Abstract
The secondary structure of a truncated P5abc subdomain (tP5abc, a 56-nucleotide RNA) of the Tetrahymena thermophila group I intron ribozyme changes when its tertiary structure forms. We have now used heteronuclear NMR spectroscopy to determine its conformation in solution. The tP5abc RNA that contains only secondary structure is extended compared with the tertiary folded form; both forms coexist in slow chemical exchange (the interconversion rate constant is slower than 1 s−1) in the presence of magnesium. Kinetic experiments have shown that tertiary folding of the P5abc subdomain is one of the earliest folding transitions in the group I intron ribozyme, and that it leads to a metastable misfolded intermediate. Previous mutagenesis studies suggest that formation of the extended P5abc structure described here destabilize a misfolded intermediate. This study shows that the P5abc RNA subdomain containing a GNRA tetraloop in P5c (in contrast to the five-nucleotide loop P5c in the tertiary folded ribozyme) can disrupt the base-paired interdomain (P14) interaction between P5c and P2.
Knowledge of a ribozyme's conformation is essential for understanding its biological functions. Extensive structural studies of ribozymes have been done by x-ray crystallography and solution NMR spectroscopy (1, 2). Folding of a single-stranded RNA into its functional form occurs first through the formation of secondary structure, and then through the formation of tertiary structure. Prediction of RNA secondary structure—the formation of base-paired helices, hairpin loops, bulges, and internal loops—is well established (3–5). However, prediction of RNA tertiary folding remains challenging (6).
Tertiary folding of large ribozymes such as the multidomain group I introns (Fig. 1) may take up to minutes to accomplish in vitro (7–10). Before tertiary folding, the RNA of a group I intron forms a metastable intermediate characterized by a misfolded catalytic core (11, 12), which is stabilized by the peripheral helices including P5c, P2, and P9 (13). Mutations that speed up folding of the ribozyme core are disruptive to the tertiary fold of the P5abc subdomain (11, 14, 15). Tertiary folding of P5abc is the earliest event in the ribozyme folding, and it takes less than 1 s before P5c becomes protected from hydroxyl radicals generated by a synchrotron x-ray source. Domain P4-P6 folds cooperatively within a few seconds, whereas the catalytic core requires minutes to fold (10). Wu and Tinoco (16) showed that tertiary folding of tP5abc caused significant changes in its secondary structure. The NMR-determined secondary structure of tP5abc is consistent with the lowest free energy form predicted by using the Zuker folding algorithm (3); we call this structure the extended form. Tertiary interactions upon folding of the extended form offset the free energy increase caused by the change in secondary structure (17).
Figure 1.
The secondary structure of the 414-nt group I intron ribozyme from T. thermophila. The catalytic core (part of P1, P3-P7, and P4-P6) is in the shaded area. The peripheral helices P2, P2.1, P9.1, and P9 wrap around the core to provide extra stability. Interdomain interactions that connect the peripheral helices are denoted by broken lines. In particular, P14 is composed of complementary base pairs between L5c and L2. The lowercase letters at the 3′ and 5′ ends are part of the exon. The figure was modified from figure 1 in ref. 13.
We have used high-resolution NMR to determine the solution conformation of the extended tP5abc. The 56-nucleotide RNA is among the largest RNA molecules whose atomic resolution structures have been determined by NMR. Its structure in the extended form is significantly different from the folded form in a crystal. The formation of an extended P5abc intermediate is consistent with a fast-folding model for the Tetrahymena group I intron suggested by kinetic studies (18).
Methods
RNA Synthesis, Labeling, and Purification.
Four RNA constructs were made that correspond to tP5abc (56-nt), P5a (29-nt), P5b (18-nt), and P5c (18-nt). The RNAs were transcribed in vitro by using T7 RNA polymerase, directed by chemically synthesized DNA templates with C2′ methoxyls at the two 5′ end nucleotides to reduce the N+1 product (19–21). 15N-labeled RNA samples were transcribed by using uniformly 15N-labeled NTPs purified from Escherichia coli cells grown in medium with 15NH4Cl (Cambridge Isotope Laboratories, Cambridge, MA) as the only source of nitrogen. 15N- and 13C-labeled RNA samples were transcribed by using uniformly double-labeled NTPs. The NTPs were either directly purchased or enzymatically phosphorylated (22, 23) from the double-labeled NMPs (recycled or from Cambridge Isotope Laboratories). RNA samples were purified by using 16% ≈ 20% denaturing PAGE followed by a 24 ≈ 48 h dialysis. Unless specified, the buffer used for all NMR experiments contained 10 mM sodium phosphate, 0.01 mM EDTA, and pH 6.4. For exchangeable proton experiments, 10% D2O was added to provide a lock signal. For nonexchangeable proton experiments, the samples were repeatedly lyophilized and redissolved in 99.96% D2O. The pH was not readjusted for samples in D2O. A typical sample concentration was 1.4 mM for a nonlabeled RNA sample, and was 0.6 mM for a labeled sample. The RNA concentration was determined by UV absorbance at 260 nm.
