Abstract
Unlike human L1 retrotransposons, the 5′ UTR of mouse L1 elements contains tandem repeats of ∼200 bp in length called monomers. Multiple L1 subfamilies exist in the mouse which are distinguished by their monomer sequences. We previously described a young subfamily, called the TF subfamily, which contains ∼1800 active elements among its 3000 full-length members. Here we characterize a novel subfamily of mouse L1 elements, GF, which has unique monomer sequence and unusual patterns of monomer organization. A majority of these GF elements also have a unique length polymorphism in ORF1. Polymorphism analysis of GF elements in various mouse subspecies and laboratory strains revealed that, like TF, the GF subfamily is young and expanding. About 1500 full-length GF elements exist in the diploid mouse genome and, based on the results of a cell culture assay, ∼400 GF elements are potentially capable of retrotransposition. We also tested 14 A-type subfamily elements in the assay and estimate that about 900 active A elements may be present in the mouse genome. Thus, it is now known that there are three large active subfamilies of mouse L1s; TF, A, and GF, and that in total ∼3000 full-length elements are potentially capable of active retrotransposition. This number is in great excess to the number of L1 elements thought to be active in the human genome.
Non-LTR LINE1 (L1) elements are non-long terminal repeat (non-LTR) retrotransposons that are capable of autonomous retrotransposition and have expanded to large copy numbers in mammalian genomes. The mouse genome contains >100,000 L1s comprising ∼10% of the genomic DNA (Hutchison et al. 1989). Most L1 elements are inactive, a consequence of 5′-end truncation, inversion, or mutation. The consensus sequence for full-length mouse L1s is ∼7 kb and, like human L1s, has two open reading frames (ORFs) and a 3′ poly(A) tail, and is flanked by short target site duplications (TSDs). Unlike human L1s, mouse L1s have a bipartite 5′ UTR consisting of tandemly repeated sequences of ∼200 bp called monomers, which are situated upstream of single-copy, nonmonomeric sequence. By linking monomers to reporter genes, it has been shown that they possess promoter activity and that increasing the number of monomers increases the level of transcription (Severynse et al. 1992; DeBerardinis and Kazazian 1999).
Phylogenetic analyses by Adey et al. (1994a) suggest that mouse L1 evolution has been dominated by a single lineage which has spawned several subfamilies of L1s differing in sequence at their 5′ ends. A-type subfamily and F-type subfamily members have monomer lengths of 208 bp and 206 bp, respectively (Fanning 1983; Loeb et al. 1986; Padgett et. al. 1988). Members of the more ancient V subfamily probably lack a 5′-repeated sequence (Jubier-Maurin et al. 1992). For some time it was thought that all F elements and most A elements had been rendered inactive by mutation, but that a small subset of A elements remained active in the genome. Evidence supporting this hypothesis includes the findings that some A-type elements are highly similar to each other, possess two intact ORFs, and are transcribed. However, their capacity for retrotransposition was not directly assayed (Shehee et al. 1987; Schichman et al. 1992; Martin 1995).
Unexpectedly, two disease-causing insertions, L1spa and L1orl, were found to be members of a large, young, and expanding subfamily of mouse L1s (Kingsmore et al. 1994; Takahara et al. 1996; Naas et al. 1998). The monomer sequence belonging to members of this TF subfamily was on average 72% identical to the consensus monomer sequence of F-type L1s. Based on the results of a retrotransposition assay of 11 randomly cloned TF-type L1s in cell culture, we estimated that a majority of full-length TF elements are potentially active (DeBerardinis et al. 1998).
Here we present evidence that a previously unreported subfamily of L1 elements exists in high copy number in the mouse genome and that many members of this novel subfamily are likely capable of retrotransposition.
RESULTS
A Unique 5′ UTR Distinguishes a New Mouse L1 Subfamily
Examination of the GenBank database revealed the presence of a previously undescribed subfamily of mouse L1 elements remarkable for their 5′ UTR regions. Like those of the previously described TF elements (DeBerardinis et al. 1998), the 5′ monomer arrays of L1s from this new subfamily are related to, but clearly distinct from, the monomers of F subfamily members. We have called these GF elements and have extracted from the database 17 members for characterization (Table 1).
Table 1.
GF-Type L1 Sequences Used in This Study
| L1 name | Accession # | GI # | Databasea | Coordinatesb | Intact ORF1/ORF2 | # full length monomersc | # truncated monomersc | % identity with body consensusd | Active w/wo CMV promoter |
|---|---|---|---|---|---|---|---|---|---|
| GF13 | AF146793 | 7684609 | nr | 114636–121199 | Y/Y | 2.2 | 2 | 99.5 | Y/N? |
| GF21 | AC021631 | 13310926 | htgs/nr | 68991–62229 | Y/Y | 3.3 | 2 | 99.5 | Y/Y |
| GF23 | AF229843 | 6862583 | nr | 42031–48206 | N/Y | 0.7 | 1 | 98.6 | |
| GF26 | AC002406 | 2981248 | nr | 19967–26229 | N/N | 1.2 | 1 | 96.1 | |
| GF27 | AC026767 | 12643015 | htgs/nr | 118223–110609 | N/Y | 2.2 | 2 | 99.4 | N/N? |
| GF36 | AC003996 | 2772523 | nr | 20312–27252 | Y/N | 1.4 | 3 | 96 | |
| GF43 | AL450393 | 11876100 | htgs | 7656–13977 | Y/Y | 1.1 | 2 | 99.6 | |
| GF46 | AL049866 | 7630118 | nr | 22185–15089 | Y/N | 5.1 | 1 | 99.3 | |
| GF56 | AC058786 | 11120825 | nr | 200048–206662 | Y/N | 2.4 | 2 | 99.6 | |
| GF62 | AC068252 | 13357510 | htgs | 161311–154202 | Y/Y | 6.1 | 1 | 99.6 | Y/Y |
| GF68 | AC006508 | 4454569 | nr | 167020–160414 | N/N | 2.2 | 2 | 97.7 | |
| GF71 | AC079221 | 10122065 | htgs | 58136–64915 | Y/N | 1.1 | 1 | 98.4 | |
| GF78 | AC007978 | 12000475 | nr | 42762–49862 | Y/N | 5.1 | 1 | 99.4 | |
| GF82 | AC083912 | 10697433 | htgs | 925–7430 | Y/Y | 2 | 2 | 99.2 | |
| GF84 | AC087134 | 11610860 | htgs | 179278–172998 | N/N | 1.7 | 0 | 92.6 | |
| GF251 | AF259071 | 8571430 | nr | 61240–55080 | Y/N | 1.1 | 0 | 96.4 | |
| GF253 | AF259073 | 8575573 | nr | 6526–256 | Y/N | 1.7 | 0 | 93.4 |
Database refers to the non-redundant or high-throughput genomic sequence databases of GenBank.
