Abstract
A large-scale BAC end-sequencing project at The Institute for Genomic Research (TIGR) has generated one of the most extensive sets of sequence markers for the mouse genome to date. With a sequencing success rate of >80%, an average read length of 485 bp, and ABI3700 capillary sequencers, we have generated 449,234 nonredundant mouse BAC end sequences (mBESs) with 218 Mb total from 257,318 clones from libraries RPCI-23 and RPCI-24, representing 15× clone coverage, 7% sequence coverage, and a marker every 7 kb across the genome. A total of 191,916 BACs have sequences from both ends providing 12× genome coverage. The average Q20 length is 406 bp and 84% of the bases have phred quality scores ≥ 20. RPCI-24 mBESs have more Q20 bases and longer reads on average than RPCI-23 sequences. ABI3700 sequencers and the sample tracking system ensure that > 95% of mBESs are associated with the right clone identifiers. We have found that a significant fraction of mBESs contains L1 repeats and ∼48% of the clones have both ends with ≥ 100 bp contiguous unique Q20 bases. About 3% mBESs match ESTs and > 70% of matches were conserved between the mouse and the human or the rat. Approximately 0.1% mBESs contain STSs. About 0.2% mBESs match human finished sequences and > 70% of these sequences have EST hits. The analyses indicate that our high-quality mouse BAC end sequences will be a valuable resource to the community.
Because of the high stability (Shizuya et al. 1992; Kim et al. 1996a,b), libraries constructed in bacterial artificial chromosome (BAC) vectors have become the standard clone sets in high-throughput genomic sequencing projects of organisms with large genomes. End sequences from BACs provide highly specific markers. A genome sequencing approach (Venter et al. 1996) has been described, in which a clone contig is extended by selecting the minimally overlapping clones in each direction by searching the finished BAC sequence against a BAC end sequence (BES) database. Because BACs (an average insert size of 150 kb) are sufficiently large to traverse most tandem arrays of homology units and repeats, BESs are useful in genome assembly and chromosome walking and have been used extensively to confirm, join, and order existing contigs (International Human Genome Sequencing Consortium 2001a). The whole-genome shotgun sequencing strategy relies on BESs as the primary scaffold onto which the end sequences from the smaller clones are assembled (Venter et al. 1998, 2001).
The mouse and the human share many fundamental biological processes. Consequently, the mouse has been used frequently in medical research and is the best model system for studying human disease. Additionally, the mouse genome sequence facilitates the accurate annotation of the human genome. As such, National Institutes of Health (NIH) launched a mouse genome-sequencing project in October, 1999 (http://www.nhgri.nih.gov/NEWS/MouseRelease.htm).
Compared with the human, significantly fewer large-scale mapping efforts have been conducted for the mouse and much less data are available to the community (Hudson et al. 1995; Dietrich et al. 1996; Schuler et al. 1996; McCarthy et al. 1997; Stewart et al. 1997; Deloukas et al. 1998; Van Etten et al. 1999; International Human Genome Mapping Consortium 2001a; Olivier et al. 2001). A large-scale BAC end-sequencing project generates an extensive set of random markers across the genome in an inexpensive and rapid fashion, and will be crucial to the success of the combined strategy of BAC-based sequencing and a moderate level of whole-genome shotgun sequencing that is being used for the mouse genome. The Institute for Genomic Research (TIGR) is the only center conducting large-scale BAC end-sequencing for the mouse, in which the aim of the project is to generate accurate BES pairs from 170,000 RPCI-23 clones (Osoegawa et al. 2000) and 130,000 RPCI-24 clones to support the mouse genome sequencing project. The same set of clones has been fingerprinted at the Genome Sequencing Centre of British Columbia Cancer Research Centre at Vancouver Canada (http://www.bcgsc.bc.ca/projects/mouse_mapping/). We have approached the goal of the project and have generated ∼450,000 sequences (http://www.tigr.org/tdb/bac_ends/mouse/bac_end_intro.html). To provide a better characterization of this valuable resource, we conducted comprehensive quality assessment and sequence analyses as described below.
RESULTS
Quantities
TIGR has been generating mBESs from the EcoRI-based library RPCI-23 (Osoegawa et al. 2000) and the MboI-based library RPCI-24 (Table 1). As of June 27, 2001, we have generated 449,234 nonredundant mBESs from 257,318 BACs, of which 274,277 were from 154,795 RPCI-23 clones and 174,957 were from 102,524 RPCI-24 clones. With a pair rate of 75%, a total of 191,916 (119,483 from RPCI-23 and 72,433 from RPCI-24) had both T7 and Sp6 ends, providing 11.6× genome coverage by paired-end clones, assuming an average BAC insert size of 197.5 kb for RPCI-23 (Osoegawa et al. 2000) and 155 kb for RPCI-24. The average edited read length was 485 bp, representing a total of 218 Mb or 7% of the mouse genome. The basic sequencing process consisted of template preparation, reaction, clean up, electrophoresis, and sequence trimming as described (Kelley et al. 1999, http://www.tigr.org/tdb/bac_ends/mouse/bac_end_intro.html), and ABI3700 capillary sequencers were used. Base calls were performed with phred (Ewing and Green 1998; Ewing et al. 1998) and the quality scores were further adjusted with Paracel TraceTuner. Sequencing trimming was conducted with the program lucy (http://www.tigr.org/softlab/) with criteria of (1) a < 2.5% overall base-call error rate; (2) a read length of > 100 bp; and (3) no vector and Escherichia coli sequences. With this standard, ∼81% sequencing attempts yielded useful reads ranging from 101 to 984 bp. The success rate with the RPCI-23 library was slightly higher than with the RPCI-24 library (83% vs. 79%) and T7 ends of RPCI-23 clones yielded a higher success rate than Sp6 ends (84% vs. 81%). Statistics indicated that most of the sequencing failures were due to sequences whose quality did not meet the trimming standard (14% for RPCI-23 and 15.6% for RPCI-24), most likely because of inadequate BAC template purity due to many possibilities. Although these RPCI libraries had higher success rates than other libraries that we have sequenced with the same protocols, it would still be useful if the percentage of the wells in which more than one type of clone population dominates were reported for the libraries. An insufficient amount of templates also resulted in sequencing failures (3% for RPCI-23 and 4% for RPCI-24), some of which were caused by empty wells (2.1% were reported for RPCI-23 at http://www.chori.org/bacpac/23framefmouse.htm and 3.35% were reported at http://www.chori.org/bacpac/mmouse24.htm). We have found < 1% vector sequence contamination and a negligible amount of E. coli sequence contamination for both libraries. Excluding low quality, vector, and E. coli sequences, for a total of 272,573 BACs that were attempted from both ends, the entire process yielded 70% clones having both ends (72% for RPCI-23 and 69% for RPCI-24), 20% having only one end (19% for RPCI-23 and 21% for RPCI-24), and 10% having no ends (9% for RPCI-23 and 10% for RPCI-24). Although the overall performance of RPCI-24 was not as nearly good as that of RPCI-23, the current success rate of this library has increased and was comparable with that of RPCI-23 with longer reads (see below).
Table 1.