NMR Spectroscopy and Restrained Molecular Dynamics (RMD) Simulation.
NMR data were acquired on a Bruker AMX-600 (Billerica, MA), a Bruker DRX-500, or a Varian Unity-800 spectrometer. All NMR data were processed with FELIX95 (Biosym Technologies). Nuclear Overhauser effect spectroscopy (NOESY) spectra (mixing times: 50 ≈ 200 ms) of exchangeable protons were recorded at temperatures ranging from 5–25°C. NOESY (mixing times: 50 ≈ 500 ms), double-quantum-filtered–correlation spectroscopy (DQF-COSY), and total correlation spectroscopy (TOCSY; mixing time: 80 ms) spectra of nonexchangeable protons were recorded at temperatures ranging from 10–40°C. A jump–return pulse (24, 25) was used to suppress the proton signal from solvent in the exchangeable proton experiments. A presaturation pulse was used to suppress the residual 1H2HO signal in the nonexchangeable proton experiments. For assignment purposes, a fast heteronuclear single quantum correlation (FHSQC) pulse sequence (26) was used to correlate a 15N nucleus to its directly attached proton [with an insensitive nuclei enhanced by polarization transfer (INEPT) delay of 2.25 ms], and to a long-range coupled proton (with an INEPT delay of 21 ms) in a 15N uniformly labeled sample. A constant time heteronuclear multiple quantum coherence (HMQC; ref. 27) experiment was used to correlate a 13C nucleus to its directly connected sugar protons in 15N and 13C uniformly labeled samples.
The structure determination was based on an x-plor (28) simulated annealing protocol of restrained molecular dynamics (RMD) simulation (29). Distance constraints were derived from NOE cross peaks as strong (1.8–3.0 Å), medium (2.0–4.0 Å), weak (2.5–5 Å), and very weak (3.0–5 Å). A distance of 3.0–7.0 Å was assigned to a NOE that might be affected by spin diffusion. Distance constraints were used to define a hydrogen bond's proton-heavy atom and heavy atom-heavy atom distances (30) in each base pair (six distances for a G⋅C, four distances for an A⋅U, a G⋅U, and an imino G⋅A pair). A distance of 10.9 Å between the two C1s within a base pair and a planarity constraint were used to maintain the A-form geometry of Watson–Crick base pairs in the stem regions. In situations where resonance overlap occurred in the spectra of tP5abc, NOE constraints from a small RNA construct (P5a, P5b, or P5c) were used, if possible. A torsion angle constraint was used to keep each amino group in the same plane as the base. Glycosidic angles of all nucleotides were constrained in the anticonformation (with a range of 50°). Each ribose sugar conformation was constrained by four torsion angles (ν0, ν1, ν2, ν3). One hundred random structures were generated initially. After the global fold and refinement, a range of the relative orientations of the three stems in tP5abc was observed; we used the lowest energy structure after refinement.
Exchange Rate Between the Extended and Folded tP5abc.
The exchange rate between two noncoupled species can be estimated from the intensity of the exchange cross peaks in a two-dimensional (2D) exchange spectrum (31). For two states in equilibrium: [Extended] ↔ [Folded], the exchange rate is kex = I/[τm(I + I0)], where I and I0 are an exchange cross peak volume and its diagonal peak volume, respectively, and τm is the mixing time. In the absence of an exchange cross peak (where I is below the noise level), a 10% signal-to-noise ratio was used to estimate an upper limit of kex. Under all conditions where both the extended and the folded tP5abc coexist, no resolved cross peaks were observed between the two species in the 2D exchange spectra with a τm ranging from 50–150 ms. This means that kex is slower than about 1 s−1.