Coordinates are the first and last nucleotides in the GenBank record delimiting the L1.
See Figure 1B for description of monomer structure.
Identity with GF consensus sequence beginning 250 bp upstream of the start of ORF1.
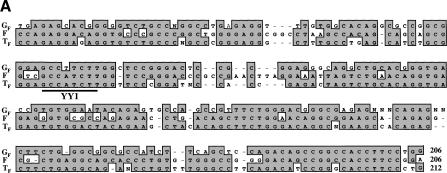
The 17 GF elements contain a variable number of monomers, averaging 2.4 with a maximum of 6. We aligned the sequences of 34 full-length monomers to obtain a consensus sequence 206 bp in length and 69% identical to the reconstructed F monomer consensus sequence described by Adey et al. (1994b) and 67% identical to the 212-bp TF monomer consensus (DeBerardinis and Kazazian 1999). Individual monomers ranged in size from 204 to 207 bp. The 3′ end of the GF consensus monomer sequence shown in Figure 1A corresponds to the point at which the monomer array joins the nonmonomeric 5′ UTR region 250 bp upstream of the start of GF ORF1. The monomer array of a TF element begins 258 bp upstream of TF ORF1. The nonmonomeric 5′ UTRs of the GF consensus and TF consensus sequences are 85% identical.
Figure 1.
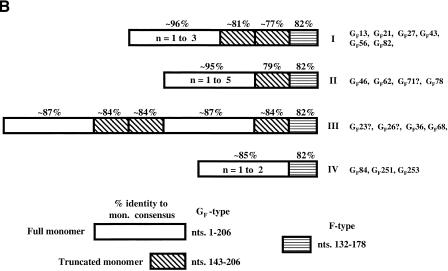
(A) Alignment of the 206-bp GF monomer consensus sequence, the 206-bp reconstructed F-consensus monomer (Adey et al. 1994b), and the 212-bp TF monomer consensus (DeBerardinis and Kazazian 1999). The location of the consensus YY1-binding sequence is underlined. (B) GF element variants (labeled I– IV) with different patterns of 5′ UTR monomer organization. The fragment shown with horizontal stripes in each element is in the nonmonomeric 5′ UTR and is 82% identical to sequence in the F-consensus monomer. GF monomers are either full-length (204–207 bp) or truncated (∼64 bp). The monomeric regions of some clones (indicated by “?”) are too short to be certain of their variant group.
The organization of the 5′ UTR of GF elements is atypical of mouse L1s. The nonmonomeric region of the 5′ UTR of all GF elements contains a 47-nt sequence which is 82% identical to a portion of the F monomer consensus (nts 132–178) (Fig. 1B). The monomer arrays are organized in four distinct patterns. At the 3′ end of many arrays are 5′ truncated monomers which exist singly (patterns II and III) or as a pair (pattern I) and which consist of the last 64 bps (nts 143–206) of a full-length monomer. Pattern III also has two additional truncated monomers upstream of the first full-length monomer. All of the abbreviated monomers are the same length and only 77%–84% identical to the consensus GF monomer (69%–75% identical to the F consensus). Most GF elements fall within patterns I and II whose full-length monomers are greater than 95% identical to the GF consensus (∼70% identical to the F consensus).
DeBerardinis and Kazazian (1999) noted that many TF L1s were truncated at their 5′ ends in the vicinity of a predicted binding site for the transcription factor YY1 (GCCATCTT, Fig. 1A). A YY1 site is also present in F monomers and in the human L1 5′ UTR (Becker et al. 1993). Twenty-six of the 31 TF sequences examined were truncated within 25 bp upstream or downstream of the monomer YY1 binding sequence (DeBerardinis and Kazazian 1999; J.L. Goodier, unpubl.). All GF monomers have a single nucleotide change which alters this sequence (GCCTTCTT), and this may explain why GF elements do not tend to truncate near this site (only 3 of 17 elements truncated within 25 nts of this sequence).
ORF1 Length Polymorphism Region (LPR)
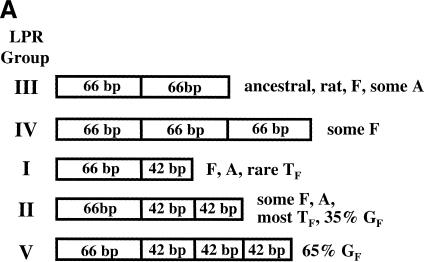
All mouse L1 subfamilies contain an LPR within the N-terminal one-third of ORF1, consisting of tandemly repeated blocks of 66 bp and/or 42 bp that do not interrupt the reading frame of the protein. These LPRs were defined by Schichman et al. (1992) and Adey et al. (1994a), and the system of nomenclature proposed by Mears and Hutchison (2001) is summarized in Figure 2A. Most A-type elements have 66-42 bp (Group I) or 66-42-42 bp (Group II) repeat structures, although some also have a 66-66 bp (Group III) pattern. F-type elements belong to Groups I–IV. Although almost all TF elements have a Group II repeat structure (DeBerardinis et al. 1998; J.L. Goodier, unpubl.), Mears and Hutchison (2001) identified a novel TF Group I element. Thirty-five percent (6/17) of GF L1s also belong to Group II. However, the majority of GF elements have an extra 42-nt repeat (i.e., 66-42-42-42 nt), a pattern not previously reported for any mouse L1 element. We have therefore placed GF elements in a new LPR group, Group V. The four LPR repeats of the GF consensus sequence are aligned in Figure 2B.
Figure 2.
(A) Classification scheme for mouse L1 ORF1 length polymorphism region (LPR) groups, as defined by Mears and Hutchison (2001). LPRs consists of tandem arrays of 66-bp and/or 42-bp blocks, and each begins with a 66-bp block. The 42-bp block is homologous with the 3′ portion of the 66-bp repeat. The novel GF variant has been designated LPR Group V. (B) Alignment of the ORF1 LPR repeats from the GF consensus sequence. As shown in the first line of the alignment, the 5′-most 24 bp of the 66-bp repeat has partial homology with the 42-bp repeat.
Features of the Body of GF Elements
We derived a nucleotide consensus sequence for the bodies of the 17 GF members of our dataset beginning at the nonmonomeric 5′ UTR. The bodies of the 17 GF elements are 92.6%–99.6% identical to their consensus sequence (Table 1). In contrast, 11 randomly cloned TF elements were on average 99.8% identical to their consensus sequence (DeBerardinis et al. 1998). This suggested that different subgroups of GF elements exist and that at least some of these are older than TF elements. We therefore performed neighbor-joining phylogenetic analyses which included both the GF members of our dataset and F, A, and TF elements of the testset used in the analysis by Mears and Hutchison (2001). Results using a portion of the nonmonomeric 5′ UTR defined as region α by Mears and Hutchison (2000) and corresponding to nucleotides 1539–1740 of L1MdA2 (Loeb et al. 1986) are shown in Figure 3A.