TIGR Mouse BAC End Sequencing Efforts (June 27, 2001)
| Library | RPCI-23 | RPCI-24 | Total |
|---|---|---|---|
| Sequencing success rate (%)a | 83 | 79 | 81 |
| Mouse BAC end sequences (mBESs) | 274277 | 174957 | 449234 |
| Clones with ≥1 end and coverageb | 154795, 10X | 102524, 5.3X | 257318, 15.3X |
| Paired-end clones and coveragec | 119483, 77%, 7.9X | 72433, 71%, 3.7X | 191916, 75%, 11.6X |
| Edited read length,d avg. ± S.D. bp | 466 ± 162 | 515 ± 188 | 485 ± 174 |
| Total bases, Mb | 128 | 90 | 218 |
| Avg. Q20 bases after trimminge, bp | 387 ± 154 | 438 ± 179 | 406 ± 166 |
| Repeat contents, %mBES and %basesf | 63 & 36.5 | 66 & 37.5 | 64 & 37 |
| Clones with ≥100 unique Q20 basesg | 65% | 66% | 66% |
| %mBESs matching mouse ESTs | 2.2 | 4.5 | 3 |
| hClone tracking accuracy | >95% | >95% | >95% |
Percent of sequencing attempts that yielded reads with an overall error rate <2.5%, an edited read length >100 bp, and free of E. coli and vector sequences.
Clones with at least one end sequence. Clone coverage was calculated assuming an average insert size of 197 kb for PRCI-23 library and 155kb for PCRI-24 library, and a genome size of 3 Gb.
Clones with both T7 and Sp6 end sequences (pairs). Clone coverage by pairs was calculated as described above. Total number of pairs, % of pairs in the database, and their coverage are shown.
Read length after sequences were trimmed. The average and standard deviation (S.D.) are shown.
Average number of bases with phred QV ≥ 20 per sequence after sequences were trimmed. The average and S.D. are shown.
Percent of mBESs that contained repeats and percent of bases that were repeats, analyzed by RepeatMasker.
Percent of paired-end clones that had at least one end with ≥100 bp contiguous unique Q20 bases.
Percent of mBESs that were associated with the correct clone identifiers.
Quality
Q20 Length Distributions
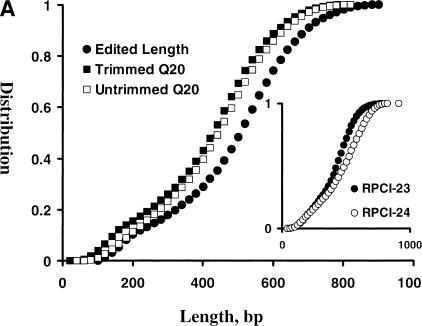
The base-call program phred (Ewing and Green 1998; Ewing et al. 1998) assigns a quality value (QV) to each base while processing the electropherogram. The sequence accuracy can be calculated by an equation as follows: error rate =10-phred QV/10, and a higher QV indicates a lower base-call error rate and a more accurate read. When a base has a phred QV ≥ 20, the error rate is ≤ 1% and the accuracy is ≥ 99%. This base is called a Q20 base or a high-quality base. It is a common practice to assess the sequence quality by the number of Q20 bases in each sequence (Q20 length). We therefore examined the phred QV of each base of mBES reads before and after trimming. For a total of 453,137 mBES traces (277,490 from RPCI-23 and 175,647 from RPCI-24) with a 388-Mb total, the Q20 length ranged from 10 to 910 bp with an average of 438 bp and a SD of 159 bp before trimming, and ranged from 3 to 839 bp with an average of 406 bp and a SD of 166 bp after trimming (Fig. 1A). The edited read length ranged from 100 to 940 bp with an average of 485 bp and a SD of 174 bp. RPCI-24 had a higher average Q20 length than RPCI-23, 461 versus 409 bp before trimming, and 438 versus 387 bp after trimming, and therefore had a longer average edited read length (515 bp versus 466 bp) (Table 1). The total bases after trimming were 219 Mb, of which 10% had phred QV < 15 and 84% had phred QV ≥ 20 (Fig. 1B). Again, RPCI-24 had an higher Q20 base fraction than RPCI-23, 86% versus 83%. Our results indicated that ∼84% of the bases had base-call error rates of ≤ 1% in each sequence on average and in the overall dataset.
Figure 1.
(A) mBES Q20 length and read-length distributions. A total of 453,137 mBES traces (277,490 with 225 Mb from RPCI-23 and 175,647 with 163 Mb from RPCI-24) were processed by a base call program phred and quality scores were further adjusted with TraceTurner from Paracel. The reads were then trimmed for low-quality bases and vector and E. coli sequences by a program lucy with criteria as follows: (1) a < 2.5% overall base-call error rate; (2) a minimum of 100 bp reads; and (3) free of vector and E. coli sequences. The phred QV of each base was examined and bases with QV ≥ 20 (Q20 bases) were counted for each sequence before and after trimming. The distributions of the Q20 length before trimming (□) and after trimming (▪), the edited read length (●) were plotted here. The Q20 length ranged from 10 to 910 bp with an average of 438 bp and a SD of 159 bp before trimming, and ranged from 3 to 839 bp with an average of 406 and a SD of 166 bp after trimming. The trimmed sequence reads ranged from 101 to 984 bp with an average of 485 bp with a SD of 174 bp. (Inset) The Q20 length distributions of the untrimmed sequences, indicating that RPCI-24 (○) had more Q20 bases than RPCI-23 (●). (B) phred QV compositions of RPCI-23 and RPCI-24 mBES databases. After the same set of mBESs were trimmed as described in Fig. 1A, phred QVs of 218,722,217 total bases (128,961,303 for RPCI-23 and 89,760,914 for RPCI-24) were examined, of which 10% had QV < 15 and 84% had QV ≥ 20. RPCI-24 (□) had a slightly higher Q20 base fraction (86%) than RPCI-23 (▪, 83%).
Repetitive DNA
Unique sequences are most useful for genome assembly and it is therefore desirable to know the repetitive DNA content in the mBES dataset. We analyzed 453,317 mBESs that consisted of 277,490 (129 Mb) RPCI-23 sequences and 175,647 (90 Mb) RPCI-24 sequences by RepeatMasker (http://ftp.genome.washington.edu/RM/RepeatMasker.html) (Smit 1996) and found that 64% of the sequences contained repeats and 37% of the bases were repeats. RPCI-24 mBESs had slightly more repeats than RPCI-23 mBESs (Table 1). The repeat sequences ranged from 20 to 925 bp with an average of 276 bp and a SD of 194 bp.
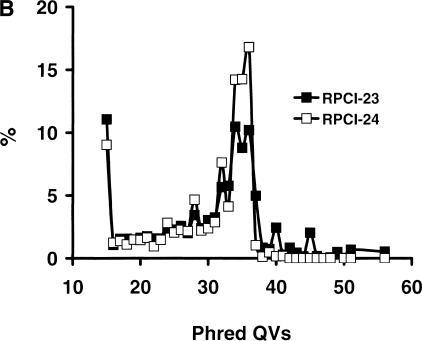
We examined the repeat composition of mBESs (Fig. 2) and found that a significant fraction were LINE1 (L1) repeats, 27.6% sequences and 21% bases for both libraries. The L1 repeats ranged from 11 to 925 bp with an average of 370 bp and accounted for 2%–100%, with an 80% average of the total bases in L1-containing mBESs, making a significant portion of mBESs less useful. To find out whether this high percentage of L1 represented the true repeat composition of the mouse genome, we examined 258-Mb mouse phase 1–3 genomic sequences and found that 30%–36% of the bases were repeats and 7%–15% were L1, depending on the sequencing phase (the lowest for phase 1 and the highest for phase 3 probably because of the sequence accuracy) and the GC content, indicating that mBESs from both libraries contained more L1 repeats than sequences that were obtained by complete sequencing of discrete mouse genomic regions up to 300 kb. To find out whether this was related to the cloning sites (EcoRI for RPCI-23 and MboI for RPCI-24) of the BAC libraries, we studied the frequency of EcoRI and MboI in L1 and L1-free sequences separated from the mouse phase 1–3 sequences. EcoRI occurred once every 3 kb in L1 repeats and once every 4 kb in L1-free sequences, thus increased by 33% in L1. Similarly, MboI occurred once every 0.3 kb in L1 and once every 0.4 kb in L1-free sequences, increasing by 33%. mBESs were similar to the phase 3 sequences in GC content, the increased occurrence of these restriction sites in L1 repeats would mostly explain the ∼33% more L1 in mBESs. To further test this hypothesis, we studied the repeat composition of 50,000 (24 Mb) plasmid ends from a 10-kb sheared DNA library submitted to dbGSS by the Utah genome center and have found that the overall repeat content was similar to that of mBESs; however, L1 only accounted for 20% of the sequences and 15% of the bases. Similar results were obtained for 50,000 trimmed mouse whole-genome shotgun reads that were randomly chosen from TraceArchive (http://www.ncbi.nlm.nih.gov/Traces/trace.cgi?). These studies further supported the hypothesis that L1 contents increased in the end sequences of clones from EcoRI and MboI partial digest libraries.
