Abstract
Animal models have been used primarily as surrogates for humans, having similar disease-based phenotypes. Genomic organization also tends to be conserved between species, leading to the generation of comparative genome maps. The emergence of radiation hybrid (RH) maps, coupled with the large numbers of available Expressed Sequence Tags (ESTs), has revolutionized the way comparative maps can be built. We used publicly available rat, mouse, and human data to identify genes and ESTs with interspecies sequence identity (homology), identified their UniGene relationships, and incorporated their RH map positions to build integrated comparative maps with >2100 homologous UniGenes mapped in more than one species (∼6% of all mammalian genes). The generation of these maps is iterative and labor intensive; therefore, we developed a series of computer tools (not described here) based on our algorithm that identifies anchors between species and produces printable and on-line clickable comparative maps that link to a wide variety of useful tools and databases. The maps were constructed using sequence-based comparisons, thus creating “hooks” for further sequence-based annotation of human, mouse, and rat sequences. Currently, this map enables investigators to link the physiology of the rat with the genetics of the mouse and the clinical significance of the human.
Over the past 200 years, animal models have been selected and used primarily as surrogates for humans. The primary selection criteria for the animal models have been disease-based phenotypic characteristic(s) similar to those of humans. Indeed, many rat and mouse models share pathobiological characteristics similar to a human condition (Desnick et al. 1982). The idea that genomic organization also tends to be evolutionarily conserved between species was postulated in the early 1900s (Castle and Wachter 1924; Haldane 1927). Studies involving banding conservation and chromosome painting (ZOO-FISH) have since shown that large stretches of DNA are conserved in mammalian species as divergent as humans and fin whales (Nash and O'Brien 1982; Sawyer and Hozier 1986; Scherthan et al. 1994; Weinberg and Stanyon 1995). Although these studies showed genome conservation, they could not show the explicit conserved gene order at high resolution; such detail can only be accomplished at the genetic/physical mapping or sequence level. Several studies evaluating genome conservation at the genetic and physical mapping level have determined that gene order does tend to be conserved between mammals (Oakey et al. 1992; Sellar et al. 1994; Stubbs et al. 1994), opening up the prospect of constructing comparative maps between multiple species based on genetic sequence and map information (Nadeau 1989; Anderson et al. 1996; DeBry and Seldin 1996; Lyons 1997).
As genetic and physical maps of human and model organisms developed with the advent of the Human Genome Project in the 1990s and as the number of identified genes increased, the number of possible integration points dramatically enhanced the potential quality and density of comparative maps (O'Brien et al. 1999). The increased number of mapped genes and expressed sequence tag (EST) sites has led to sequence comparisons to identify orthologous genes (homologous genes in different species evolving from the same common ancestral gene; Clark 1999; Fitch 2000). When mapped in both species, these orthologs serve as anchors that are useful in identifying conserved segments between species. However, until absolute phylogeny of the genes is truly known, the ortholog assignments between these species must be considered preliminary; thus, it is prudent to assign gene-based anchors using the more conservative homolog relationships. The Mouse Genome Informatics (MGI) group at The Jackson Laboratories (http://www.informatics.jax.org/; Blake et al. 2000) has curated and assigned 2105 rat-mouse (R-M), 1950 rat-human (R-H), and 5603 mouse-human (M-H) orthologs. However, fewer of these genes have been mapped across all three species, limiting the number of anchors for building comparative maps. Several lower-resolution comparative maps have been generated between rat, mouse, and human using fluorescence in situ hybridization (Levan et al. 1991; Scalzi and Hozier 1998; Grutzner et al. 1999) and combined genetic/radiation hybrid (RH) maps (Watanabe et al. 1999), the later identifying 522 anchor points between rat and human and/or mouse. The combined genetic/RH maps identified 41 conserved segments (identified by containing at least two homologous genes) between rat and mouse and 89 between rat and human (Watanabe et al. 1999). Using the analytical methodology developed by Nadeau and Taylor (1984), Watanabe et al. (1999) predicted the number of evolutionarily conserved segments between rat and human to be 152+21 and between rat and mouse to be 49+7.