Figure 3.
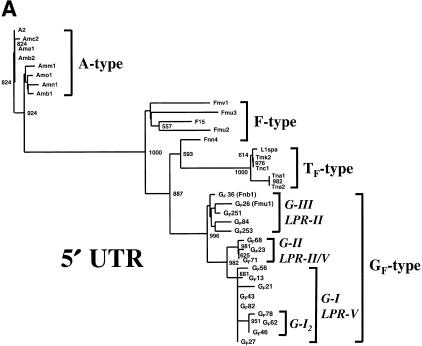
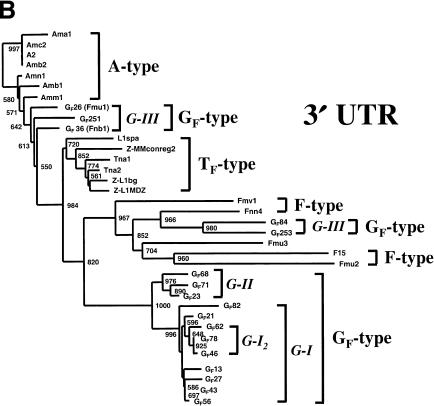
(A) Phylogenetic analysis of that portion of the 5′ UTR of mouse L1s defined as region α by Mears and Hutchison (2001) and corresponding to nucleotides 1539–1740 of the A-type element L1Md-A2. The unrooted tree was constructed by the nearest neighbor-joining method (Saitou and Nei 1987) and includes the 17 members of the GF dataset and members of the dataset used by Mears and Hutchison (2001). GF clades identified by the analysis are named G-I, G-II, and G-III, and their corresponding LPR groups are indicated. Significant bootstrap values (of 1000 replicates) are shown at the nodes. (B) Phylogenetic analysis of 3′ UTR region corresponding to nts 6701–7333 of L1MdA2. Also included in this analysis are L1s classified by Hardies et al. (2000) as members of the “Z subfamily”: MMconreg2, L1bg (Perou et al. 1997), and L1MdZ (Kraft et al. 1992). These group with TF-type elements.
Three distinct GF clades (labeled G-I, G-II, and G-III) are evident and are supported by 1000 rounds of bootstrap analysis. Most of the elements (9 of 17) belong to clades G-I and are on average 99.1% identical to each other and 94.0% identical to the TF consensus (DeBerardinis et al. 1998). These elements also have monomers closest in sequence to the consensus and 5′ UTR structures as depicted in Figure 1B, patterns I and II. The existence of a subset of clade G-I, designated G-I2, is weakly supported across the length of the element. Members of clade G-II are on average 97.9% identical to each other, and members of clade G-III are most dissimilar (94.8%) and have 5′ UTR patterns III and IV (Fig. 1B). Only GF68 (Clade G-II) and members of clade G-III belong to ORF1 LPR Group II; all others belong to the novel LPR Group V (66-42-42-42 bp). Two members of clade G-III, GF36 (Fnb1) and GF26 (Fmu1), were previously defined as representatives of a young clade of F elements (F-II2) by Mears and Hutchison (2001). While these authors did not describe the monomer structure of these elements, it is now evident that the monomers are more closely related to the GF (87% identity) than the F (74% identity) monomer consensus.
Phylogenetic analysis of a portion of the 3′ UTR (nts 6701–7333 of L1MdA2 and region θ as defined by Mears and Hutchison 2001) supports clades G-I and G-II which were established by the 5′ UTR analysis (Fig. 3B). However, clade G-III splits into two subgroups whose members have undergone separate recombination events. As proposed by Mears and Hutchison (2001), GF36 (Fnb1) and GF26 (Fmu1), together with GF251, have been formed by recombination with either an A-type Group II LPR element or a young F-type element donating the 3′ end. On the other hand, GF84 and GF253 group with older F-type elements at their 3′ ends. Phylogenetic analysis of the set of subsequences spanning the entire L1 used by Mears and Hutchison (2001) reveal that the former recombination breakpoint occurred in the central portion of ORF2 (regions ɛ, nts 2798–4115, or ξ, nts 4116–5455), whereas the latter occurred closer to the N-terminus of ORF2 (region ɛ). Clades G-I and G-II are supported by analyses of regions spanning the length of the L1, and these clades do not contain recombinant members. Included in the analysis of region θ are L1s designated as members of a new “Z subfamily” by Hardies et al. (2000). These L1s are not GF elements and, as previously reported by Mears and Hutchison (2001), are in fact members of the TF subfamily.
GF Elements Transduce 3′ Flanking DNA
Retrotransposing L1s can occasionally transduce nonL1 DNA flanking their 3′ ends to new genomic locations. This is a consequence of the inherent weakness of the L1 polyadenylation signal which during transcription is occasionally bypassed in favor of a stronger signal downstream. We previously identified (in the mouse genome database) two transduction events involving A-type elements which in both instances mobilized 3 kb of sequence (Goodier et al. 2000). Two GF elements in our dataset have transduced flanking DNA (Fig. 4). Element GF71 transduced 558 nts, while the 3′ flank of element GF27 is the final product of two consecutive transduction events, an initial movement of 24 nts followed by a second retrotransposition event and the transduction of an additional 1033 nts.
Figure 4.
Two GF elements, GF27 and GF71, have transduced DNA flanking their 3′ ends to new genomic locations. Consecutive retrotransposition events have created the composite transduction of GF27. TSDs are shown in bold lowercase letters, and putative hexanucleotide poly(A) signals are in bold uppercase letters.
GF Elements Have Been Recently Active in the Mouse
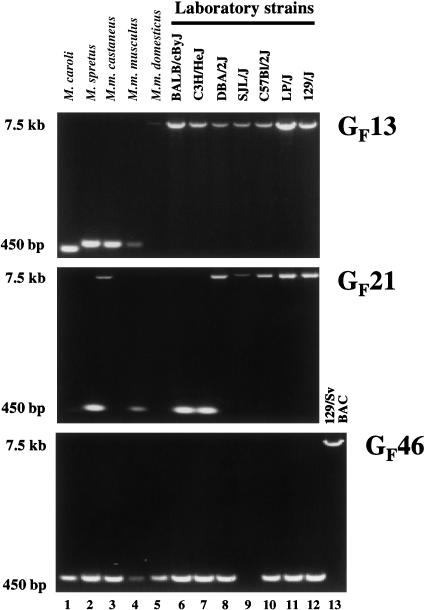
The high degree of sequence similarity among some of the GF elements implies that these elements may have recently dispersed within the mouse genome. To test this, we examined the genomes of Mus subspecies and laboratory strains for the presence or absence of three GF elements (Fig. 5). We designed PCR primers flanking the elements which would amplify a 7.5-kb filled site or a 450-bp empty site. None of the GF elements were detected in Mus spretus or Mus musculus genomes. GF13 was detected in laboratory strains only, whereas GF21 was present in M. m. castaneus but absent from some laboratory strains. Surprisingly, GF46 was absent from all samples tested. However, it was possible to amplify GF46 DNA from the bacmid library clone sequenced for the GenBank entry. The library was generated from an embryonic stem cell library of strain 129/Sv. We confirmed the presence of the L1 sequence in this clone by PCR amplification and sequencing.