Figure 2.
Mouse repeats by RepeatMasker. The repeat contents were analyzed by RepeatMasker for 277,490 (129 Mb) RPCI-23 mBESs and 175,647 (90 Mb) RPCI-24 mBESs, 258-Mb mouse phase 1–3 sequences, 50,000 (24 Mb) end sequences of a 10-kb sheared DNA plasmid library from dbGSS, and 50,000 (36 Mb) mouse whole-genome shotgun (WGS) reads randomly chosen from the TraceArchive site. mBESs contained a significantly higher fraction of LINE (nearly all were L1); and sheared DNA plasmid ends and WGS reads contained a higher fraction of satellite repeats. DNA = MER1_type + MER2_type; LINE = LINE1 (L1) + LINE2 + L3/CR1; LTR = MaLRs + ERVL + ERV classI + ERV classII; SINE = B1s + B2 − B4 + IDs + MIRs.
Unique Q20 Bases
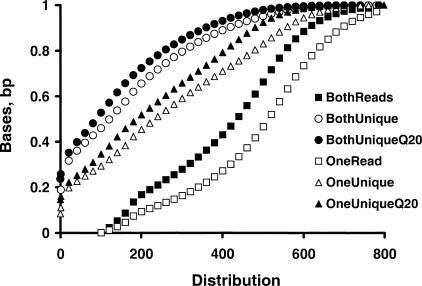
Genome assembly requires high-quality unique sequences; we therefore studied the Phred QVs of the unique sequences of mBESs. After 219 Mb of the same mBES dataset described above were repeat masked by RepeatMasker, we identified 124 Mb (57%) unique sequences, among which similar phred QV composition was observed as the entire dataset shown in Figure 1B, and a total of 105 Mb had QVs ≥ 20 (the unique Q20 bases). RPCI-24 dataset had ∼1% lower unique base, but similar unique Q20 base fractions compared with RPCI-23 dataset, and T7 ends had 2% lower unique base, and 1% lower unique Q20 base fractions than Sp6 ends, possibly because of their higher average Q20 length. Clones with paired ends (two-end clones) were more useful than clones with only one end. Therefore, we examined the distribution of various types of bases for two-end clones (119,483 from RPCI-23 and 72,433 from RPCI-24) (Fig. 3). By use of 100 bp as the cut off length, all clones had both ends above 100 bp, 71% of the clones had at least one end, and 54% had both ends, with ≥ 100 bp contiguous unique sequences; 66% had at least one end, and 48% had both ends, with ≥ 100-bp contiguous unique Q20 bases. The last fraction (48%) of the clones was most useful in genome assembly. Similar distributions were observed for both libraries, but RPCI-24 had on average ∼20 bp more unique Q20 bases than RPCI-23.
Figure 3.
Length distributions of sequence reads, contiguous unique sequences, and contiguous unique Q20 bases of mBES paired ends. The unique bases and associated phred QVs were examined for 383,832 repeat masked mBESs (186 Mb) that consisted of paired ends from 119,483 RPCI-23 clones and 72,433 RPCI-24 clones. The numbers of read length, contiguous unique bases, and contiguous unique Q20 bases were counted for each mBES. The analyses indicated that all clones have their two BESs above 100 bp [(□) read-length distribution of the two BESs of each clone, OneRead]; [(▪) read length distribution of the shorter BES of each clone, BothReads]; 71% of the clones had at least one end with ≥ 100 bp contiguous unique sequences [(▵) OneUnique] and 54% had both ends with ≥ 100 bp contiguous unique sequences [(○) BothUnique]; 66% had at least one end with ≥ 100 bp contiguous unique Q20 bases [(▴) OneUniqueQ20] and 48% had both ends with ≥ 100 bp contiguous unique Q20 bases [(●) BothUniqueQ20].
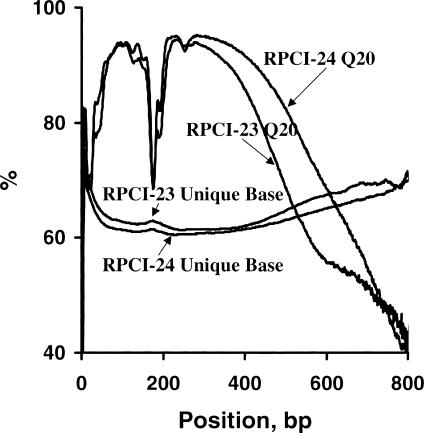
We examined the phred QV composition at each base position of mBESs because this information is useful for applications such as primer design. Our analyses indicated two high-quality regions with Q20 base fractions above 90%, bases 63–151 and 208–367 for RPCI-23, and 54–162 and 201–430 for RPCI-24 (Fig. 4). Lower quality regions included the first 30–40 bases and those after base 450 (RPCI-23) or 530 (RPCI-24), as well as a middle region (bases 168–184) that was possibly caused by a dye blob resulting from increased reaction cycles required to sequence BAC ends. RPCI-24 reads had higher quality than those of RPCI-23 at most positions, especially toward the 3′ end. For the same reasons discussed above, it is useful to associate the sequence quality with the repeat information. We therefore studied the repeat content at each base position and have observed similar profiles for both ends and for both libraries, except that repeat contents at most positions were higher in T7 ends than Sp6 ends, and for RPCI-24 than RPCI-23. The total repeat content increased from 19% at base 1 to above 36% at base 40 (RPCI-23) or 35 (RPCI-24), and remain above 36% until base 500 (RPCI-23) or 550 (RPCI-24). Unique base profiles were the reverse of those of repeats and were in an inverse relationship with those of Q20 bases (Fig. 4), indicating that more repeats can be identified from more accurate reads. Therefore, for a large dataset of random genome-wide sequences, a higher repeat content generally indicates a higher sequence quality. This was also supported by the bigger difference between RPCI-23 and RPCI-24 in both Q20 base and unique-base contents toward the 3′ end (Fig. 4). On the basis of our analyses, for studies such as primer design for RH mapping (Oliver et al. 2001) using mBESs, unique sequences from bases 63–151 and 208–367 for RPCI-23; and 54–162 and 201–430 for RPCI-24 were the best because of the high quality.
Figure 4.
Q20 base and unique sequence profiles of RPCI-23 and RPCI-24 mBESs. A total of 277,490 RPCI-23 mBESs with 129 Mb and 175,647 RPCI-24 mBESs with 90 Mb were examined at each base position for Q20 base and unique sequence contents. The unique sequence fraction was in an inverse relationship with the Q20 base fraction. Two high-quality regions were identified as follows: bases 63–151 and 208–367 of RPCI-23; and bases 54–162 and 201–430 of RPCI-24 had Q20 base fractions above 90%. These regions had lower unique base contents (< 65%). RPCI-24 had a higher Q20 base content (5%–10%) and lower unique base contents (1%–2%) at some regions, especially after base 450 bp.