The emergence of the RH maps in human, rat, and mouse (Gyapay et al. 1996; Steen et al. 1999; VanEtten et al. 1999), coupled with the development of large numbers of UniGenes and ESTs for all three species, has revolutionized the way comparative maps can be built and maintained, before the complete genome sequencing of all three species. Indeed, the mapping approach described here can easily be extended to other mammals with significant EST libraries and RH maps and with entire genome sequences that will not likely be determined. There are many advantages of using the RH maps over curated or integrated genetic maps. First, RH mapping facilitates the integration of genetic markers, genes, and ESTs onto a single backbone map. Second, anchor (homology and map) assignments (based on sequence alignment, UniGene assemblies of ESTs, and map information) between species provide large numbers of hooks on and between the RH maps of rat, mouse, and human, which are useful for further sequence-based annotation of finished sequence from any source and, in particular, annotation of gene function based on results in animal models. Finally, the backbone of the maps has been developed and constructed using sequence-based comparison assignments coupled to a sophisticated scoring algorithm to choose the most likely homologies, thus providing an algorithm for de novo construction of comparative maps as the fundamental EST, gene assembly (UniGene or other), and RH map data sets mature. As the genomic sequence for human and mouse are in finishing and the sequencing of the rat is underway (Marshall 2000; Pennisi 2000a,b), such an RH-based scaffold becomes a powerful tool for early rat physical mapping, sequencing, and annotation of function. Comparative maps as described here provide a powerful platform for the integration of physiological and pharmacological information in the rat with genetic information in the mouse and clinical information in the human.
RESULTS
We have used publicly available rat, mouse, and human data to identify genes and ESTs with interspecies sequence identity (Table 1) and have coupled the information on sequence alignment (homolog) with both gene assemblies (UniGene) and their RH map positions to build comparative maps. Our threshold for positive EST sequence alignment is 85% sequence identity over at least a 100-bp stretch after masking interspersed repeats (e.g., long interspersed elements [LINEs]) and low-complexity regions, criteria we experimentally established (see Methods) and that are supported in studies by Makalowski and Boguski (1998) and used in the National Center for Biotechnology Information (NCBI) HomoloGene algorithm (Zhang et al. 2000). Results of sequence identity testing between multi-organism gene and EST sequences were then subject to an algorithm that compresses them into homologous UniGene objects (see Compression and Scoring Algorithm). The objects are then scored to predict unique (one-to-one in each species) homologous UniGene anchors that have high affinity for each other based on the gene and EST sequence alignment(s). If map information is available, these anchors are then assigned a consensus map position. This algorithm identified 18,901 R-H, 84,680 R-M, and 28,973 M-H putative UniGene homologies (using UniGene builds 115 for human, 77 for rat, and 78 for mouse), of which 8012 R-H, 14,370 R-M, and 9164 M-H were classified as unique homologous UniGene anchors using the algorithm. Those unique homologous UniGenes with consistently mapped ESTs were used as the anchor points for the comparative maps. We conclude that the majority of these anchors (gene pairs) resulting from our algorithm are in fact orthologous genes.
Table 1.
UniGene, EST, and Map Summary for Human, Mouse, and Rat
| UniGenes | Sequences | Mapped ESTs (RH panel) | UniGene Build (date of build) | |
|---|---|---|---|---|
| Human | 81,979 | 1,677,192 | 45,741 (GB4) | 115 (6/28/00) |
| Mouse | 87,512 | 894,559 | 2,158 (T31)MGC | 78 (6/28/00) |
| Rat | 36,953 | 169,843173,027 | 6.102 (T55) | 77 (6/28/00) |
UniGene and EST information as reported by NCBI (http://www.ncbi.nlm.nih.gov/UniGene/). Human map information is from GeneMap 99 (http://www.ncbi.nlm.nih.gov/genemap99/; Deloukas et al. 1998). Mouse map information is from the Mouse Genome Center (MGC; http://websql.har.mrc.ac.uk/mps/maps/0/LOD_7/graphic.html, 2000a). Rat map information is from RGD (http://rgd.mcw.edu/maps/; Steen et al. 1999).