Figure 5.
Some GF elements are recent insertions. Shown are PCR reaction products which verify the presence (7.5-kb product) or absence (450-bp product) in the genomes of individual mice of different subspecies and laboratory strains. A faint, ∼7.5-kb product is present in lane 5 (M. m. domesticus) of GF21 which we have not been able to confirm as L1-related by sequencing. In the case of GF46, no PCR product is seen in lane 9 (strain SJL/J), although in other reactions we have been able to amplify an obvious 450-bp product. The 7.5-kb product labeled 129/Sv BAC (lane 13 of GF46) was amplified from bacmid DNA of CITB/Research Genetics clone #437P9.
Some A and GF Subfamily Members Have Retrotransposition Capability
We previously tested A-type L1s in the cell culture retrotransposition assay (DeBerardinis et al. 1998). From phage library clones we PCR-amplified ten A-type elements without their monomers and cloned these into a retrotransposition vector under transcriptional control of a CMV promoter (Moran et al. 1996). One of the ten cloned elements supported retrotransposition. However, the possibility existed that PCR errors introduced during cloning could have inactivated otherwise potentially active elements. Therefore, we directly isolated eight A-type elements (including four elements tested in the previous experiment and four new elements) from bacteriophage and inserted each without their monomers into a retrotransposition vector in which both the monomer array of the TF element L1spa and the CMV promoter directed expression. In all, two (A101 and A102) of a total of 14 different A elements from both experiments (14%) were capable of supporting retrotransposition (Table 2). Although transcription of these A-type elements was not driven by their endogenous promoters, Severynse et al. (1992) showed that a single A monomer is sufficient to direct transcription. Since A101 possesses 3.5 monomers and A102 possesses at least two monomers, it is probable that these two elements are capable of transcription and retrotransposition in vivo. Saxton and Martin (1998) estimated that the diploid genome of mouse strain 129/Sv contains 6500 full-length A-type elements. If 14% are active, then over 900 potentially active A elements reside in the genome.
Table 2.
GF Elements Retrotranpose in Human TK− 143B Cells and HeLa Cells
| Name | Strain Clone #) | Retrotransposition frequency | |||
|---|---|---|---|---|---|
| 143B cells | HeLa cells | ||||
| with CMV | w/o CMV | with CMV | w/o CMV | ||
| GF13 | 120/Ola (19–37) | 1 | <1 | ||
| C57Bl/2J (14–25) | 0 | <1 | |||
| 129/Ola (19) | 12 | 26 | |||
| C57Bl/6J (15) | 3 | 6 | |||
| C57Bl/6J (14) | 6 | 4 | |||
| 129/Sv+C/+P (22) | 0 | nk | |||
| GF21 | C57Bl/6J (18–17) | 1482 | 720 | ||
| C57Bl/6J (35–10) | 1418 | 1683 | |||
| C57Bl/6J (18–36) | 1150 | 1523 | |||
| C57Bl/6J (35–37) | 727 | 2942 | |||
| C57Bl/6J (35–92) | 394 | 2240 | |||
| SJL/J (86–39) | 380 | 816 | |||
| GF27 | C57Bl/6J (55–62) | 0 | 0 | ||
| C57Bl/6J (55–48) | 0 | 2 | |||
| GF62 | LP/J (92–59) | 35 | 1 | ||
| LP/J (92–45) | 65 | 69 | |||
| A101 | 332 | 792 | |||
| A102 | 682 | 3767 | |||
| TF9.1 | 557 | 1173 | |||
| L1spa | 121 | 352 | |||
| L1spa ORF2 D709Y | 0 | 0 | |||
| L1RP | 1884 | ∼7300 | |||
| JM105 ORF2 D709Y | 0 | 0 | |||
Shown for each construct is the average retrotransposition frequency (G418R foci/106 hygromycin-resistant cells) for at least three independent transfections. Cloned GF constructs were obtained by PCR amplification of DNA from a single individual from several different laboratory strains of mouse. Clonal differences in activity may be due to PCR errors. DNA from strains 129/Ola and 129/Sv+C/+P was obtained from pluripotent embryonic stem cells (ATCC CRL-1821 (ES-E14TG2a) and ATCC CRL-1934 (ES-D3), respectively). The TF subfamily member, TF9, is described in DeBerardinis et al. (1998). L1RP, a human L1 discovered as a de novo insertion in the RP2 gene of a retinitis pigmentosa patient, is the most active L1 that has so far been tested in the cell culture assay (Kimberland et al. 1999). As negative controls, we used reverse-transcriptase defective alleles of the TF element, L1spa (ORF2 D709Y), and the human element, JM105 (ORF2 D709Y) (Moran et al. 1996; Naas et al. 1998).
Seven of the 14 A-type elements tested in the retrotransposition assay have a Group II ORF1 LPR (66-42-42 nts). Both active elements have a Group I (66-42 nts) LPR and belong to a younger transcribed A-type subset whose members possess >99.5% identity with each other in their 5′ UTR and ORF1 regions (Schichman et al. 1992). RNP-58B, a cDNA copy of an L1 RNA isolated by Martin (1995) from a ribonucleoprotein (RNP) particle, is also a member of this subset. L1 RNPs are comprised of ORF1 protein bound to L1 RNA and are thought to be intermediates in the retrotransposition process. The ORF1 sequences of the active elements A101 and A102 are identical, while RNP-58A differs at a single position (R22C). Only four residues in the ORF2 of RNP-58A differ from those of both A101 and A102: S86L in the endonuclease domain, T359K situated between the endonuclease and reverse transcriptase domains, and R761W and V/A928D, which are not conserved among mammalian species. Therefore, it is likely that RNP-58A is also an active L1 element.