Self-Comparison
To study the randomness of the mBES dataset, we searched each of 274,285 repeat-masked RPCI-23 mBESs against the database containing the same sequences and selected matches having identity ≥ 90%, length ≥ 100 bp, and unmatched bases at either 5′ or 3′ end (overhang) < 50 bp. We identified 19% mBESs matching other mBESs with an average identity of 96.6% and an average length of 283 bp. Among these mBESs, > 90% hit < 5 other sequences and under 1% hit > 50 other sequences. We did the same analyses with 171,858 mBES from RPCI-24 and found that 9% had hits with similar parameters. Because the average sequence identity was only 96.6%, the higher self-hit rate obtained for RPCI-23 was likely due to more false positives caused by repeat matches and might indicate that RPCI-23 mBESs contained more repeats that yet need to be identified. Only a small fraction of mBESs was identified in this study, indicating that mBESs were fairly randomly distributed on the genome.
Comparison with Finished Mouse Sequences
Paired-End Coverage
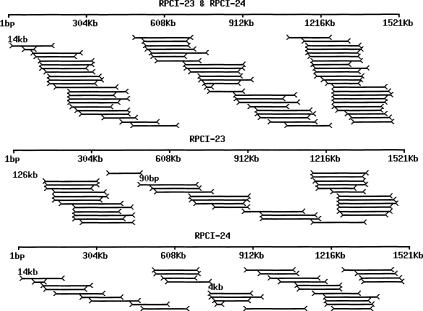
To study the effective coverage of the genome by paired-end clones, we matched a total of 191,916 mBES pairs (119,482 from RPCI-23 and 72,433 from RPCI-24) to two mouse contigs as follows: 1.5 Mb NT_002588 of chromosome 17 (C17) and 0.6 Mb NT_026540 of chromosome 5 (C5). We chose these contigs because they were big and seemed to be more representative of the genome than other contigs, as the overall repeat content was ∼30% closer to that of the whole genome shotgun reads (34%). Performing searches as described in the legend to Figure 5, we identified 69 pairs with 7.9× coverage and one 14-kb gap on C17 (Fig. 5) and 26 pairs with 7.9× coverage and one 4-kb gap on C5. The average insert size of matched pairs on the two contigs was calculated to be 193 kb with a SD of 29.7 kb for RPCI-23 and 162 kb with a SD of 36.7 kb for RPCI-24, similar to the reported values [(http://www.chori.org/bacpac/23framefmouse.htm and http://www.chori.org/bacpac/mmouse24.htm, Osoegawa et al. (2000)]. The gaps were located at the beginning of the contigs and were the result of one end of the potentially matched pairs matching outside of the contigs. Among the identified pairs on C17, 36 with 4.6× coverage were from RPCI-23 and 33 with 3.4× coverage were from RPCI-24 (Fig. 5). Similar coverage was observed for C5. The observed coverage here was lower than the expected 7.8× for RPCI-23, showing 119,482 pairs and an insert size of 197 kb, and was closer to the expected 3.4× for RPCI-24, showing 72,433 pairs and an insert size of 155 kb, possibly because of more false negatives caused by repeat masking for RPCI-23 and likely indicating that RPCI-24 was more random.
Figure 5.
Effective clone coverage and gaps by paired ends of RPCI-23 and RPCI-24. The 1.5-Mb mouse chromosome 17 contig NT_002588|Mm17_2723 was searched against a database consisting of 119,482 paired ends from RPCI-23 (7.8× clone coverage assuming an average insert size of 197 kb) and 72,433 paired ends from RPCI-24 (3.7× clone coverage assuming an average insert size of 155 kb). The searches were done in two steps as follows: (1) The contig sequence was searched against repeat-masked BESs by BLASTN; (2) the same sequence was researched against a mini-database consisting of unmasked paired ends from the clones identified by step one. Multiple hits were excluded and matches (1) with above, identity ≥ 95%; (2) match length ≥ 3/4 mBES length; (3) paired ends pointing toward each other; and (4) insert size ≤ 400 kb were selected. We have identified a total of 36 RPCI-23 pairs with a clone coverage of 4.6× and an average insert size of 191.6 kb (± 28.5 kb); and 33 RPCI-24 pairs with a clone coverage of 3.4× and an average insert size of 156 kb (± 39.7 kb). The paired-ends matches were plotted with the top for RPCI-23 and RPCI-24 (69 pairs with 7.9× coverage and one 14-kb gap), the middle for RPCI-23, and the bottom for RPCI-24. Bars represent the contig. Lines with arrows at both ends represent the matched pairs. Numbers represent the gaps.
Sequence Identity
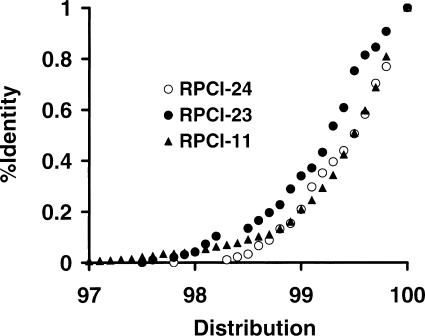
Together with Q20 length, mBES sequence accuracy can also be assessed by examining their identities to finished sequences. We therefore studied the identities of mBESs to the mouse contigs and found an average of 99.4% for RPCI-24 and 99.2% for RPCI-23. RPCI-24 mBESs were more identical to finished sequences than RPCI-23 mBESs (Fig. 6), consistent with the Q20-length results. As a comparison, Figure 6 also showed identities of TIGR human BES pairs from library RPCI-11 to human finished chromosome 21 sequences and the distribution was almost identical to that of RPCI-24 mBES. The high-sequence identities to finished sequences showed that mBESs from both RPCI-23 and RPCI-24 were sufficiently accurate for any applications involved.
Figure 6.
Identity distributions between finished sequences and BESs for RPCI-23, RPCI-24, and RPCI-11. The sequence comparison between mBESs and mouse contig sequences were done as described in Fig. 5. The average identity was 99.2% with a SD of 0.6% for RPCI-23 (●), and 99.4% with an SD of 0.5% for RPCI-24 (○). The same analyses were performed by comparing a total of 62,226 human BES pairs (3.4× coverage) from RPCI-11 library with finished human chromosome 21 sequence (34 Mb). A total of 779 pairs (3.4× coverage) were identified matching the chromosome. The average identity was 99.4% with a SD of 0.6% (▴).
Sequence Coverage
Upon examining the match length of mBESs to the mouse contigs C5 and C17, we found a 482-bp average or 2.2% sequence coverage for RPCI-23, and a 475-bp average or 2.1% coverage for RPCI-24. These were lower than the expected 3.7% coverage for RPCI-23 and similar to the expected 2.5% for RPCI-24, possibly because repeat masking obscured more of the real matches for RPCI-23 and RPCI-24 was more random.
Clone Tracking
Along with a high-sequencing success rate, a good clone tracking system is required for large-scale BAC end sequencing. The process involves hundreds of thousands of clones and mistakes can occur at any steps, such as library replication, clone picking, cell growth, template preparation, cycling reaction, electrophoresis, and database loading. TIGR is one of the two centers that conducted large-scale human BAC end sequencing and our tracking analyses showed that > 90% of the human BESs were associated with the right clones (Zhao et al. 2000). The analyses also indicated that > 90% tracking errors originated from lane mistracking with the ABI377 sequencers (the other 10% were caused by errors such as plate mishandling, primer mislabeling, etc.). We switched to the ABI3700 machines for the mouse project. Taking advantage of these capillary sequencers, we have incorporated into the process a complete tracking check that, for each 96-well plate, involved sequencing two ends of a clone directly picked from the 384-well master plate and matching the sequences to the corresponding BESs obtained from the large-scale process. More automation has also been introduced to the process. To find out how much improvement has been achieved, we conducted a comprehensive tracking analysis involving both external and internal data, which not only evaluated the effectiveness of the modified process and helped to identify places in which further improvement is required, but also provided BES users information on clone fidelity in the dataset.
