN1 -10154G>T showed the higher proportion of linoleic acid (18:2ω-6, LA) (P = 0.002) and lower proportion of stearic acid (18:0, SA) (P = 0.045), dihomo-γ-linolenic acid (20:3ω-6, DGLA) (P = 0.038) and arachidonic acid (20:4ω-6, AA) (P < 0.001) in serum phospholipids than the GG homozygotes. The minor G allele carriers of FADS2 rs174575C>G had the higher proportion of LA (P = 0.001) and lower proportion of SA (P = 0.009) than the CC homozygotes. The minor T allele carriers of FADS2 rs2727270C>T showed the higher proportion of LA (P = 0.026), and lower proportions of DGLA (P = 0.035) and AA (P < 0.001) than the CC homozygtes. The minor T allele carriers of FADS3 rs1000778C>T had the lower proportion of AA (P = 0.022) than the CC homozygotes. Among FA ratios which indicate FA desaturase activity, AA/LA were lower in the subjects carrying the minor allele of FEN1 -10154G>T, FADS2 rs174575C>G, FADS2 rs2727270C>T and FADS3 rs1000778C>T than the major allele homozygote, respectively. DGLA/LA were also were lower in the subjects carrying the minor allele of FEN1 -10154G>T, FADS2 rs2727270C>T and FADS3 rs1000778C>T than the major allele homozygote, respectively. Additionally, DGLA/LA were lower in the subjects carrying the minor allele of FEN1 -10154G>T and FADS2 rs2727270C>T than the major allele homozygote, respectively.
Correlation among fatty acid composition in serum phospholipids together with HOMA-IR
Among the FA composition in serum phospholipids, DGLA was very highly correlated with AA (r = 0.684, P =< 0.001) and LA (r = 0.533, P =< 0.001). LA was also highly correlated with AA (r = 0.550, P =< 0.001). In addition, HOMA-IR positively correlated with LA (r = 0.107, P = 0.011) and DGLA (r = 0.129, P = 0.002), and negatively with AA/DGLA (r = -0.140, P = 0.001) in total subjects. HOMA-IR also correlated with anthropometric and basic biochemical parameters such as BMI (r = 0.339, P < 0.001), waist (r = 0.305, P < 0.001), systolic blood pressure (r = 0.229, P < 0.001), diastolic blood pressure (r = 0.174, P < 0.001), triglyceride (r = 0.335, P < 0.001), total cholesterol (r = 0.083, P = 0.048), HDL-cholesterol (r = -0.197, P < 0.001), nonHDL-cholesterol (r = 0.154, P < 0.001), glucose (r = 0.4479, P < 0.001) and insulin (r = 0.967, P < 0.001).
Association of FADS gene SNPs with insulin resistance according to the proportion of FA in serum phospholipids
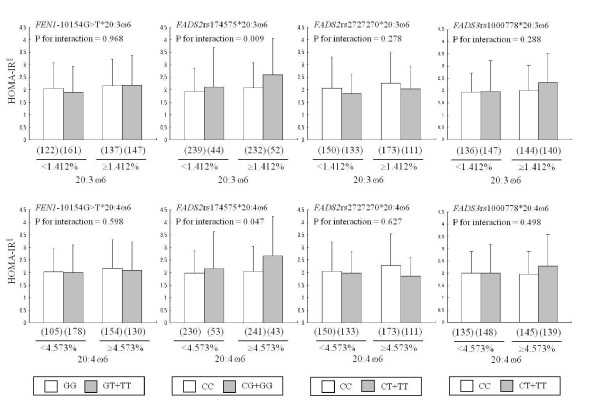
Based on the correlation results between HOMA-IR and FA compositions mentioned above, study subjects were subdivided into two groups according to the proportion of LA, DGLA or AA in serum phospholipids: upper median (LA≥12.396%, DGLA≥1.412% and AA≥4.573%. of total FA composition in serum phospholipids) and lower median (LA < 12.396%, DGAL < 1.412% and AA < 4.573%). Then, we investigated the interaction effect of genetic variants and proportion of LA, DGLA or AA on their HOMA-IR levels (Figure 1). Interestingly, HOMA-IR was significantly higher in minor G allele carriers of rs174575C>G than the CC homozygotes, when the proportion of DGLA or AA were higher than median value (Interaction P = 0.009 and P = 0.047, respectively). However, significant interaction effect was neither observed with other SNPs (FEN1 -10154G>T, FADS2 rs2727270C>T or FADS3 rs1000778C>T) nor with LA.