Results
Tertiary Folding of tP5abc in Mg2+.
In the presence of Mg2+, tP5abc folds into a compact tertiary complex. The Mg2+-dependent UV thermal melting profiles of tP5abc reveal a sharp transition at approximately 5.5 mM Mg2+ (Fig. 2a), indicating a highly cooperative folding. A similar transition was observed by NMR and by nondenaturing PAGE in a variety of RNA concentrations (Fig. 2 b and c). Tertiary folding cannot be induced by Co(III)(NH3)6, consistent with Mg2+ being directly coordinated in the tertiary folded complex. The extended and folded forms coexist in slow chemical exchange at an exchange rate of less than 1 s−1 characterized by magnetization transfer measured in a 2D NOESY NMR experiment. A nonfolding tP5abc mutant, A186U, was synthesized as a control (15). It forms a similar extended form as the wild-type tP5abc in the absence of Mg2+, as shown by its imino spectrum. But contrary to the wild-type tP5abc, the imino proton spectrum of A168U does not change in the presence of up to 5 mM Mg2+, indicating that A168U does not undergo tertiary folding (Fig. 2c).
Figure 2.
(a) The magnesium-dependent UV thermal melting profiles of tP5abc. The UV thermal melting curves of tP5abc were recorded on a Gilford 250 spectrometer with four cell compartments. Samples were heated to 90°C and cooled to 4°C several times at a rate of 0.1°C/min. The increased UV absorbance as temperature increases indicates the breaking of secondary and tertiary structure of the RNA. The melting temperature of tP5abc does not change with the RNA concentration, showing that the thermal melting event is unimolecular. (b) The magnesium-dependent gel mobility of tP5abc in nondenaturing PAGE. The higher mobility of the wild-type tP5abc in 2 mM Mg2+ and 10 mM Na+ indicates that it folds into a more compact conformation than A186U, a mutant that prevents the tertiary interactions. (c) One-dimensional imino NMR spectra of tP5abc wild-type and A186U. The wild-type molecule folds in 2 mM Mg2+, whereas the A186U mutant does not fold, as shown by the unchanged imino spectra up to 5 mM Mg2+. Six arrows point to the imino protons of the three G⋅U base pairs in the extended tP5abc that disappear on tertiary folding. *, the imino proton of G169 within the GCAA tetraloop that also disappears when P5c rearranges. In contrast, imino proton resonances of the three G⋅U base pairs of A186U do not change in the presence of Mg2+, indicating no change in the secondary structure.
Resonance Assignments of the Extended tP5abc.
A D2O NOESY displaying the aromatic to H1′ proton region is shown in Fig. 3. Most NOE cross peaks in this spectrum were assigned based on the NOE sequential connectivity, heteronuclear NMR experiments, and comparison with spectra of the individual P5a, P5b, and P5c stem loops. The assignments in the 3-helix junction were particularly challenging because of the spectral overlap and the lack of a comparison spectrum from a smaller molecule construct. Sequential NOE connectivities were observed in all three stems and in the continuous strand from P5a to P5b (except the 5′ strand of P5c because of dynamics). The GNRA tetraloops in P5b and P5c were identified by their unique NOE patterns (32, 33) characterized by a large up-field shifted resonance (at ≈3.8 ppm) of the H1′ of the 3′ closing nucleotide (A173 and G158 of P5c and P5b, respectively). In the 3-helix junction, the resonance of A139 H2 is up-field shifted (at ≈7.0 ppm), characteristic of an adenine H2 within stacked bases in an A-form helix. It also has NOE cross-peaks to the H1′ of the next residue, A140, and to the cross-strand G180, also consistent with an H2 in an A-form helix. The chemical shifts of the H1′ of A140 and of G180 are degenerate at 25°C, so only one cross-peak was observed to align with A139 H2. The spectrum recorded at 10°C is shown as an Inset in Fig. 3, in which the chemical shifts of the H1's of A140 and G180 were resolved. Assignments of the adenine H2 resonances were confirmed by their attached 13C chemical shifts in a proton–carbon correlated heteronuclear multiple quantum coherence (HMQC) spectrum.
Figure 3.