Comparing BES to the External Data
Overlapping BESs Between Laboratories
One way to assess the clone-tracking accuracy is to examine the discrepancy of BESs from a statistically significant number of clones that were independently sequenced by TIGR and another center. This analysis was reported for the human BAC end-sequencing project (Zhao et al. 2000). With the mouse, no data were available for such an analysis as no other genome centers were conducting BAC end sequencing on a large-scale for mice. Only a small number of RPCI-23 and RPCI-24 clones have been end sequenced in individual laboratories for studies such as mapping. In one such case, only 3 of 51 mBESs did not match (M. Bucan, pers. comm.) and the match rate was 94%.
Matching to BAC Sequences of the Same Clones
Another possible method for tracking assessment is to match the BESs to the BAC sequences of the same clones whose entire insert have been sequenced as a result of the Human Genome Project (BAC Resource Consortium 2001; International Human Genome Sequencing Consortium 2001; Zhao 2001a). We examined ∼1000 RPCI-23 phase 1–3 clones and found that the match rates ranged from 68% to 77%. The fundamental problem with this analysis was that many such clones did not contain end sequences because of the following: (1) the low-sequence coverage and gaps, especially for phase 1 clones, and (2) the trimming of overlapping clone ends (the end sequences of some phase 3 clones were trimmed because they overlapped with part of the clones whose sequences have been deposited in GenBank). Therefore, this was not a valid way to assess the clone-tracking accuracy.
Matching to Large Contigs
Another way to assess clone tracking is to compare BES pairs with a large contig and select those having both ends matching the contig with correct orientation and reasonable insert sizes. Excluding false positives that arise from repeat matches and false negatives that are caused by low-quality sequences, a higher fraction of pairs with such matches generally indicates a more accurate clone tracking. We compared ∼100,000 mBES pair-end clones to mouse chromosome 17 contig (1.5 Mb) and found that 85% of the identified clones had two ends matching to the contig. The same analyses were conducted by matching ∼100,000 human BES pairs to chromosomes 21 (Hattori et al. 2000) and 22 (Dunham et al. 1999); the match rate was 80%. Although we used BESs that were masked with known repeats (Smit 1996), there were still matches caused by the genome-wide or chromosome-specific repeats that still need to be identified (especially for the mouse). Therefore, the actual tracking accuracy should be higher than 85% for mBESs.
The comparisons of TIGR BESs to the external data indicated an improved performance for the mouse. To look at our site only, we conducted the following analyses with the internal data that included all clones whose end(s) has been successfully sequenced from both the large-scale operation and another independent process.
Internal Sequence Comparisons
In the course of the project, a small fraction of clones were additionally end sequenced by processes that were independent of the large-scale operation and included the following: (1) each corner clone of 384-well plates was sequenced twice to ensure that the re-arraying from 384-well plates to 96-well plates was carried out accurately before clones entering the large-scale pipeline (re-array check sequencing); (2) for each 96-well plate, a clone picked directly from the 384-well master plate was end sequenced and matched to the corresponding large-scale BESs to verify the accuracy of the entire operation (final tracking check sequencing); (3) a number of clones were resequenced for reasons such as per BES user's request, suspicious tracking, the unusually low sequencing success rate, or testing new protocols (priority clone sequencing). The comparison results of BESs from these processes to the corresponding large-scale BESs follows. Of a total of 1033 two-end clones, 93% had both-ends-match and 5% had one-end-match; of 2321 total one-end clones, 95% matched. Overall, 95% BESs matched. The same analysis was conducted for the human RPCI-11 clones for comparison purposes; of 1092 total two-ends clones, 88% had both-ends-match and 9% had one-end-match; of 4711 total one-end clones, 90% matched; and 91% BES matched overall.
Another set of data to assess the tracking came from sequencing clones that were provided to us by a collaborator. The clones were from a different copy of the same library (RPCI-23) and therefore provided a good source to evaluate the entire process from library replicating to sequence generation. We compared these mBESs with the corresponding large-scale sequences and obtained the following results: for 418 two-end clones, 91% had both-ends-match and 5% had one-end-match; for 216 total one-end clones, 89% matched, and the overall matching rate was 92% of mBESs. The rates here were somewhat lower than the values obtained above, possibly because these clones have been through more growth and selection cycles that potentially could introduce more problems such as human errors.
The clone tracking for our large-scale process should be more accurate than the apparent sequence match rate of 95% obtained here. This is because unlike the large-scale operation in which clones were picked by a robot, the clones in the processes described above were generally picked by hand and more human errors could be introduced. Beside the match rates, it would be useful to know the percentages in which clear mistracking happened. However, such analyses were more complicated. The phred quality scores of the external sequences used in the analyses were not available and therefore we could not distinguish false negatives due to the low quality data. For internal sequence comparisons, we only found very few cases (< 10) in which mismatches were clearly due to mishandling of the large-scale process (clone mispicking, plates misaligned, primer mislabeled, etc.). More studies need to be done to find out other reasons such as more than one type of BAC population in a well.
Sequence Analyses
EST Contents
A total of 453,137 repeat-masked mBESs (277,490 RPCI-23 and 175,647 RPCI-24) were searched against human, mouse, and rat EST databases at TIGR (Adams et al. 1991, 1995; Quackenbush et al. 2000) and matches with identity ≥ 95% and score ≥ 300 were selected. The results indicated that a larger fraction of mBESs matched mouse ESTs with a higher identity and a longer match length on average, 3% mBESs matched mouse ESTs with a 98% identity and a 200-bp length, whereas 0.1% mBESs matched human ESTs with a 97% identity and a 182-bp length, and 0.2% mBESs matched rat ESTs with a 97% identity and a 167-bp length. A higher percentage of RPCI-24 mBESs was found to match ESTs (Table 1). Matched ESTs were involved in signal transduction, cell defense, gene expression, structure, metabolism, and other functions. A significant fraction of the matches were found to be conserved between the species, 71% between the human and the mouse, 70% between the mouse and the rat, 36% between the human and the rat, and 27% among the three species. Most of the conserved matches were not classified (80%), whereas others included heat shock proteins, transcription factors, and ribosome proteins. We did the same analyses with the Unigene database and obtained similar results.
STS Contents
By running e-PCR (Schuler 1998) with 210,412 STSs on the same set of mBESs as above, we identified 1198 matches that involved 1130 mBESs and 882 STSs. The analyses made chromosome assignments to 933 BACs (531 for RPCI-23 and 402 for RPCI-24), with more on chromosomes 9 and 11 and fewer on chromosomes 14 and X. Only two BACs were assigned to more than one chromosome.