Figure 1.
Insulin resistance according to the proportion of dihomo-γ-linolenic acid (20:3ω-6, DGLA) or arachidonic acid (20:4ω-6, AA) in serum phospholipid and FADS gene polymorphisms. Means ± S.D. (n), §tested by log-transformed, P for interaction was tested by multiple logistic regression analysis after adjusting for age and body mass index. HOMA-IR: homeostasis model assessment of insulin resistance
Stepwise multiple regression analysis to find the major contributors to HOMA-IR among basic parameters and FADS gene SNPs
Stepwise multiple linear regression analysis was performed to determine whether FADS gene SNPs are the independent influencing factors on HOMA-IR. Based on the results of Pearson correlation coefficients (between HOMA-IR and anthropometric or basic biochemical parameters) and those of the LD test, we put HOMA-IR as a dependent variable, and body mass index, triglyceride, blood pressure and 4 SNPs (FEN1-10154 rs174537G>T, FADS2 rs174575 C>G, FADS2 rs2727270C>T and FADS3 rs1000778C>T) as independent variables in the multiple stepwise linear regression model. Among the 3 SNPs, we found that FADS2 rs174575 C>G (β-coefficient ± S.E.: 0.253 ± 0.099, p < 0.001) remained as a significant predictor of HOMA-IR together with body mass index, triglyceride and blood pressure (R2 = 0.435, P < 0.001). In addition, when we put the 4 SNPs in the model by the forms of 'the homozygotes for a given allele' (major homozygotes) and 'the carrier of the alternative allele' (heterozygotes and minor homozygotes), we found that FADS2 rs174575C>G (β-coefficient ± S.E.: 0.259 ± 0.110, p = 0.019) and FADS2 rs2727270C>T (β-coefficient ± S.E.: -0.207 ± 0.083, p = 0.013) remained as significant predictors of HOMA-IR together with other variables (R2 = 0.446, P < 0.001).
Discussion
In this present study, we found that HOMA-IR as well as serum FA composition and FA ratios were associated with the variants of FADS SNPs. This result is meaningful because it was first reported in Korean men but already done in Caucasians [17,18]. We also confirmed that HOMA-IR was related to FA composition in serum phospholipids, particularly LA, DGLA and AA/DGLA. Interestingly, we found the interactive effect between FADS2 rs174575 and the proportion DGLA or AA in serum phospholipids on HOMA-IR: FADS2 rs174575G allele carriers showed the remarkably higher HOMA-IR levels than the major CC homozygotes, when they had a higher proportion of DGLA (≥1.412% of total FA in serum phospholipids) or AA (≥4.573%. of total FA composition in serum phospholipids) in serum phospholipids.
So far, many studies have researched on the relationship of serum phospholipid FA composition or dietary fat with IR or metabolic disorder [1-5]. For example, Vessby and his colleague reported that people with IR or MetS had higher levels of palmitic acid and lower levels of LA in serum phospholipids [4,5]. Warensjö et al and others showed the significant positive relationship between the metabolic disorder (ie. IR and obesity) and the proportion of palmitoleic acid (16:1) and DGLA in serum phospholipids FAs [1,4,10]. They also showed the inverse association of IR with AA/DGLA which indicates the D5D activity. In our study, we found that HOMA-IR was positively associated with DGLA and negatively with AA/DGLA. However, we additionally found that HOMA-IR was positively related with LA, which is different from others. It may be related to the subject characteristics or the sources of FA. First, our study subjects were all healthy without MetS nor obesity [1,4,10]. In addition, LA and α-linolenic acid (18:3ω-3, ALA) in serum phospholipids are generally known as biomarkers of long-term essential FA intake [6-9], but palmitoleic acid and DGLA are synthesized endogenously by a sequence of desaturation and elongation reactions rather than they reflect dietary intake of those FAs [1]. It may indicate that DGLA or D5D are more relevant to explain the relationship with IR or metabolic disorder.