Using these data, we generated comparative framework maps between rat, human, and mouse. After compressing the EST alignments into unique homologous UniGenes, we identified 1244 mapped R-H homologs, 368 mapped R-M homologs, and 569 mapped M-H homologs, corresponding to 2155 homologous UniGenes mapped in more than one species, ∼6% of all mammalian genes (Adams et al. 2000). The map information was obtained from publicly available rat (http://rgd.mcw.edu; http://ratEST.uiowa.edu; Steen et al. 1999; Sheetz et al. 2001), human (http://www.ncbi.nlm.nih.gov/genemap99/; Deloukas et al. 1998), and mouse (http://websql.har.mrc.ac.uk/mps/maps/0/LOD_7/graphic.html; VanEtten et al. 1999) RH maps. From these comparative maps, we have identified 107 conserved segments with at least 2 anchors between rat and human and 37 between rat and mouse. The average conserved segment length between rat and human is 94.25 cR, with a range of 0.2 to 483 cR; between rat and mouse, 326.4 cR, with a range of 0.2 to 867 cR. It is important to note that although these numbers reflect conserved segments, many of them are interrupted by intrachromosomal rearrangements, that is, local gene order has not been as well conserved. This trend has been more and more evident with the increasing resolution of comparative maps (Carver and Stubbs 1997; Thomas et al. 2000). For example, two comparative maps between human and chromosome 7 mouse have reported 14 and 13 conserved segments, respectively, when using genetic map information in the mouse and either cytogenetic or genomic sequence information for the human (DeBry and Seldin 1996; Lander et al. 2001). However, a refined map for this chromosome that used mouse sequence tag site (STS) maps (rather than consensus genetic maps of the mouse) and human genomic sequence maps identified 20 conserved segments, only half of which correspond to nonadjacent regions (Thomas et al. 2000). Therefore, the remaining 50% of the conserved segments were produced by intrachromosomal rearrangements. This may affect the previous estimates of conserved segment length, as these calculations assume when a segment is defined by two or more syntenic orthologous genes, gene order is conserved between those anchors (Nadeau and Taylor 1984). Indeed, for human chromosome 7, previous conserved segment length estimates were >50% than those determined in the refined comparative maps using STS and sequence maps. More detailed sequence-based comparisons, resulting from the human, mouse, and rat genomic sequence, will serve to better determine whether this phenomenon is specific to human chromosome 7 or whether it is a general trend.
We also identified 200 singleton segments (defined by a single homologous anchor) between rat and human and 84 singleton segments between rat and mouse. Although some of these singletons could be short conserved segments, they may also be caused by incorrect assignments of orthologs and/or incorrect mapping information. The homology maps between human and mouse also detect a large representation of singleton segments, ranging from 141 (Lander et al. 2001) to 223 (http://www.ncbi.nlm.nih.gov/Homology/), and it has been suggested that nearly 50% of these are likely true conserved segments. To avoid some of the complexities caused by ambiguities in RH placement position, we define ESTs within a 20-cR bin interval as a single UniGene placement. We anticipate a reduction in the total number of singletons as more map information is made available, UniGene rebuilds improve, genomic sequence for all three species is available, and singleton segments are subsumed into adjoining newly mapped syntenic regions.
The generation of high-resolution comparative maps is an iterative and labor-intensive exercise as new ESTs, RH map iterations, and ongoing UniGene rebuilds are produced. Therefore, we have developed a series of computer tools (data not shown) based on our algorithm that, on a quarterly basis, identify unique homologous UniGene anchors between rat, human, and mouse; develop and annotate the comparative map information; and build and display the comparative framework maps in printable and online clickable formats using the most current available data. Figure 1, produced as the poster enclosed with this issue, shows static R-H and R-M comparative maps generated using our computer tool; all of these maps, along with those using mouse and human as backbone species, have been generated and are available (http://rgd.mcw.edu/VCMAPS). The displayed version of these maps was generated to give a visually pleasing picture of the comparative maps but, because of the density of markers on the maps, does not display all available information. All conserved segments are displayed (having at least two homologous anchors), but not all detail is included at the whole-chromosome level. For instance, lines are drawn between homologous UniGenes only if the homologous anchors have been displayed in both organisms. The backbone species is in the middle of each map, with the corresponding species on either side. The backbone map is drawn to scale; however, the corresponding homologous regions in the other two species are not, rather they are displayed to span the length of the backbone map. However, because the maps are clickable, more detailed mapping and homology information is also available in a tabular format (Table 2) via a direct link. A user can either click on a colored bar of the backbone map or directly enter the desired interval to display all anchor data for that interval, including framework markers in the backbone map to aid in orientation. Each UniGene anchor is also clickable, displaying more detailed alignment and mapping information and providing direct links to UniGene information at NCBI (http://www.ncbi.nlm.nih.gov/UniGene/), which can then be navigated for additional information. These maps are the first example to our knowledge that, in an automated fashion, provides comprehensive comparative information in a single source for rat, mouse, and human. We have incorporated these maps into the Rat Genome Database (RGD; http://rgd.mcw.edu/), where they will be maintained and serve as an integration point for genomic and physiological data in the rat and a direct tie into human and mouse genome information. This integration allows for direct queries using marker, UniGene IDs, or accession numbers as well as desired map location within any of the backbones. As the iterative process of EST sequencing, UniGene builds, and RH map density increases, and as genomic sequence is annotated, identified anchor points, conserved segments, and resulting comparative maps will reflect the increased information. Some conserved segments will merge, and some additional segments may be identified. New builds will be performed and released by RGD on a quarterly basis, starting with the first release in June 2001.