Comparison with Human Finished Sequences
tBLASTX compares query nucleotide sequences with a nucleotide database on the protein level by six-frame translation. Using tBLASTX, we compared 197,099 repeat-masked RPCI-23 mBESs with 2355 human contigs (544 Mb total), ranging from 1.8 kb to 34 Mb from GenBank and selected matches with identity ≥ 90% and match length ≥ 50 bp. We have found 922 matches by 350 mBESs with a 95% identity and a 269-bp length on average. The match frequency was one every 590 kb overall and one every 1–1.5 Mb on finished chromosomes 20, 21, and 22. The higher frequency on other chromosomes might be due to non-randomness of the finished sequences or the selection of gene-dense regions for sequencing. We have found 71% of the identified mBESs had hits to ESTs of the mouse (67%), human (37%), or rat ( 33%). The majority of these ESTs were not categorized, and those characterized included ribosomal proteins, ubiquitin, and other abundant proteins. We currently are repeating the analyses to map all mBESs from libraries RPCI-23 and RPCI-24 to the assembled human genomes, both the public version (GoldenPath, NCBI) and the private version (www.celera.com).
One concern with tBLASTX was speed, and it took months to finish the analyses. We therefore looked for alternatives such as BLASTN. With appropriate parameters and matrix (http://sapiens.wustl.edu/∼ikorf/mmhs/index.html), BLASTN can achieve a high sensitivity with tremendous gains on speed, as it compares sequences on the nucleotide level. We compared matches identified by tBLASTX using BLASTN and found that all had nucleotide identities above 76% and a majority (95%) were above 80% with an average of 90%. The lower identity on the nucleotide level was due to third-base wobble. An effective approach to place mBESs onto the human genome might be a two-step comparison by searching the entire database first with liberal criteria by BLASTN and then using tBLASTX to search the smaller dataset of potential candidates identified by BLASTN.
DISCUSSION
The goal of a large-scale BAC end-sequencing project is to generate dense and accurate end-sequence pairs that are randomly distributed across the genome. With this goal in mind, we have been end sequencing mouse BACs on a large-scale to support the mouse genome project and have generated ∼450,000 mBESs from ∼260,000 RPCI-23 and RPCI-24 BACs with 75% of the clones having paired ends. We have improved the protocol used for sequencing our human BAC ends and increased the success rate to > 80%. As a result, for the 273,000 clones that were attempted at both ends, 70% have two ends, 20% have one end, and 10% have no ends. The sequencing is performed with ABI3700 capillary sequencers that have greatly improved the sample tracking accuracy and > 95% of the sequences are from the right clones (below). With an average read length of 485 bp, the sequences are slightly longer than our human BAC ends (Zhao et al. 2000) and add up to 219 Mb or 7% of the genome. With a 406-bp average Q20 length and 84% of the bases having phred QV ≥ 20, mBESs match mouse finished sequences with an average identity of > 99%. The project therefore generates accurate genome-wide sequence pairs and provides the dense markers supporting the mouse genome project.
We have end sequenced the entire RPCI-23 library (170,000 clones) and are approaching our goal of 130,000 clones for the RPCI-24 library. Although the current success rate with both libraries is similar, both the average Q20 length and the average read length are longer with RPCI-24. Consequently, a higher percentage of RPCI-24 mBESs hit repeat database, mouse finished sequences, ESTs, and STSs with a longer length and a higher identity on average. Although mBESs from both libraries seem to be fairly randomly distributed on the mouse genome based on the analyses, RPCI-24 sequences seem to be more random and unique because the observed pair and sequence coverage are closer to the expected, and fewer sequences have hits in the mBESs database itself. Therefore, it seems to be more useful to end sequence more clones from BAC libraries that were made with the 4-base cutter MboI.
Repeats present problems in genome assembly and we have found that 65% of mBESs and 36% of the bases contain known genome-wide repeats. The most useful end-sequenced clones are those having both ends with ≥ 100-bp unique Q20 bases, which are found to be 48% in the database. Compared with the mouse phase 1–3 genomic sequences, as well as the end sequences from a sheared DNA plasmid library and the mouse whole-genome shotgun reads, mBESs contain significantly more L1 repeats as a result of more frequent occurrence of EcoRI and MboI sites in this type of repeats. We believe that end sequencing multiple BAC libraries with different cloning sites and with sheared DNA will make the resource more useful, as the sequences will better represent the genome.
The success of large-scale BAC end sequencing requires not only a high-sequencing success rate but also an accurate clone tracking. The linkage between the sequence data and the clones is critical because the data are only as useful as the corresponding clone is retrievable. BAC end sequencing involves hundreds of thousands of clones and mistakes can happen at many stages of the process; a good tracking system is therefore required. Our sample tracking involves both automation and a laboratory information management system (LIMS) that is based around a set of databases implemented in Sybase and uses bar codes at several stages of the sequencing process. Prior to the mouse project, TIGR had generated 300,000 BESs from 180,000 human clones and the evaluation indicated that > 90% of human BESs are associated with the right clone identifiers (Zhao et al. 2000). The analyses have revealed several sources of tracking error, however, > 90% of the errors were found to originate from lane mis-tracking with ABI377 sequencers. We have switched to the ABI3700 machines and introduced a complete control step into the process by taking advantage of the capillary sequencers, one clone's correct tracking ensures the correct tracking of the entire plate of clones. In addition, more automation has been introduced into the process, which has potentially reduced human errors. All tracking analyses with both the external and internal data indicate a more accurate performance for the mouse. We are confident that at least 95% of mouse ends are associated with the right clone identifiers.
To better characterize this valuable resource, we compared mBESs with the finished human sequences by tBLASTX on the protein level and found ∼70% of the identified mBESs have EST hits, indicating that the majority of the conserved regions are transcribed. Gene densities vary with chromosomes (Crollius et al. 2000; Ewing and Green 2000; Liang et al. 2000) and chromosomes 17, 19, and 22 are gene rich, whereas chromosomes 4, 18, 21, and X are gene poor. Our match frequency also varies with chromosomes and is ∼1 every 1–1.5 Mb on chromosome 21 and 22. The mBESs dataset used in the study accounts for 3% of the genome, which would indicate 1 hit per 30–45-kb human sequences by mBESs with 100% sequence coverage. This somewhat supports 1 gene per 85 kb for the human genome (Ewing and Green 2000). We found that 3%, 0.1%, and 0.2% of mBESs match mouse, human, and rat ESTs, respectively, and a majority of the matches are conserved between these species, further supporting the hypothesis that transcribed sequences are more conserved. Approximately 0.1% mBESs contain STS markers, which made chromosome assignment to > 900 BACs. Our analyses indicate that the mouse BAC ends resource will be even more useful than the human BAC ends resource for many research fields.
METHODS
BAC Libraries
Mouse BAC libraries RPCI-23 and RPCI-24 were purchased from BACPAC RESOURCES at Children's Hospital Oakland Research Institute (http://www.chori.org/bacpac/orderingframe.htm). RPCI-23 was made by cloning the EcoR1/EcoRI methylase partially digested female C57BL/6J DNA in the pBACe3.6 cloning vector at the EcoR1 site (Osoegawa et al. 2000). RPCI-24 was made by cloning the MboI partially digested male C57BL/6J DNA in the pTARBAC1 cloning vector at the BamH1 site.