Recently, as examined in our study, the relationship between FA composition in serum phospholipids and the variants of FADS gene cluster has been studied [13-16]. Among the SNPs in FADS gene cluster, FEN1-10154G>T near FADS1 was reported to be most significantly associated with plasma FA composition; the minor T allele homozygotes of the FEN1-10154G>T are associated with lower levels of plasma AA than the major GG homozygotes [16]. In our study, the minor allele carriers of FEN1 -10154G>T, rs2727270C>T and FADS3 rs1000778C>T showed significantly lower proportion of AA. Moreover, the proportions of DGLA, DGLA/LA and AA/DGLA (D5D activity) were lower in the minor allele carriers of FEN1 -10154G>T and FADS2 rs2727270 SNPs. In addition, HOMA-IR levels were lower in the FADS2 rs2727270T allele carriers compared with the major CC homozytoges. All of these results were still retained after the adjustment for confounders followed by Bonferroni's correction for multiple comparisons. According to Rodriguez et al., AA or long-chain ω-6 PUFA were inversely associated with fasting insulin levels which might be due to the reduced desaturation-elongation cascade under IR condition furthermore, this phenomenon may worsen as the degree of IR increases [28]. Taken all together, our results may indicate that subjects with the major homozygotes had the impaired FA metabolism which may cause the accumulation of DGLA, and in consequence increase IR.
In addition, we found that rs2727270C>T (negative direction) and particularly FADS2 rs174575C>G (positive direction) were significant influent factor on HOMA-IR together with anthropometric or basic biochemical parameters. On the other hand, we found that HOMA-IR levels were higher in the FADS2 rs174575 G allele carriers compared with the major CC homozytoges, which were different from the results shown in FADS2 rs2727270C>T. These results were still retained after the adjustment for confounders followed by Bonferroni's correction for multiple comparisons. Furthermore, the correlation between HOMA-IR and serum PUFA composition were more clearly found in major C homozygotes (data not shown). Interestingly the higher HOMA-IR in FADS2 rs174575G carriers were more remarkable when the proportion of DGLA or AA in serum phospholipids was higher than median levels (interaction P = 0.009 and P = 0.047, respectively). These results were also still retained after the adjustment for confounders followed by Bonferroni's correction for multiple comparisons. Regarding this discordant result, we can assume that FADS2 rs174575 SNP differently from other FADS SNPs may affect the FA metabolism and that rs174575 may interact with other genes, such as the nuclear receptor peroxisome proliferator-activated receptor-γ (PPAR-γ) or the adipocyte C1q and collagen-domain-containing gene (ADIPOQ). PPAR-γ is a transcriptional regulator which directly interacts with the ADIPOQ promoter, both of them are closely associated with IR, MetS and diabetes mellitus [29-32]. It is known that PUFA acts as ligands of PPAR-γ [31], thus the pharmacologic activation with PPAR-γ agonists may lead to the increased plasma adiponectin [30,32], which may affect the glucose and insulin metabolism. This part needs to be further studied.
Our study has several limitations. First, study subjects were all healthy Korean men; thus, the results may not be applicable to women, other ethnic groups, or patients with cardiometablic syndrome whose lifestyle habits or biochemical characteristics may differ from those in our subjects. For example, Warensjö et al demonstrated that serum FA composition [i.e palmitoleic acid, γ-linolenic acid (18:3ω-6, GLA), DGLA and AA] and D6D activity were different between men and women [1]. Second, this study was performed in a cross-sectional design. It is not suitable for assessing the time sequential associations because the exposure and outcomes are collected at one point in time. Third, we need to consider the involvement of other genetic backgrounds which are associated in FA metabolism, for example PPAR-γ and ADIPOQ genes as mentioned above.