Using PCR, we directly amplified DNA of three GF elements with two complete ORFs from the genomes of several Mus m. domesticus strains. We also tested GF27, which has a frameshift mutation 16 amino acid residues from the end of ORF1, but found this element to be inactive in human 143B cells and barely active when driven by a CMV promoter in HeLa cells. All elements belonged to clade G-I (Fig. 3A). They were cloned with their entire 5′ UTRs intact into retrotransposition vectors both with and without an exogenous CMV promoter and tested for retrotransposition activity. All three of the cloned elements with intact ORFs were capable of retrotransposition in cell culture when linked with a CMV promoter (Table 2). GF21 and GF62 were clearly active when driven by their endogenous promoters alone, while GF13 may be active at a very low level. GF21 and the A-type element, A102, are the most active mouse L1s tested in the retrotransposition assay to date. After screening a mouse strain 129/OlaHsd genomic library with a GF monomer probe, we estimated that ∼1500 full-length GF elements reside in the diploid mouse genome. This number was confirmed by probing GenBank's high-throughput genomic sequence (htgs) mouse database with amino acid sequence from the N-terminus of the GF consensus ORF1. It is likely that 440 full-length GF elements have two complete ORFs (since 5 of the 17 members of our dataset have intact ORFs). These data suggest that ∼400 potentially active full-length GF elements may exist in the mouse genome.
DISCUSSION
GF elements belong to a novel subfamily of mouse L1s with unique monomeric 5′ UTR sequence. At different times in evolution, murine L1s have acquired novel 5′ sequences, perhaps by recombination or by the capture of upstream promoter sequence. The shuffling of L1 fragments, whether by recombination or template switching, is a leit motif in the evolution of murine subfamilies. In Rattus norvegicus, the youngest subfamily, L1mlvi2, acquired its ORF1 from an ancestral L1 (Hayward et al. 1997). Flood et al. (1998) identified unusual hybrid L1s in the mouse which possess fused 3′ sequence homologous to a fragment in the first intron of Cɛ immunoglobulin. Five hybrid elements have poly(A) signals and A-rich 3′ ends, and one has TSDs, evidence of retrotransposition. (Since the C-terminus of ORF2 was displaced by the fusion, transcomplementation by ORF2 protein from an intact L1 may have allowed this hybrid element to multiply). In the mouse, Saxton and Martin (1998) and Mears and Hutchison (2001) have shown that TF elements are recombinant, deriving 5′ UTR sequence from an “older” F-type element and 3′ sequence from a “young” F-type or an A-type element. Similarly, by phylogenetic reconstruction we show that 5 of the 17 GF elements in our dataset are recombinant. Three of these elements, GF36, GF26, and GF251, are members of a subclade which Mears and Hutchison (2001) designated F-II2 and estimated to be 1.4–2 million years old.
In addition to a novel 5′ UTR, a majority of GF elements also contain a unique length polymorphism in ORF1. The LPR is contained within the N-terminal one-third of ORF1, a region which is highly divergent in sequence among different mammalian L1 families. In this region, human L1 ORF1 contains a putative leucine zipper domain implicated in protein–protein interaction (Holmes et al. 1992; Hohjoh and Singer 1996). Mouse elements do not contain a leucine zipper. However, Martin et al. (2000) predicted that the N-terminal region of mouse ORF1 is capable of forming a coiled-coil structure, and they demonstrated that this region mediates protein multimerization. Furthermore, employing two-hybrid assays, those authors showed that ORF1 protein of the TF element L1spa dimerizes more strongly than does protein of an A-type element, and they speculate that this might be due to the longer LPR region of the TF ORF. It would be interesting to determine whether those GF elements with the long Group V LPR show even stronger ORF1 interactions. However, the finding that two very active A-type elements have the short Group II LPR indicates that LPR length does not correlate with retrotransposition capacity.
Three ORF1 protein variants, p41, p43, and p43.5, have been detected in mouse F9 cells by Kolosha and Martin (1995). A-type and TF-type elements have been identified with the first two forms, respectively. Phosphatase treatment suggested that the 43.5 kD protein was a phosphorylated form of p43. Furthermore, the 43.5 kD form is too small to represent ORF1 protein expressed from GF elements with the Group V LPR (expected size 44.5 kD). Previous RT-PCR analyses also failed to detect ORF1 sequence with a Group V LPR (Schichman et al. 1992; Kolosha and Martin 1995), although the primers used in those studies likely would not have detected GF RNA.
It would appear that the dominant active elements in the mouse genome are members of the TF subfamily which have accounted for at least five of the seven known mutagenic insertions. These include L1spa (Kingsmore et al. 1994; Mulhardt et al. 1994), L1orl (Takahara et al. 1996), and insertions into the beige (Perou et al. 1997), black-eyed white (Yajima et al. 1999), and disabled1 (Kojima et al. 2000) genes. The disabled gene L1 lacks a poly(A) tail, 3′ end, and TSDs, and thus may not be a bona fide retrotransposition event. The remaining insertions into the sodium channel gene Scn8a (Kohrman et al. 1996) and the copper-transporting ATPase gene Atp7a (Cunliffe et al. 2001) are too short to assign to a subfamily (Kohrman et al. 1996). However, the number of elements characterized to date is still small, and we expect that A and GF insertions will eventually be detected. Phylogenetic analysis of several GF elements reveals their relatively recent expansion in the Mus genus. All three GF elements tested here were absent from the M. spretus and M. m. musculus (CZECH II/Ei) genomes. M. spretus diverged from the M. musculus lineage 1–3 million years ago (Thaler 1986), and M. musculus subspecies shared a common ancestor 350,000 years ago (She et al. 1990). We have also shown that element GF46 is a de novo insertion specific to a particular mouse substrain or perhaps even a single individual.
Three distinct L1 subfamilies possessing unique promoter regions are simultaneously capable of retrotransposition in the mouse genome. Approximately 400 GF elements and 900 A elements are potentially active in the mouse. DeBerardinis et al. (1998) estimated that mouse strain 129/OlaHsd contains ∼4800 full-length TF elements, while Saxton and Martin (1998) reported that the diploid genome of strain 129/Sv contains 2500 full-length elements. Examination of GenBank's mouse htgs database with amino acid sequence from the N-terminus of ORF1 suggested ∼3000 as the number of TF elements in the mouse genome. Assuming that 64% of TF elements are retrotranspositionally competent (DeBerardinis et al. 1998), ∼1800 of these elements may be present in the diploid genome. Thus the total number of potentially active L1 retrotransposons from the A, TF, and GF subfamilies in the diploid genome is roughly 3000. This is in sharp contrast to 40–70 active L1s in the human genome, most belonging to a single subfamily, Ta (Skowronski et al. 1988; Sassaman et al. 1997).
The large number of potentially active elements in the mouse accounts for the much higher rate of mutation due to L1 retrotransposition. It has been estimated that L1 insertions make up ∼2.5% of spontaneous mutations in the mouse, and only 0.07% in humans (Kazazian and Moran 1998). Therefore, it is probable that retrotransposition has been a factor in driving the high rate of evolution in the mouse (She et al. 1990).