The aromatic to sugar H1′ region of a D2O NOESY (300 ms mixing time) spectrum of tP5abc recorded at 25°C. The spectrum was constructed by overlapping the identical regions of both sides of the diagonal (cross-peaks are colored in purple and black). The aromatic to H1′ sequential NOE connectivity walks colored in red, green, and blue correspond to P5c, P5a, and P5b, respectively. The free induction decay was recorded on an 800-MHz Varian NMR spectrometer and processed by using FELIX95 (Biosym Technologies). The small spectrum insert at around 7.0 ppm (D1) and 5.9 ppm (D2) is the identical region of a D2O NOESY (300 ms mixing time) spectrum recorded at 10°C. The H1′ resonance frequencies of residue A140 and G180 are degenerate at 25°C, but clearly resolved at 10°C. Because the two cross-peaks in the spectrum insert are well separated from the crowed region, the assignment is unambiguous.
Solution Conformation of the Extended tP5abc.
The extended tP5abc consists of three A-form stems (P5a, P5b, and P5c) that are connected by a 3-helix junction. A 5-nt A-rich bulge forms an ≈90° turn located in P5a. Stems P5b and P5c are terminated with a GAAA and a GCAA tetraloop, respectively. Sugars that are not in pure 3′-endo conformations are mostly located in the junction and loop regions (indicated by italics in Fig. 4a). Significant structural differences were observed for these regions in comparison with the crystal structure of P4-P6 (ref. 34; Fig. 4b). The structure shown is the lowest energy structure determined by NOE distance-restrained molecular dynamics (RMD) simulations in combination with torsion angle restraints derived from NMR spectra, and ideal geometry restraints for A-form helices.
Figure 4.
(a) The extended and tertiary-folded secondary structures of tP5abc. A magnesium ion core forms on tertiary folding and causes extensive rearrangement of P5c and the adenine-rich bulge (15). Each solid bar denotes a Watson–Crick base pair, and each hollow bar denotes a non-Watson–Crick base pair. The green dots in the folded form represent five magnesium ions revealed in the crystal of P5-P6 that directly coordinate (thick green lines) to G or phosphate oxygen (p), or indirectly coordinate to phosphate mediated by a water molecule (thin green lines). Mutations (colored in blue) enable the catalytic core of a group I intron to become protected faster from RNase H digestion during magnesium-ion induced folding (11). These mutations do not disrupt the stability of an extended P5abc (or tP5abc): A186U, A183U, and U167C do not disrupt the base pairing; A171G converts the GCAA tetraloop into a GCGA tetraloop, which belongs to the same GNRA tetraloop family and should have similar structure and stability (33). NMR experiments showed that mutant A186U of tP5abc folds into a similar secondary structure as the wild-type tP5abc and that it does not fold in the presence of Mg2+ (Fig. 3). Mutant + 174G is an insertion mutant that can form a stable GNAAA 5-nt loop that is analogous to a GNRA tetraloop (38). (b) Comparison of the extended tP5abc with the tertiary folded tP5abc in the P4-P6 crystal. The folded tP5abc is generated by removing four base pairs in the P5b stem from the native structure of P4-P6 (34). The folded tP5abc is highly compact at the 3-helix junction. The extended tP5abc generated from NMR constraints was the lowest energy structure obtained from one hundred random starting structures.
From the NOESY, we conclude that base stacking of the 5′ strand from P5a to P5b is continuous within the 3-helix junction. The base stacking was characterized by base to base NOE cross peaks from C138 to G141. Base A178 is not paired, it stacks on top of the G164⋅U177 pair and protects the imino protons of G164 and U177 from exchange with the solvent protons. This base stacking was supported by the NOE cross-peaks between A178 H8 and H6 of U177 and it explains why the imino peaks of G164 and U177 were so sharp. Although no imino peak was observed for U179, it is likely that A139 and U179 forms a Watson–Crick A⋅U pair. The missing U179 imino peak is presumably due to fast exchange with solvent protons, consistent with the conformation of the 3-helix junction shown in Fig. 4b.
The conformation of P5c in the extended form is dramatically different from a tertiary folded P5c in a crystal (34). The 5′ side of the stem slides one nucleotide toward the junction, adding two more base pairs to the P5c stem and forming a GCAA tetraloop closed by the A173⋅U168 pair. The chemical shifts of both the G169 imino proton and its directly attached nitrogen (acquired in an HSQC spectrum) are consistent with a sheared G169⋅A172 pair (2). The line widths of protons from the 5′ side of P5c (residue G164 to C170) are broadened and the sequential NOEs are very weak, indicating dynamic motion. This conformational flexibility may facilitate base pair sliding on tertiary folding.