Despite these limitations, the present study shows that HOMA-IR as well as serum FA composition and FA ratios were associated with the variants of FADS SNPs. Furthermore, it suggested that HOMA-IR may be modulated by the interaction between the FADS genetic variant and the proportion of FA, particularly DGLA in serum phospholipids.
Conclusion
HOMA-IR and serum FA composition or FA ratios were associated with the FADS SNPs. Interestingly, FADS2 rs174575 and the proportion DGLA or AA in serum phospholipids may interactively affect HOMA-IR even in healthy people.
List of abbreviations
DGLA: dihomo-γ-linolenic acid; AA: arachidonic acid; Apo: apolipoprotein; BMI: body mass index; DBP: diastolic blood pressure; FADS: fatty acid desaturase; FEN: flapstructure specific endonuclease; HDL: high density lipoprotein cholesterol; HOMA: homeostasis model assessment; HWE: Hardy Weinberg Equilibrium; IR: insulin resistance; LA: linoleic acid; LD: Linkage Disequilibrium; LDL: low density lipoprotein; MetS: metabolic syndrome; MUFA: monounsaturated fatty acid; PUFA: polyunsaturated fatty acids; SBP: systolic blood pressure; SFA: saturated fatty acid; TCI: Total calorie intake; TEE: Total energy expenditure; TG: triglyceride
Competing disclosure
The authors declare that they have no competing interests.
Authors' contributions
All the authors were involved in the development of the study protocol and the experimental design. Sample collection and experiments were performed by OYK, HHL, JSC, and YJ. Data were analyzed by OYK and HHL OYK and JHL provided the research funding and wrote the manuscript. All the authors read, commented on, and contributed to the submitted manuscript.
Supplementary Material
Genotype and haplotype distribution of study population. It included genotype and haplotype distribution of 4 SNPs (FEN1-10154 rs174537G>T, FADS2 rs174575C>G, FADS2 rs2727270C>T and FADS3 1000778C>T) in the whole study subjects. The selected 4 SNPs satisfied the HWE (P > 0.05): minor T allele frequency of FEN1-10154 rs174537G>T was 0.312, minor G allele frequency of FADS2 rs174575C>G was 0.082, minor T allele frequency of FADS2 rs2727270C>T was 0.24, and minor T allele frequency of FADS3 1000778C>T was 0.293. This file also included the haplotype distribution of rs174537-rs174575-rs2727270-rs1000778: GCCC was the most highly frequent haplotype and the nonGCCC frequency was 0.450.
Macro-nutrient intake and energy expenditure of study population according to FEN1 -10154G>T, FADS2 rs174575, FADS2 rs2727270 and rsFADS3 1000778C>T. It included the information of Macro-nutrient intake and energy expenditure according to the genotypes of the FEN1 -10154G>T, FADS2 rs174575C>G, FADS2 rs2727270C>T and FADS3 rs1000778C>T, respectively. No significant genotype-associated differences were observed for total energy intake, proportions of energy intake derived from carbohydrates and fat in each of 4 FADS SNPs.
Contributor Information
Oh Yoen Kim, Email: ohkite@yonsei.ac.kr.
Hyo Hee Lim, Email: joyhh0819@yonsei.ac.kr.
Long In Yang, Email: longin@yonsei.ac.kr.
Jey Sook Chae, Email: jeysookchae@yonsei.ac.kr.
Jong Ho Lee, Email: jhleeb@yonsei.ac.kr.
Acknowledgements
We thank the research volunteers who participated in the studies described in this report. We also thank the researchers at DNA Link Ltd. for their technical help in DNA extraction and genotyping.
Financial support
This work was supported by the National Research Foundation, Ministry of Education, Science and Technology (Mid-career Researcher Program: 2010-0015017, 2010-0011003, M10642120002-06N4212-00210 and C00048), Republic of Korea.