METHODS
Isolation of L1s, Subcloning and Retrotransposition Assay
Using GF monomer sequence to search GenBank entries, we examined the highest scoring contigs in both the nonredundant (score >150) and high-throughput genomic sequence (score >200) databases and extracted from these 17 GF elements (Table 1). Only full-length elements having obvious TSDs were considered.
To isolate full-length A elements, we screened a phage genomic library of the strain 129/OlaHsd embryonic stem cell line E14TGa with a monomer from the A-type element L1Md-A2 (Loeb et al. 1986). A-type elements were isolated from the phage and cloned by two methods. First, we amplified ten elements by PCR (primers: L1ANOT5P, CGTACGCGGCCGC TGGTTCGAACACCAGATATCTGGG; L1ABSTZ3P, ATACG TATACATTTCCAATGCTATACCAAAAG). These were directly cloned into our retrotransposition vector under the control of a CMV promoter (Moran et al. 1996). Secondly, we excised eight A-element sequences from purified phage DNA (four were duplicates of those cloned by PCR amplification and four were unique) and inserted them into the retrotransposition construct in the same manner as described for cloning TF elements (DeBerardinis et al. 1998). Briefly, we swapped SmaI-SfiI fragments (including 296 bp of the A-type nonmonomeric 5′ UTR and 495 bp of the 3′ UTR) for the corresponding fragment of the TF element L1spa. Since the resulting constructs contained both a CMV promoter and L1spa monomers, we could assay A-type element ORF protein activity only. Complete sequences of the two active A elements have been deposited in GenBank (accession numbers AY053455 and AY053456).
To test GF elements, we selected from our dataset four elements, three with two intact ORFs, and we PCR-amplified these from genomic DNA of several mouse strains using primers closely flanking the TSDs. These elements were cloned into the retrotransposition vector with or without a CMV promoter. All cell culture retrotransposition assays were performed in either human 143B TK− osteosarcoma (ATCC# CRL-8303) or HeLa cells as described (Moran et al. 1996). In two previous papers (Naas et al. 1998; DeBerardinis et al. 1998) we reported testing TF element retrotransposition in mouse LTK− cells. It recently came to our attention that the cells assayed were not mouse LTK− cells but rather human TK− 143B cells. We have determined that L1s are also active in LTK− cells but at a level two orders of magnitude lower than reported.
Analysis of GF Polymorphism
Mouse genomic DNA was obtained from the Jackson Laboratories. Primers used for genomic amplification of the following elements were as follows. GF13: GTCTGCGTAAGGCCT GTGCTTGC (1AF1465P) and GCAAGTTTGATCTTCACCAT CAGG (2AF1463P); GF21: TTCCTGATATGAAGCCTATG TACC (3AC02163P5) and TCTCTGAATGTTACATGATTTGGC (4AC02163P3); GF46: GCCTGTGCTCTAAATCGCCAACAC (5AL049ES5P) and AGAGAAGTACCTGCGTGGCCCACC (6AL049ES3P). The M. spretus sample is SPRET/Ei, the M. m. castaneus sample is CAST/Ei, the M. m. musculus sample is CZECH II/Ei, and the M. m. domesticus sample is WSB/Ei. Amplifications were performed with the Expand Long PCR system (Roche), and PCR conditions were optimized for the generation of both empty site (∼450 bp) and filled site (7.1–7.6 bp) products using an MJ Research PTC-200 Peltier Thermal Cycler. DNA mixing studies showed that our PCR conditions could detect in a single reaction both empty and filled sites (data not shown). Despite this ability, no individual mice heterozygotic for the presence and absence of an L1 were discovered.
Many PCR products were gel purified and directly sequenced to confirm the presence or absence of the L1. The L1 GF46 was also amplified from the bacmid clone originally used to generate the GenBank sequence entry (CITB/Research Genetics clone #437P9, originating from mouse strain 129/Sv).
Estimation of GF Copy Number
We screened the mouse genomic phage library with the monomer array from element GF46. The array was PCR-amplified from the bacmid clone DNA (CITB/Research Genetics clone #437P9) using the 5′ primer, TAAGGAATTCCATC TATTTCGAGGGGGTAAAG (3AL049ECOR), and 3′ primer, TAAGCTCGAGTCCCAGAAGCTGTGTTGCTTTG (2PROBEGXHO). The GenBank entry (GI# 7630118) shows GF46 to have 5.1 monomers. However, our PCR product was 600 bp longer than expected, suggesting that GF46 in reality contains about eight monomers and that the GenBank sequence was misassembled. We confirmed that our probe TF contained no non-L1 DNA by end-sequencing and by analysis with restriction enzymes cutting only once within each monomer. The PCR product was cloned into XhoI/EcoRI sites of pBS KS− and reexcised for use as a hybridization probe to screen nylon filters lifted from five plates containing about 5 × 104 plaques each. Hybridization and washing conditions were high-stringency under standard conditions (Sambrook and Russell 2001). We confirmed by slot blot analysis that the probe would not cross-hybridize to the TF element L1spa, an A-monomer probe from L1Md-A2 (accession #M13002; Loeb et al. 1986), or to an F-monomer probe from Padgett et al. (1988) but would hybridize to clones GF21 and GF62. Knowing the average insert size of the library (16 kb) and the mouse diploid genome size (6 × 109), we estimated the GF copy number to be ∼1500 elements.
We also examined GenBank's high-throughput genomic (htgs) mouse database to confirm our estimate of the number of full-length GF or TF elements, by using the first 63 amino acid residues of consensus ORF1 as query sequence in a TBLASTN search. This region of ORF1 was used because it contains both GF and TF subfamily-specific residues and because L1s that contain the 5′ end of the ORF1 coding region are likely to represent full-length elements. At the time the searches were performed, the htgs database represented 9.6% of the total mouse genome. After counting the number of sequence hits, we extrapolated to determine the total number of GF and TF elements in the diploid genome (1500 and 3000 elements, respectively).
DNA Sequence and Phylogenetic Analyses
Sequences were aligned with MacVector 6.53 (Oxford Molecular Group) or ClustalW 1.8 and consensus sequences were determined with MacVector. Phylogenetic analyses were performed with the ClustalW program using the neighbor joining algorithm of Saitou and Nei (1987) with exclusion of gaps. Significance was determined by 1000 bootstrap analyses. Unrooted maximum parsimony analyses were also performed as confirmation (using the DNAPARS program in PHYLIP ver. 3.5c; Felsenstein 1993). Trees produced by the phylogenetic analyses were viewed and manipulated with TreeView.
Acknowledgments
We thank R.J. DeBerardinis for critical reading of the manuscript, E. Luning-Prak for mouse genomic DNA, and K. Kaestner for supplying the mouse genomic library. The work was supported by a grant from the NIH to H.H.K and a Howard Hughes Medical Institute Predoctoral Fellowship to E.M.O.