BAC End Sequencing
BAC end sequencing and trimming were performed following the basic procedure as described (Kelley et al. 1999) with a few modifications. The sequencing was conducted on the ABI3700 capillary sequencers. The process included BAC template preparation, cycling reactions, electrophoresis, and sequence trimming. Detailed protocols can be found at http://www.tigr.org/tdb/bac_ends/mouse/bac_end_intro.html. Briefly, BAC template preparation was based on the 96-well format. BAC libraries were received in 384-well plates and were arrayed to 96-well plates with a robot (Flexys) before sequencing. BAC clones were innoculated into one 96-deep well block containing 1.5 mL/well medium, and grown overnight in an oxygenated growth chamber (HI-GRO from GeneMachines) at 37°C. The cells were harvested by centrifugation and the BACs were purified by use of a 96-well BAC purification kit from QIAGEN. The cells were lysed by a standard alkaline lysis protocol, and passed over a QIAGEN Turbo filter plate using vacuum filtration. BAC DNA was precipitated with isopropanol and washed with 70% ethanol. The DNA was then resuspended in 35 μL of resuspension buffer (1 mM Tris at pH 8.0) and 10 μL was used per sequencing reaction once the quality of DNA was checked on agarose gels. Sequencing reactions were performed using 0.7-strength Big Dye terminator chemistry (ABI PN 4303154), 0.1–0.5 μg of template DNA, 4 pmoles of primer, 1× CSA sequencing buffer (ABI PN 361028C), and MgCl2 added to 1.4 mM. The cycling reactions were conducted with MJ Tetrad Thermal Cyclers under the following conditions: 96°C – 2 min; cycle 74× 96°C – 10 sec, 54°C – 10 sec, 60°C – 4 min; 4°C – hold. The standard T7 primer and a custom-designed primer (CTGGCCGTCGA CATTTAGG) at the SP6 end were used. The reaction mixture was then cleaned up by isopropanol precipitation followed by 70% ethanol wash. Electrophoresis of the reaction mix was carried out with the ABI 3700 Automated DNA Sequencers using POP5 polymer. Sequence trimming was conducted by processing the traces using base-calling software phred (Ewing and Green 1998; Ewing et al. 1998) and the quality scores were further adjusted with TraceTuner from Paracel that was specifically trained for our ABI3700 data with POP5, and the sequences were then trimmed by a locally written software lucy (http://www.tigr.org/softlab/) with criteria of overall base-call error < 2.5%, reads > 100 bp, and free of vector and E. coli sequences.
Clone Tracking
The linkage between the sequences and the clones was tracked using a laboratory information management system (LIMS) that was based around a set of databases implemented in Sybase and used barcodes at several stages of the sequencing process. In addition, two more tracking control steps were built into the process. (1) For template preparation, a re-array check was conducted to ensure that 96-well plates were arrayed (by a robot) correctly from their 384-well master plates. This involved picking clones from the corners of 384-well plates and their corresponding 96-well plates, and sequencing to verify that the clones were identical. (2) A final complete clone tracking check was conducted to verify the accuracy of the entire process. For each 96-well plate, one clone was picked directly from the 384-well master plate, sequenced from both ends, and compared with its corresponding sequences from the large-scale process.
Data Source
Sequences other than BAC ends used in the analyses were downloaded from GenBank. BAC end sequences can be searched by clone or by sequence at http://www.tigr.org/tdb/humgen/bac_end_search/bac_end_search.html and the entire database can be ftped at ftp://ftp.tigr.org/pub/data/m_musculus/bac_end_sequences/.
Sequence Searches
Sequence comparisons were performed using programs BLAST series (Altschul et al. 1990; Altschul and Gish, 1996) and the BLAST version used for all searches was WU-BLAST2.0 from Washington University (http://blast.wustl.edu/). BLAST outputs were parsed out in a tab-delimited format using the program btab (http://www.tigr.org/softlab/). A multiple FASTA file was searched against a database one by one using the program sx from NCBI. The searches were performed with a parallel virtual machine Linux cluster consisting of 14 nodes, each with a 450 MHZ Pentium II, 512 MB RAM and 18 GB storage space.
Sequence Repeats Masking
Sequences were repeat masked by either RepeatMasker (http://ftp.genome.washington.edu/RM/RepeatMasker.html) or its faster version MaskerAids (http://sapiens.wustl.edu/maskeraid/) that replaces crossmatch with BLASTN.
Sequence Analyses and Graphical Presentations
Perl and shell scripts were used extensively for the analyses. Graphical plots were generated using either a Perl module GD.pm, Microsoft Excel, or PowerPoint.
Acknowledgments
We are grateful for the excellent sequencing work provided by all present and past members of TIGR BAC Ends Team; to Michael Heaney, Michael Holmes, Susan Lo, Eddy Arnold, Mark Sengamalay, Billy Lee, and other informatics members at TIGR for their database support; to all the genome centers producing the human and mouse sequence data used in the analyses; and to Mark Adams, Warren Gish, Allan Bradley, Richard Gibbs, Maja Bucan, Kristi Berry, John Gill, Kazu Osegowa, Pieter de Jong, Ken Dewar, Marvin Stodolsky, Adam Felsenfeld, Doug Smith, John Quankenbush, Bruce Roe, Arian Smit, and Jerzy Jurka for their critical comments and useful discussion. This work was supported by Grant U01-HG02137 from NIH to S.Z. and W.C.N.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL szhao@tigr.org; FAX (301) 838-0208.
Article published on-line before print: Genome Res., 10.1101/gr.179201.
Article and publication are at http://www.genome.org/cgi/doi/10.1101/gr.179201.
REFERENCES
- Adams MD, Kelley JM, Gocayne JD, Dubnick M, Polymeropoulos MH, Xiao H, Merril CR, Wu A, Olde B, Moreno RF, et al. Complementary DNA sequencing: Expressed sequence tags and human genome project. Science. 1991;252:1651–1656. doi: 10.1126/science.2047873. [DOI] [PubMed] [Google Scholar]
- Adams MD, Kerlavage AR, Fleischmann RD, Fuldner RA, Bult CJ, Lee NH, Kirkness EF, Weinstock KG, Gocayne JD, White O, et al. Initial assessment of human gene diversity and expression patterns based upon 83 million nucleotides of cDNA sequence. Nature. 1995;377:3–174. [PubMed] [Google Scholar]
- Altschul SF, Gish W. Local alignment statistics. Methods Enzymol. 1996;266:460–480. doi: 10.1016/s0076-6879(96)66029-7. [DOI] [PubMed] [Google Scholar]
- Altschul S, Gish W, Miller W, Myers E, Lipman D. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- BAC Resource Consortium. Integration of cytogenetic landmarks into the draft sequence of the human genome. Nature. 2001;409:953–958. doi: 10.1038/35057192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crollius RH, Jaillon O, Bernot A, Dasilva C, Bouneau L, Fischer C, Fizames C, Wincker P, Brottier P, Quetier F, et al. Estimate of human gene number provided by genome-wide analysis using Tetraodon nigroviridis DNA sequence. Nat Genet. 2000;25:235–238. doi: 10.1038/76118. [DOI] [PubMed] [Google Scholar]