References
- Warensjö E, Ohrvall M, Vessby B. Fatty acid composition and estimated desaturase activities are associated with obesity and lifestyle variables in men and women. Nutr Metab Cardiovasc Dis. 2006;16(2):128–136. doi: 10.1016/j.numecd.2005.06.001. [DOI] [PubMed] [Google Scholar]
- Riccardi G, Giacco R, Rivellese AA. Dietary fat, insulin sensitivity and the metabolic syndrome. Clin Nutr. 2004;23:447–456. doi: 10.1016/j.clnu.2004.02.006. [DOI] [PubMed] [Google Scholar]
- Vessby B. Dietary fat and insulin action in humans. Br J Nutr. 2000;83(Suppl 1):S91e6. doi: 10.1017/s000711450000101x. [DOI] [PubMed] [Google Scholar]
- Vessby B. Dietary fat, fatty acid composition in plasma and the metabolic syndrome. Curr Opin Lipidol. 2003;14:15–19. doi: 10.1097/00041433-200302000-00004. [DOI] [PubMed] [Google Scholar]
- Leeson CP, Mann A, Kattenhorn M, Deanfield JE, Lucas A, Muller DP. Relationship between circulating n-3 fatty acid concentrations and endothelial function in early adulthood. Eur Heart J. 2002;23:216–222. doi: 10.1053/euhj.2001.2728. [DOI] [PubMed] [Google Scholar]
- Katan MBJP, van Birgelen AP, Penders M, Zegwaard M. Kinetics of the incorporation of dietary fatty acids into serum cholesteryl esters, erythrocyte membranes, and adipose tissue: an 18-month controlled study. J Lipid Res. 1997;38:2012–2022. [PubMed] [Google Scholar]
- Dougherty RM, Galli C, Ferro-Luzzi A, Iacono JM. Lipid and phospholipid fatty acid composition of plasma, red blood cells, and platelets and how they are affected by dietary lipids: a study of normal subjects from Italy, Finland, and the USA. Am J Clin Nutr. 1987;45:443–455. doi: 10.1093/ajcn/45.2.443. [DOI] [PubMed] [Google Scholar]
- Glatz JF, Soffers AE, Katan MB. Fatty acid composition of serum cholesteryl esters and erythrocyte membranes as indicators of linoleic acid intake in man. Am J Clin Nutr. 1989;49:269–276. doi: 10.1093/ajcn/49.2.269. [DOI] [PubMed] [Google Scholar]
- Ma J, Folsom AR, Shahar E, Eckfeldt JH. Plasma fatty acid composition as an indicator of habitual dietary fat intake in middle-aged adults. The Atherosclerosis Risk in Communities (ARIC) Study Investigators. Am J Clin Nutr. 1995;62:564–571. doi: 10.1093/ajcn/62.3.564. [DOI] [PubMed] [Google Scholar]
- Vessby B, Gustafsson IB, Tengblad S, Boberg M, Andersson A. Desaturation and elongation of fatty acids and insulin action. Ann N Y Acad Sci. 2002;967:183–195. doi: 10.1111/j.1749-6632.2002.tb04275.x. [DOI] [PubMed] [Google Scholar]
- Emken EA, Adlof RO, Gulley RM. Dietary linoleic acid influences desaturation and acylation of deuterium-labeled linoleic and linolenic acids in young adult males. Biochim Biophys Acta. 1994;1213:277–288. doi: 10.1016/0005-2760(94)00054-9. [DOI] [PubMed] [Google Scholar]
- Di Stasi D, Bernasconi R, Marchiali R, Marfisi RM, Rossi G, Rognoni G, Sacconi MT. Early modifications of fatty acid composition in plasma phospholipids, platelets and mononucleates of healthy volunteers after low doses of n-3 polyunsaturated fatty acids. Eur J Clin Pharmacol. 2004;60:183–190. doi: 10.1007/s00228-004-0758-8. [DOI] [PubMed] [Google Scholar]
- Marquardt A, Stöhr H, White K, Weber BH. cDNA cloning, genomic structure, and chromosomal localization of three members of the human fatty acid desaturase family. Genomics. 2000;66:175–183. doi: 10.1006/geno.2000.6196. [DOI] [PubMed] [Google Scholar]