Footnotes
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked advertisement in accordance with 18 USC section 1734 solely to indicate this fact.
E-MAIL jgoodier@mail.med.upenn.edu, kazazian@mail.med.upenn.edu; FAX (215) 573–7760.
Article published on-line before print: Genome Res., 10.1101/gr.198301.
Article and publication are at http://www.genome.org/cgi/doi/10.1101/gr.198301.
REFERENCES
- Adey NB, Schichman SA, Graham DK, Peterson SN, Edgell MH, Hutchison CA., III Rodent L1 evolution has been driven by a single dominant lineage that has repeatedly acquired new transcriptional regulatory sequences. Mol Biol Evol. 1994a;11:778–789. doi: 10.1093/oxfordjournals.molbev.a040158. [DOI] [PubMed] [Google Scholar]
- Adey NB, Tollefsbol TO, Sparks AB, Edgell MH, Hutchison CA., III Molecular resconstruction of an extinct ancestral promoter for mouse L1. Proc Natl Acad Sci. 1994b;91:1569–1573. doi: 10.1073/pnas.91.4.1569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker KG, Swergold GD, Ozato K, Thayer RE. Binding of the ubiquitous nuclear transcription factor YY1 to a cis regulatory sequence in the human LINE-1 transposable element. Hum Mol Genet. 1993;2:1697–1702. doi: 10.1093/hmg/2.10.1697. [DOI] [PubMed] [Google Scholar]
- Cunliffe P, Reed V, Boyd Y. Intragenic deletions at Atp7a in mouse models for Menkes disease. Genomics. 2001;74:155–162. doi: 10.1006/geno.2001.6529. [DOI] [PubMed] [Google Scholar]
- DeBerardinis RJ, Kazazian HH., Jr Analysis of the promoter from an expanding mouse retrotransposon subfamily. Genomics. 1999;56:317–323. doi: 10.1006/geno.1998.5729. [DOI] [PubMed] [Google Scholar]
- DeBerardinis RJ, Goodier JL, Ostertag EM, Kazazian HH., Jr Rapid amplification of a retrotransposon subfamily is evolving the mouse genome. Nat Genet. 1998;20:288–290. doi: 10.1038/3104. [DOI] [PubMed] [Google Scholar]
- Fanning TG. Size and structure of the highly repetitive BamHI element in mice. Nucleic Acids Res. 1983;11:5073–5091. doi: 10.1093/nar/11.15.5073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felsenstein J. PHYLIP (Phylogeny Inference Package) Version 3.5c, Distributed by the author. Dept. of Genetics, University of Washington; 1993. [Google Scholar]
- Flood WD, Rogozin IB, Ruvinsky A. A novel subfamily of LINE-derived elements in mice. Mamm Genome. 1998;9:881–885. doi: 10.1007/s003359900887. [DOI] [PubMed] [Google Scholar]
- Goodier JL, Ostertag EM, Kazazian HH., Jr Transduction of 3′-flanking sequences is common in L1 retrotransposition. Hum Mol Genet. 2000;9:653–657. doi: 10.1093/hmg/9.4.653. [DOI] [PubMed] [Google Scholar]
- Hardies SC, Wang L, Zhou L, Zhao Y, Casavant NC, Huang S. LINE-1 (L1) lineages in the mouse. Mol Biol Evol. 2000;17:616–628. doi: 10.1093/oxfordjournals.molbev.a026340. [DOI] [PubMed] [Google Scholar]
- Hayward BE, Zavanelli M, Furano AV. Recombination creates novel L1 (LINE-1) elements in Rattus norvegicus. Genetics. 1997;146:641–654. doi: 10.1093/genetics/146.2.641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hohjoh H, Singer MF. Cytoplasmic ribonucleoprotein complexes containing human LINE-1 protein and RNA. EMBO J. 1996;15:630–639. [PMC free article] [PubMed] [Google Scholar]
- Holmes SE, Singer MF, Swergold GD. Studies on p40, the leucine zipper motif-containing protein encoded by the first open reading frame of an active human LINE-1 transposable element. J Biol Chem. 1992;267:19765–19768. [PubMed] [Google Scholar]
- Hutchison CA, Hardies SC, Loeb DD, Shehee WR, Edgell MH. LINEs and related retroposons: Long interspersed sequences in the eucaryotic genome. In: Berg DE, Howe MM, editors. Mobile DNA. Washington, D.C.: ASM Press; 1989. pp. 593–617. [Google Scholar]
- Jubier-Maurin V, Cuny G, Laurent A-M, Paquereau L, Roizes G. A new 5′ sequence associated with mouse L1 elements is representative of a major class of L1 termini. Mol Biol Evol. 1992;9:41–55. doi: 10.1093/oxfordjournals.molbev.a040707. [DOI] [PubMed] [Google Scholar]
- Kazazian HH, Jr, Moran JV. The impact of L1 retrotransposons on the human genome. Nat Genet. 1998;19:19–24. doi: 10.1038/ng0598-19. [DOI] [PubMed] [Google Scholar]
- Kimberland ML, Divoky V, Prchal J, Schwahn U, Berger W, Kazazian HH., Jr Full-length human L1 insertions retain the capacity for high frequency retrotransposition in cultured cells. Hum Mol Genet. 1999;8:1557–1560. doi: 10.1093/hmg/8.8.1557. [DOI] [PubMed] [Google Scholar]
- Kingsmore SF, Giros B, Suh D, Bieniarz M, Caron MG, Seldin MF. Glycine receptor beta-subunit gene mutation in spastic mouse associated with LINE-1 element insertion. Nat Genet. 1994;7:136–141. doi: 10.1038/ng0694-136. [DOI] [PubMed] [Google Scholar]
- Kohrman DC, Harris JB, Meisler MH. Mutation detection in the med and medJ of the sodium channel Scn8a. J Biol Chem. 1996;271:17576–17581. doi: 10.1074/jbc.271.29.17576. [DOI] [PubMed] [Google Scholar]
- Kojima T, Nakajima K, Mikoshiba K. The disabled1 gene is disrupted by a replacement with L1 fragment in yotari mice. Mol Brain Res. 2000;75:121–127. doi: 10.1016/s0169-328x(99)00313-7. [DOI] [PubMed] [Google Scholar]
- Kolosha VO, Martin SL. In vitro properties of the first ORF protein from mouse LINE-1 support its role in ribonucleoprotein particle formation during retrotransposition. Proc Natl Acad Sci. 1997;94:10155–10160. doi: 10.1073/pnas.94.19.10155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kraft R, Kadyk L, Leinwand LA. Sequence of organization of variant mouse 4.5 S RNA genes and pseudogenes. Genomics. 1992;12:255–566. doi: 10.1016/0888-7543(92)90448-2. [DOI] [PubMed] [Google Scholar]