- Deloukas P, Schuler G, Gyapay G, Beasley EM, Soderlund C, Rodriguez-Tome P, Hui L, Matise TC, et al. A physical map of 30,000 human genes. Science. 1998;282:744–746. doi: 10.1126/science.282.5389.744. [DOI] [PubMed] [Google Scholar]
- Dietrich WF, Miller J, Steen R, Merchant MA, Damron-Boles D, Husain Z, Dredge R, Daly MJ, Ingalls KA, O'Connor TJ, et al. A comprehensive genetic map of the mouse genome. Nature. 1996;380:149–152. doi: 10.1038/380149a0. [DOI] [PubMed] [Google Scholar]
- Dunham I, Shimizu N, Roe BA, Chissoe S, Hunt AR, Collins JE, Bruskiewich R, Beare DM, Clamp M, Smink LJ, et al. The DNA sequence of human chromosome 22. Nature. 1999;402:489–495. doi: 10.1038/990031. [DOI] [PubMed] [Google Scholar]
- Ewing B, Green P. Base-calling of automated sequencer traces using phred. II. Error probabilites. Genome Res. 1998;8:186–194. [PubMed] [Google Scholar]
- ————— Analysis of expressed sequence tags indicates 35,000 human genes. Nat Genet. 2000;25:232–234. doi: 10.1038/76115. [DOI] [PubMed] [Google Scholar]
- Ewing B, Hillier L, Wendl MC, Green P. Base-calling of automated sequencer traces using phred. I. Accuracy assessment. Genome Res. 1998;8:175–185. doi: 10.1101/gr.8.3.175. [DOI] [PubMed] [Google Scholar]
- Fleischmann RD, Adams MD, White O, Clayton RA, Kirkness EF, Kerlavage AR, Bult CJ, Tomb JF, Dougherty BH, Merrick JM, et al. Whole-genome random sequencing and assembly of Haemophilus influenzae Rd. Science. 1995;269:496–512. doi: 10.1126/science.7542800. [DOI] [PubMed] [Google Scholar]
- Hattori M, Fujiyamam A, Taylor TD, Watanabe H, Yada T, Park HS, Toyoda A, Ishii K, Totoki Y, Choi DK, et al. The DNA sequence of human chromosome 21. The chromosome 21 mapping and sequencing consortium. Nature. 2000;405:311–319. doi: 10.1038/35012518. [DOI] [PubMed] [Google Scholar]
- Hudson TJ, Stein LD, Gerety SS, Ma J, Castle AB, Silva J, Slonim DK, Baptista R, Kruglyak L, Xu SH, et al. An STS-based map of the human genome. Science. 1995;270:1945–1954. doi: 10.1126/science.270.5244.1945. [DOI] [PubMed] [Google Scholar]
- International Human Genome Sequencing Consortium. Initial sequencing and analysis of the human genome. Nature. 2001a;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- International Human Genome Mapping Consortium. A physical map of the human genome. Nature. 2001b;409:934–941. doi: 10.1038/35057157. [DOI] [PubMed] [Google Scholar]
- Kelley JM, Field CE, Craven MB, Bocskai D, Kim U, Rounsley SD, Adams MD. High throughput direct end sequencing of BAC clones. Nucleic Acids Res. 1999;27:1539–1546. doi: 10.1093/nar/27.6.1539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim UJ, Birren B, Sheng YL, Slepak T, Mancino V, Boysen C, Kang HL, Simon MI, Shizuya H. Construction and characterization of a human Bacterial Artificial Chromosome library. Genomics. 1996a;34:213–218. doi: 10.1006/geno.1996.0268. [DOI] [PubMed] [Google Scholar]
- Kim UJ, Shizuya H, Kang H-L, Choi SS, Garrett LL, Smink LJ, Birren BW, Korenberg JR, Dunham I, Simon MI, et al. A bacterial artificial chromosome-based framework contig map of human chromosome 22q. Proc Natl Acad Sci. 1996b;93:6297–6301. doi: 10.1073/pnas.93.13.6297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang F, Holt I, Pertea G, Karamycheva S, Salzbergm S,L, Quackenbush J. Gene index analysis of the human genome estimates approximately 120,000 genes. Nat Genet. 2000;25:239–240. doi: 10.1038/76126. [DOI] [PubMed] [Google Scholar]
- Marra M, Kucaba T, Dietrich N, Green ED, Brownstein B, Wilson RK, McDonald KM, Hillier LW, McPherson JD, Waterson RH, et al. High throughput fingerprint analysis of large-insert clones. Genome Res. 1997;7:1072–1084. doi: 10.1101/gr.7.11.1072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarthy LC, Terrett J, Davis ME, Knights CJ, Smith AL, Critcher R, Schmitt K, Hudson J, Spurr NK, Goodfellow PN. A first-generation whole genome-radiation hybrid map spanning the mouse genome. Genome Res. 1997;7:1153–1161. doi: 10.1101/gr.7.12.1153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olivier M, Aggarwal A, Allen J, Almendras AA, Bajorek ES, Brady SD, Bushard JM, Bustos VI, Chu A, Chung TR, et al. A high-resolution radiation hybrid mMap of the human genome draft sequence. Science. 2001;291:1298–1302. doi: 10.1126/science.1057437. [DOI] [PubMed] [Google Scholar]
- Osoegawa K, Tateno M, Woon PY, Frengen E, Mammoser AG, Catanese JJ, Hayashizaki Y, de Jong PJ. Bacterial artificial chromosome libraries for mouse sequencing and functional analysis. Genome Res. 2000;10:116–128. [PMC free article] [PubMed] [Google Scholar]
- Quackenbush J, Liang F, Holt I, Pertea G, Upton J. The TIGR gene indices: Reconstruction and representation of expressed gene sequences. Nucleic Acids Res. 2000;28:141–145. doi: 10.1093/nar/28.1.141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuler GD. Electronic PCR: Bridging the gap between genome mapping and genome sequencing. Trends Biotechnol. 1998;16:456–459. doi: 10.1016/s0167-7799(98)01232-3. [DOI] [PubMed] [Google Scholar]
- Schuler GD, Boguski MS, Stewart EA, Stein LD, Gyapay G, Rice K, White RE, Rodriguez-Tome P, Aggarwal A, Bajorek E, et al. A gene map of the human genome. Science. 1996;274:540–546. [PubMed] [Google Scholar]
- Shizuya H, Birren B, Kim UJ, Mancino V, Slepak T, Tachiiiri Y, Simon M. Cloning and stable maintenance of 300-kilobase-pair fragments of human DNA in Escherichia coli using an F-factor-based vector. Proc Natl Acad Sci. 1992;89:8794–8797. doi: 10.1073/pnas.89.18.8794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smit AF. The origin of interspersed repeats in the human genome. Curr Opin Genet Dev. 1996;6:743–748. doi: 10.1016/s0959-437x(96)80030-x. [DOI] [PubMed] [Google Scholar]
- Stewart EA, McKusick KB, Aggarwal A, Bajorek E, Brady S, Chu A, Fang N, Hadley D, Harris M, Hussain S, et al. An STS-based radiation hybrid map of the human genome. Genome Res. 1997;7:422–433. doi: 10.1101/gr.7.5.422. [DOI] [PubMed] [Google Scholar]
- Van Etten WJ, Steen RG, Nguyen H, Castle AB, Slonim DK, Ge B, Nusbaum C, Schuler GD, Lander ES, Hudson TJ. Radiation hybrid map of the mouse genome. Nat Genet. 1999;22:384–387. doi: 10.1038/11962. [DOI] [PubMed] [Google Scholar]
- Venter, J.C., Smith, H., and Hood, L.A. 1996. New strategy for genome sequencing. [DOI] [PubMed]
- Zhao S. Human BAC ends. Nucleic Acids Res. 2000;28:129–132. doi: 10.1093/nar/28.1.129. . 1996. Nature 381: 364–366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venter JC, Adams MD, Sutton GG, Kerlavage AR, Smith HO, Hunkapiller M. Shotgun sequencing of the human genome. Science. 1998;280:1540–1542. doi: 10.1126/science.280.5369.1540. [DOI] [PubMed] [Google Scholar]
- Venter JC, Adams MD, Myers EW, Li PW, Mural RJ, Sutton GG, Smith HO, Yandell M, Evans CA, Holt RA, et al. The sequence of the human genome. Science. 2001;291:1304–1351. doi: 10.1126/science.1058040. [DOI] [PubMed] [Google Scholar]
- Zhao S. A comprehensive BAC resource. Nucleic Acids Res. 2001;29:141–143. doi: 10.1093/nar/29.1.141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao S, Malek J, Mahairas G, Fu L, Nierman W, Venter JC, Adams MD. Human BAC rnds quality assessment and sequence analyses. Genomics. 2000;63:321–332. doi: 10.1006/geno.1999.6082. [DOI] [PubMed] [Google Scholar]