- Cho HP, Nakamura MT, Clarke SD. Cloning, expression, and nutritional regulation of the mammalian delta-6 desaturase. J Biol Chem. 1999;274:471–477. doi: 10.1074/jbc.274.1.471. [DOI] [PubMed] [Google Scholar]
- Cho HP, Nakamura M, Clarke SD. Cloning, expression, and fatty acid regulation of the human delta-5 desaturase. J Biol Chem. 1999;274:37335–37339. doi: 10.1074/jbc.274.52.37335. [DOI] [PubMed] [Google Scholar]
- Tanaka T, Shen J, Abecasis GR, Kisialiou A, Ordovas JM, Guralnik JM, Singleton A, Bandinelli S, Cherubini A, Arnett D, Tsai MY, Ferrucci L. Genome-wide association study of plasma polyunsaturated fatty acids in the InCHIANTI Study. PLoS Genet. 2009;5(1):e1000338. doi: 10.1371/journal.pgen.1000338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dupuis J, Langenberg C, Prokopenko I, Saxena R, Soranzo N, Jackson AU, Wheeler E, Glazer NL, Bouatia-Naji N, Gloyn AL. et al. New genetic loci implicated in fasting glucose homeostasis and their impact on type 2 diabetes risk. Nature Genetics. 2010;43:105–120. doi: 10.1038/ng.520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingelsson E, Langenberg C, Hivert MF, Prokopenko I, Lyssenko V, Dupuis J, Mägi R, Sharp S, Jackson AU, Assimes TL. et al. Detailed physiologic characterization reveals diverse mechanisms for novel genetic Loci regulating glucose and insulin metabolism in humans. Diabetes. 2010;59(5):1266–75. doi: 10.2337/db09-1568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinelli N, Girelli D, Malerba G, Guarini P, Illig T, Trabetti E, Sandri M, Friso S, Pizzolo F, Schaeffer L, Heinrich J, Pignatti PF, Corrocher R, Olivieri O. FADS genotypes and desaturase activity estimated by the ratio of arachidonic acid to linoleic acid are associated with inflammation and coronary artery disease. Am J Clin Nutr. 2008;88(4):941–949. doi: 10.1093/ajcn/88.4.941. [DOI] [PubMed] [Google Scholar]
- Schaeffer L, Gohlke H, Muller M, Heid IM, Palmer LJ, Kompauer I, Demmelmair H, Illig T, Koletzko B, Heinrich J. Common genetic variants of the FADS1 FADS2 gene cluster and their reconstructed haplotypes are associated with the fatty acid composition in phospholipids. Hum Mol Genet. 2006;15:1745–1756. doi: 10.1093/hmg/ddl117. [DOI] [PubMed] [Google Scholar]
- Malerba G, Schaeffer L, Xumerle L, Klopp N, Trabetti E, Biscuola M, Cavallari U, Galavotti R, Martinelli N, Guarini P, Girelli D, Olivieri O, Corrocher R, Heinrich J, Pignatti PF, Illig T. SNPs of the FADS gene cluster are associated with polyunsaturated fatty acids in a cohort of patients with cardiovascular disease. Lipids. 2008;43(4):289–299. doi: 10.1007/s11745-008-3158-5. [DOI] [PubMed] [Google Scholar]
- Mathews DR, Hosker JP, Rudenski AS, Naylor BA, Treacher DF, Turner RC. Homeostasis model assessment: Insulin resistance and b-cell function from fasting plasma glucose and insulin concentrations in man. Diabetologia. 1985;28:412–419. doi: 10.1007/BF00280883. [DOI] [PubMed] [Google Scholar]
- Folch J, Lees M, Sloane Stanley GH. A simple method for the isolation and purification of total lipids from animal tissues. J Biol Chem. 1957;226:497–509. [PubMed] [Google Scholar]