- Loeb DD, Padgett RW, Hardies SC, Sheehee WR, Comer MB, Edgell MH, Hutchinson CA., III The sequence of a large L1Md element reveals a tandemly repeated 5′ end and several features found in retrotransposons. Mol Cell Biol. 1986;6:168–182. doi: 10.1128/mcb.6.1.168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin SL. Characterization of a LINE-1 cDNA that originated from RNA present in ribonucleoprotein particles: Implications for the structure of an active mouse LINE-1. Gene. 1995;53:261–266. doi: 10.1016/0378-1119(94)00785-q. [DOI] [PubMed] [Google Scholar]
- Martin SL, Li J, Weisz JA. Deletion analysis defines distinct functional domains for protein–protein and nucleic acid interactions in the ORF1 protein of mouse LINE-1. J Mol Biol. 2000;304:11–20. doi: 10.1006/jmbi.2000.4182. [DOI] [PubMed] [Google Scholar]
- Mears ML, Hutchison CA., III The evolution of modern lineages of mouse L1 elements. J Mol Evol. 2001;52:51–62. doi: 10.1007/s002390010133. [DOI] [PubMed] [Google Scholar]
- Moran JV, Holmes SE, Naas TP, DeBerardinis RJ, Boeke JD, Kazazian HH., Jr High frequency retrotransposition in cultured mammalian cells. Cell. 1996;87:917–927. doi: 10.1016/s0092-8674(00)81998-4. [DOI] [PubMed] [Google Scholar]
- Moran JV, DeBerardinis RJ, Kazazian HH., Jr Exon-shuffling by L1 retrotransposition. Science. 1999;283:1530–1534. doi: 10.1126/science.283.5407.1530. [DOI] [PubMed] [Google Scholar]
- Mulhardt C, Fischer M, Gass P, Simon-Chazottes D, Guenet JL, Kuhse J, Betz H, Becker CM. The spastic mouse: Aberrant splicing of glycine receptor beta subunit mRNA caused by intronic insertion of L1 element. Neuron. 1994;13:1003–1015. doi: 10.1016/0896-6273(94)90265-8. [DOI] [PubMed] [Google Scholar]
- Naas TP, DeBerardinis RJ, Moran JV, Ostertag EM, Kingsmore SF, Seldin MF, Hayashizaki Y, Martin SL, Kazazian HH., Jr An actively retrotransposing, novel subfamily of mouse L1 elements. EMBO J. 1998;17:590–597. doi: 10.1093/emboj/17.2.590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Padgett RW, Hutchinson CA, III, Edgell MH. The F-type 5′ motif of mouse L1 elments: A major class of L1 termini similar to the A-type in organization but unrelated in sequence. Nucl Acids Res. 1988;16:739–749. doi: 10.1093/nar/16.2.739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perou CM, Pryor RJ, Naas TP, Kaplan J. The bg allele mutation is due to a LINE1 element retrotransposon. Genomics. 1997;42:366–8. doi: 10.1006/geno.1997.4740. [DOI] [PubMed] [Google Scholar]
- Saitou N, Nei M. The neighbour-joining method: A new method for reconstructing phylogenetic trees. Mol Biol Ecol. 1987;4:406–435. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- Sambrook J, Russell DW. Molecular Cloning, A Laboratory Manual. New York: Cold Spring Harbor Laboratory Press; 2001. [Google Scholar]
- Sassaman DM, Dombroski BA, Moran JV, Kimberland ML, Naas TP, DeBerardinis RJ, Gabriel A, Swergold GD, Kazazian HH., Jr Many human L1 elements are capable of retrotransposition. Nat Genet. 1997;16:37–43. doi: 10.1038/ng0597-37. [DOI] [PubMed] [Google Scholar]
- Saxton JA, Martin SL. Recombination between subtypes creates a mosaic lineage of LINE-1 that is expressed and actively retrotransposing in the mouse genome. J Mol Biol. 1998;280:611–622. doi: 10.1006/jmbi.1998.1899. [DOI] [PubMed] [Google Scholar]
- Schichman SA, Severynse DM, Edgell MH, Hutchison CA., III Strand-specific LINE-1 transciption in mouse F9 cells originates from the youngest phylogenetic subgroup of LINE-1 elements. J Mol Biol. 1992;224:559–574. doi: 10.1016/0022-2836(92)90544-t. [DOI] [PubMed] [Google Scholar]
- Severynse DM, Hutchinson CA, III, Edgell MH. Identification of transcriptional regulatory activity within the 5′ A-type monomer sequence of the mouse LINE-1 retroposon. Mamm Genome. 1992;2:41–50. doi: 10.1007/BF00570439. [DOI] [PubMed] [Google Scholar]
- She JX, Bonhomme F, Boursot P, Thaler L, Catzeflis F. Molecular phylogenies in the genus Mus: Comparative analysis of electrophoretic, scnDN5, and mtDNA RFLP data. Biol J Linnean Soc. 1990;41:83–103. [Google Scholar]
- Shehee WR, Chao S, Loeb DD, Comer MB, Hutchison CA, III, Edgell MH. Determination of a functional ancestral sequence and definition of the 5′ end of A-type mouse L1 elements. J Mol Biol. 1987;196:757–767. doi: 10.1016/0022-2836(87)90402-5. [DOI] [PubMed] [Google Scholar]
- Skowronski J, Fanning TG, Singer MF. Unit-length line-1 transcripts in human teratocarcinoma cells. Mol Cell Biol. 1988;8:1385–1397. doi: 10.1128/mcb.8.4.1385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahara T, Ohsumi T, Kuromitsu J, Shibata K, Sasaki N, Okazaki Y, Shibata H, Sato S, Yoshiki A, Kusakabe M, et al. Dysfunction of the Orleans reeler gene arising from exon skipping due to transposition of a full-length copy of an L1 sequence into the skipped exon. Hum Mol Genet. 1996;5:989–993. doi: 10.1093/hmg/5.7.989. [DOI] [PubMed] [Google Scholar]
- Thaler L. Origin and evolution of mice: An appraisal of fossil evidence and morphological traits. Curr Topics Microbiol Immunol. 1986;127:3–11. doi: 10.1007/978-3-642-71304-0_1. [DOI] [PubMed] [Google Scholar]
- Yajima I, Sata S, Kimura T, Yasumoto K, Shibahara S, Goding CR, Yamamoto H. An L1 element intronic insertion in the black-eyed white (Mitfmi-bw) gene: The loss of a single Mitf isoform responsible for the pigmentary defect and inner ear deafness. Hum Mol Genet. 1999;8:1431–1441. doi: 10.1093/hmg/8.8.1431. [DOI] [PubMed] [Google Scholar]