The conformation of the A-rich bulge in tP5abc is similar to that previously determined in a 25-nt fragment of P5abc (35). Differences are observed for base U185, which was poorly defined previously, but well stacked on A184 in the context of tP5abc. This A-rich bulge forms an approximately 90° bend between the two A-form stems.
The relative orientation of the three stems in tP5abc is not well defined, due to the intrinsic inability to obtain long-range distance constraints by NMR. The structure we present is a best fit to the existing experimental data. Further structure refinement that takes advantage of residual dipolar couplings measured in partially oriented samples should provide a better determination of the global arrangement of tP5abc.
Discussion
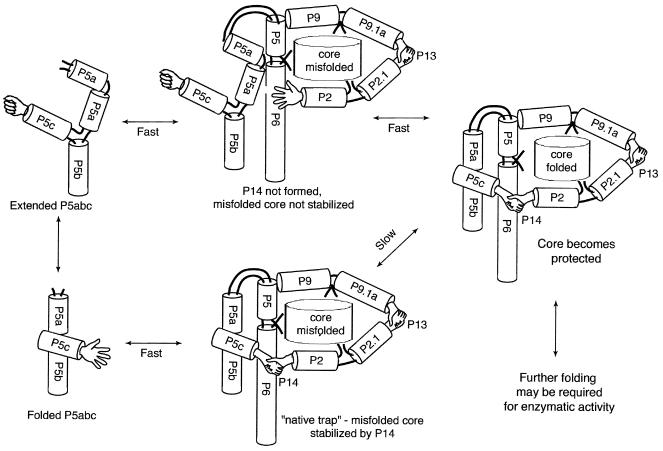
By using NMR, we have shown that P5abc can adopt two distinct forms: the tertiary folded form as seen in the x-ray structure, and the extended form (Fig. 4). In an active ribozyme, P5abc is folded and loop L5c of P5c interacts with loop L2 of P2 (the P14 interdomain interaction) through complementary base pairs to stabilize the catalytic core (36). P14 may equally stabilize a misfolded catalytic core that leads to a metastable intermediate (13). We envision the loops L5c and L2 as two open hands ready for contact in a folded ribozyme. The structure of the extended P5abc with a stable (closed) GNRA tetraloop in L5c is not suitable for base pairing in P14 (Fig. 5). We propose that formation of an extended P5abc intermediate can destabilize a misfolded catalytic core as a result of disrupting P14, and thus give rise to alternative folding pathways (18, 37). This hypothesis is consistent with known experimental results.
Figure 5.
A folding model of the group I intron ribozyme and a possible role of the GNRA tetraloop in an extended P5abc. Interdomain interactions P14 and P13, represented by two pairs of interacting hands, can stabilize the misfolded core (18). P14 is an interaction of the hands from L5c and L2. When all hands in the peripheral helices are closed, the catalytic core of the ribozyme is tightly locked into a fixed conformation (this could be either a misfolded or a correctly folded conformation). In the lower path interdomain interactions form a “native trap” that stabilizes a misfolded catalytic core (11, 13). In an alternative (upper path) folding pathway (18), the extended form of P5abc capped by a GNRA tetraloop switches the open hand of L5c to a closed hand. This conformational change in L5c abolishes P14 and consequently facilitates the catalytic core rearrangement. Further folding may be required after the catalytic core becomes protected (9).
First, a single mutation that abolishes a base pair in P14 increases the overall rate of folding, whereas the compensatory mutation that restores the base pair in P14 restores the slower folding rate (13). Second, mutants that allow faster formation of a catalytic core protected from RNase H digestion (9, 11) disrupt the folded P5abc, but do not disrupt the extended P5abc (Fig. 4a). Third, the extended P5abc is the thermodynamically most stable form in the absence of tertiary interactions, and it can be further stabilized by mutations. NMR and nondenaturing gel data of two tP5abc mutants A186U (Fig. 2 b and c) and U167C (17) have shown that both mutants adopt the extended form and do not fold in Mg2+. Finally, the slow exchange rate between the folded and extended P5abc measured by NMR also suggests that an extended P5abc can exist as a stable structural intermediate.