- Lepage G, Roy CC. Direct transesterification of all classes of lipids in a one-step reaction. J Lipid Res. 1986;27:114–120. [PubMed] [Google Scholar]
- Shim JS, Oh KW, Suh I, Kim MY, Shon CY, Lee EJ, Nam CM. A study on validity of a 299 semiquantitative food frequency questionnaire of Korean adults. Kor J Community Nutr. 2002;7:484–494. [Google Scholar]
- Christian JL, Greger JL. Nutrition for Living. Vol. 111. Redwood City, CA: Benjamin/Cummings; 1994. [Google Scholar]
- The American Dietetic Association. Handbook of clinical dietetics. 2. New Haven, CT: Yale University Press; 1992. pp. 5–39. [Google Scholar]
- Rodriguez Y, Christophe AB. Long-chain ω6 polyunsaturated fatty acids in erythrocyte phospholipids are associated with insulin resistance in non-obese type 2 diabetes. Clin Chim Act. 2005;354:195–199. doi: 10.1016/j.cccn.2004.11.018. [DOI] [PubMed] [Google Scholar]
- Fu Y, Luo N, Klein RL, Garvey WT. Adiponectin promotes adipocyte differentiation, insulin sensitivity, and lipid accumulation. J Lipid Res. 2005;46:1369–1379. doi: 10.1194/jlr.M400373-JLR200. [DOI] [PubMed] [Google Scholar]
- Krey G, Braissant O, L'Horset F, Kalkhoven E, Perroud M, Parker MG, Wahli W. Fatty acids, eicosanoids, and hypolipidemic agents identified as ligands of peroxisome proliferator-activated receptors by coactivator-dependent receptor ligand assay. Mol Endocrinol. 1997;11:779–791. doi: 10.1210/me.11.6.779. [DOI] [PubMed] [Google Scholar]
- Iwaki M, Matsuda M, Maeda N, Funahashi T, Matsuzawa Y, Makishima M, Shimomura I. Induction of adiponectin, a fat-derived antidiabetic and antiatherogenic factor, by nuclear receptors. Diabetes. 2003;52:1655–1663. doi: 10.2337/diabetes.52.7.1655. [DOI] [PubMed] [Google Scholar]
- Miyazaki Y, Mahankali A, Wajcberg E, Bajaj M, Mandarino LJ, DeFronzo RA. Effect of pioglitazone on circulating adipocytokine levels and insulin sensitivity in type 2 diabetic patients. J Clin Endocrinol Metab. 2004;89:4312–4319. doi: 10.1210/jc.2004-0190. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Genotype and haplotype distribution of study population. It included genotype and haplotype distribution of 4 SNPs (FEN1-10154 rs174537G>T, FADS2 rs174575C>G, FADS2 rs2727270C>T and FADS3 1000778C>T) in the whole study subjects. The selected 4 SNPs satisfied the HWE (P > 0.05): minor T allele frequency of FEN1-10154 rs174537G>T was 0.312, minor G allele frequency of FADS2 rs174575C>G was 0.082, minor T allele frequency of FADS2 rs2727270C>T was 0.24, and minor T allele frequency of FADS3 1000778C>T was 0.293. This file also included the haplotype distribution of rs174537-rs174575-rs2727270-rs1000778: GCCC was the most highly frequent haplotype and the nonGCCC frequency was 0.450.
Macro-nutrient intake and energy expenditure of study population according to FEN1 -10154G>T, FADS2 rs174575, FADS2 rs2727270 and rsFADS3 1000778C>T. It included the information of Macro-nutrient intake and energy expenditure according to the genotypes of the FEN1 -10154G>T, FADS2 rs174575C>G, FADS2 rs2727270C>T and FADS3 rs1000778C>T, respectively. No significant genotype-associated differences were observed for total energy intake, proportions of energy intake derived from carbohydrates and fat in each of 4 FADS SNPs.