Table 2.
Anchors between Rat, Human, and Mouse in the Region Spanning 260.5 cR and 306.7 cR of Rat Chromosome 13
| cR | Rat | Human | Mouse |
|---|---|---|---|
| *260.5 | d13rat94 | ||
| 261.8 | Rn. 19095 | Mm. 38399 | |
| *263.1 | d13rat75 | ||
| 269.2 | Rn. 16225 | Mm. 44881 | |
| *274.7 | d13rat52 | ||
| *280.1 | d13rat22 | ||
| 287.2 | Rn. 2022 | Hs. 77495 KIAA0242 (Chr. 02:479.27) | Mm. 22397 |
| 288.3 | Rn. 28734 | Mm. 27637 | |
| 290.2 | Rn. 799 | Hs. 189954 | Mm. 24109 |
| 293 | Rn. 18841 | Mm. 20236 | |
| 295 | Rn. 15837 | Mm. 87652 | |
| 297.2 | Rn. 11151 | Hs. 99886 C4BPB (Chr. 01:684.23) | |
| 297.5 | Rn. 6758 Ctsea | Hs. 1355 CTSE (Chr. 01:682.48) | Mm. 33671 Ctse (Chr. 01:1899.4) |
| 297.6 | Rn. 10408 C4bpa | Hs. 1012 C4BPA (Chr. 01:686.04) | Mm. 14087 C4pb (Chr. 01:1885.52) |
| 297.6 | Rn. 11774 | Mm. 24634 | |
| 302.8 | Rn. 18567 | Mm. 37672 | |
| 303.6 | Rn. 12700 | Hs. 5003 KIAA0456 (Chr. 01:682.48) | Mm. 34134 |
| *306.3 | d13rat21 | ||
| 306.7 | Rn. 39004 | Hs. 7309 (Chr. 01:678.07) | Mm. 37703 |
Link from displayed comparative map by entering a region of interest in cR or by clicking on the map backbone. On the right is the backbone RH map, with unique homologous UniGene anchors in corresponding species listed in the columns to the left of the backbone map. RH framework markers are displayed for orientation and integration to genetic mapping data. UniGene entries are hotlinked to the UniGene web site at NCBI. Official gene symbols are included when available. Chromosomal location in cR distances for human and mouse homologous UniGenes given in parentheses. RH framework markers begin with an asterisk.
To address the accuracy of the automated maps, we compared the R-H maps generated by our algorithms with those generated by Watanabe et al. (1999), which are based on curated orthologs and combined RH/cytogenetic maps. We found 80 conserved segments in common between the R-H. We identified 18 conserved segments that were not identified by Watanabe et al. Conversely, they identified 24 conserved segments that we did not. One important difference between the maps, however, is the fact that we did not consider singleton anchors in our calculations, whereas the previous study defined conserved segments with a single mapped anchor. Of the additional segments identified by Watanabe et al., 19 of them appear to be segments based on singleton anchors, and four cases resulted in an interchromosomal interruption in an otherwise conserved segments. Overall, there was remarkably good consistency between the two maps, particularly given the different methodologies and data sets used to generate them.