Kinetic studies of the folding of the group I intron ribozyme have suggested alternative folding pathways, which may not be limited by the misfolded catalytic core (18). Fig. 5 demonstrates a hypothetical folding model of the group I intron ribozyme that provides a simple explanation of how disruption of P14 by mutations, or by conformational changes, can speed up the rearrangement of a misfolded catalytic core. During the initial folding, P5abc can adopt either the folded or the extended form. A slow folding pathway is characterized by a “native trap” stabilized by P14. In an alternative folding pathway (of either a mutant or a minor conformational species of the wild-type molecule), a misfolded catalytic core can rearrange quickly into a correctly folded form because of the lack of a constraining P14. The disruption of P14 could be caused by the extended P5abc species characterized here.
Acknowledgments
We thank Ms. Barbara Dengler for managing the laboratory, Mr. David Koh for synthesizing the DNA templates, Dr. Jeffrey Pelton for NMR advice, and Prof. Joseph Puglisi for instrument time on the 800-MHz NMR at Stanford University and for help from his group. We also thank Mr. Daniel Treiber, Prof. James Williamson, and Prof. Sarah Woodson for helpful discussions, and for reading the manuscript. This research was supported by National Institutes of Health Grant GM 10840, by DOE Grant DE-FG03-86ER60406, and through instrumentation grants from the Department of Energy (DE-FG05-86ER75281) and the National Science Foundation (DMB 8609305). M.Z. and M.W. were supported by postdoctoral fellowships from PMMB (Program in Mathematics and Molecular Biology).
Abbreviation
- NOESY
nuclear Overhauser effect spectroscopy
References
- 1.Batey R T, Rambo R P, Doudna J A. Angew Chem Int Ed. 1999;38:2327–2343. doi: 10.1002/(sici)1521-3773(19990816)38:16<2326::aid-anie2326>3.0.co;2-3. [DOI] [PubMed] [Google Scholar]
- 2.Zheng M, Tinoco I., Jr . In: Ribozyme Biochemistry and Biotechnology. Krupp G, Gaur R, editors. Natick, MA: Eaton; 2000. pp. 297–314. [Google Scholar]
- 3.Zuker M. Science. 1989;244:48–52. doi: 10.1126/science.2468181. [DOI] [PubMed] [Google Scholar]
- 4.Mathews D H, Sabina J, Zuker M, Turner D H. J Mol Biol. 1999;288:911–940. doi: 10.1006/jmbi.1999.2700. [DOI] [PubMed] [Google Scholar]
- 5.Gutell R R. Curr Opin Struct Biol. 1993;3:313–322. doi: 10.1016/s0959-440x(02)00339-1. [DOI] [PubMed] [Google Scholar]
- 6.Tinoco I, Jr, Bustamante C. J Mol Biol. 1999;293:271–281. doi: 10.1006/jmbi.1999.3001. [DOI] [PubMed] [Google Scholar]
- 7.Russell R, Millett I S, Doniach S, Herschlag D. Nat Struct Biol. 2000;7:367–370. doi: 10.1038/75132. [DOI] [PubMed] [Google Scholar]
- 8.Treiber D K, Williamson J R. Curr Opin Struct Biol. 1999;9:339–345. doi: 10.1016/S0959-440X(99)80045-1. [DOI] [PubMed] [Google Scholar]
- 9.Rook M S, Treiber D K, Williamson J R. Proc Natl Acad Sci USA. 1999;96:12471–12476. doi: 10.1073/pnas.96.22.12471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sclavi B, Sullivan M, Chance M R, Brenowitz M, Woodson S A. Science. 1998;279:1940–1943. doi: 10.1126/science.279.5358.1940. [DOI] [PubMed] [Google Scholar]
- 11.Treiber D K, Rook M S, Zarrinkar P P, Williamson J R. Science. 1998;279:1943–1946. doi: 10.1126/science.279.5358.1943. [DOI] [PubMed] [Google Scholar]
- 12.Pan J, Woodson S A. J Mol Biol. 1998;280:597–609. doi: 10.1006/jmbi.1998.1901. [DOI] [PubMed] [Google Scholar]
- 13.Pan J, Woodson S A. J Mol Biol. 1999;294:955–965. doi: 10.1006/jmbi.1999.3298. [DOI] [PubMed] [Google Scholar]
- 14.Treiber D, Williamson J. J Mol Biol. 2001;305:11–21. doi: 10.1006/jmbi.2000.4253. [DOI] [PubMed] [Google Scholar]
- 15.Cate J H, Hanna R L, Doudna J A. Nat Struct Biol. 1997;4:553–558. doi: 10.1038/nsb0797-553. [DOI] [PubMed] [Google Scholar]
- 16.Wu M, Tinoco I., Jr Proc Natl Acad Sci USA. 1998;95:11555–11560. doi: 10.1073/pnas.95.20.11555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Silverman S K, Zheng M, Wu M, Tinoco I, Jr, Cech T R. RNA. 1999;5:1665–1674. doi: 10.1017/s1355838299991823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pan J, Deras M L, Woodson S A. J Mol Biol. 2000;296:133–144. doi: 10.1006/jmbi.1999.3439. [DOI] [PubMed] [Google Scholar]
- 19.Kao C, Zheng M, Rüdisser S. RNA. 1999;5:1268–1272. doi: 10.1017/s1355838299991033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Milligan J F, Groebe D R, Witherell G W, Uhlenbeck O C. Nucleic Acids Res. 1987;15:8783–8798. doi: 10.1093/nar/15.21.8783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wyatt J R, Chastain M, Puglisi J D. BioTechniques. 1991;11:764–769. [PubMed] [Google Scholar]
- 22.Nikonowicz E P, Sirr A, Legault P, Jucker F M, Baer L M, Pardi A. Nucleic Acids Res. 1992;20:4507–4513. doi: 10.1093/nar/20.17.4507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Batey R T, Inada M, Kujawinski E, Puglisi J D, Williamson J R. Nucleic Acids Res. 1992;20:4515–4523. doi: 10.1093/nar/20.17.4515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Plateau P, Gueron M. J Am Chem Soc. 1982;104:7310–7311. [Google Scholar]
- 25.Sklenar V, Bax A. J Magn Reson. 1987;74:469–479. [Google Scholar]
- 26.Mori S, Abeygunawardana C, Johnson M O, Vanzijl P C M. J Magn Reson. 1995;108:94–98. doi: 10.1006/jmrb.1995.1109. [DOI] [PubMed] [Google Scholar]
- 27.Marino J P, Diener J L, Moore P B, Griesinger C. J Am Chem Soc. 1997;119:7361–7366. [Google Scholar]
- 28.Brünger A T. x-plor, A System for Crystallography and NMR. New Haven, CT: Yale Univ.; 1993. , Version 3.1. [Google Scholar]
- 29.Wimberly B. Ph.D. thesis. Berkeley: Univ. of California; 1992. [Google Scholar]
- 30.Saenger W. Principles of Nucleic Acid Structure. New York: Springer; 1984. [Google Scholar]
- 31.Ernst R R, Bodenhausen G, Wokaun A. Principles of Nuclear Magnetic Resonance in One and Two Dimensions. Oxford: Clarendon; 1987. [Google Scholar]
- 32.Jucker F M, Heus H A, Yip P F, Moors E H, Pardi A. J Mol Biol. 1996;264:968–980. doi: 10.1006/jmbi.1996.0690. [DOI] [PubMed] [Google Scholar]
- 33.Heus H A, Pardi A. Science. 1991;253:191–194. doi: 10.1126/science.1712983. [DOI] [PubMed] [Google Scholar]
- 34.Cate J H, Gooding A R, Podell E, Zhou K, Golden B L, Kundrot C E, Cech T R, Doudna J A. Science. 1996;273:1678–1685. doi: 10.1126/science.273.5282.1678. [DOI] [PubMed] [Google Scholar]
- 35.Luebke K J, Landry S M, Tinoco I., Jr Biochemistry. 1997;36:10246–10255. doi: 10.1021/bi9701540. [DOI] [PubMed] [Google Scholar]
- 36.Lehnert V, Jaeger L, Michel F, Westhof E. Chem Biol. 1996;3:993–1009. doi: 10.1016/s1074-5521(96)90166-0. [DOI] [PubMed] [Google Scholar]
- 37.Zhuang X, Bartley L E, Babcock H P, Russell R, Ha T, Herschlag D, Chu S. Science. 2000;288:2048–2051. doi: 10.1126/science.288.5473.2048. [DOI] [PubMed] [Google Scholar]
- 38.Legault P, Li J, Mogridge J, Kay L E, Greenblatt J. Cell. 1998;93:289–299. doi: 10.1016/s0092-8674(00)81579-2. [DOI] [PubMed] [Google Scholar]