A second test for map accuracy was to annotate anchors on the rat chromosome 3 RH backbone using either the HomoloGene database or protein similarity data reported in the UniGene database to identify their predicted human orthologous UniGenes and incorporating human RH map location, using GeneMap99 links from the UniGene Web site. We then compared the results with those generated using the current iteration R-H comparative maps for rat chromosome 3. Of 142 anchor comparisons, 77% were identified by both methods, 19% were identified only by the manual annotation using HomoloGene and protein prediction data, and 4% were found only by the algorithm. Importantly, no cases revealed a discrepancy in ortholog assignment in the comparison. Furthermore, given the extensive time involved in manually annotating the maps and the ever-increasing number of genes and ESTs in the UniGene builds and RH maps, we propose that our algorithm and tool set can be used in place of manual builds of the comparative maps for the whole genome. Investigators interested in a given region may wish to conduct a manual search until the sequences of the human, mouse, and rat genomes are completed.
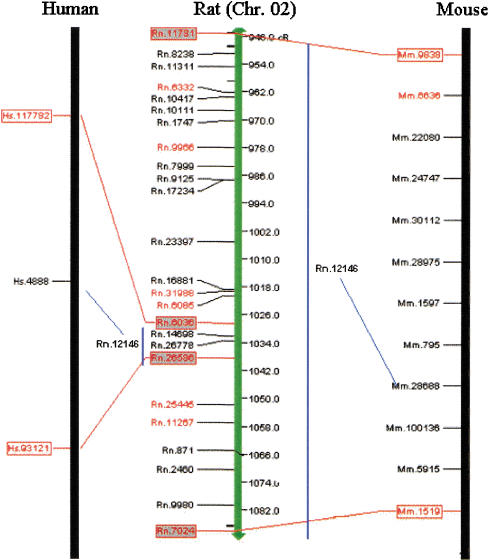
The density of the anchors and the completion of the comparative maps, on a theoretical level, suggested that the maps could be used to predict EST and gene locations (virtual mapping) in advance of wet-lab mapping or in instances in which the EST cannot be RH mapped by the wet-lab because of cross-species amplification between the donor species and hamster. Our experience is that only ∼50% of all ESTs produce a vector that can be RH mapped using a single set of polymerase chain reaction primers. However, we have identified 8012 R-H, 14,370 R-M, and 9164 M-H unique homologous UniGene anchors that can be used to increase the density of the comparative maps. We have established conserved segments by identifying at least two anchors on each segment; we can use information from UniGene anchors mapped in at least one species within that conserved segment to predict the placement of its homologous UniGene in another species, given that gene order has been conserved in that segment (Fig. 2). For instance, we determined that Rn.6036, Hs.117782, and Mm.9838 are mapped homologous UniGene anchors and are in the same conserved segment as Rn.26586, Hs.93121, and Mm.1519. Another group of homologous UniGene anchors—Rn.12146, Hs.4888, and Mm.28688—have available map information in human and mouse but lack map location in the rat. However, given that Hs.4888 maps between Hs.117782 and Hs.93121 and given that Mm.28688 maps between Mm.9838 and Mm.1519, we predict that Rn.12146 will also map between the flanking anchors Rn.6036 and Rn.26586, indicated by the blue lines connecting to the respective map. Using this approach, we were able to predict the placement of an additional 2604 rat UniGenes, 3730 mouse UniGenes, and 266 human UniGenes, assuming conserved linkage between two flanking UniGenes in other species (Table 3). Furthermore, we sought to use map information upstream and/or flanking anchors that define a conserved segment to better define the evolutionary breakpoint and potentially extend the segment by prioritizing that UniGene for wet-lab mapping. We could predict the placement of an additional 1061 rat UniGenes, 1313 mouse UniGenes, and 182 human UniGenes upstream or downstream of a conserved segment (this is a region that contains an evolutionary breakpoint), based on the map position of homologous UniGene anchors in the other species (Table 3). For this prediction, we included those breakpoints represented by a single anchor to give the opportunity to experimentally refute or confirm that conserved segment by, for example, RH mapping. The virtually mapped UniGenes have also been integrated into the online clickable maps by querying the particular region of interest, in centiRay distance on the backbone map and displaying them in a separate table, immediately following the tabular detailed comparative map information. Table 3 summarizes the virtual mapping predictions of UniGenes in rat, human, and mouse. The upstream and downstream predicted UniGenes, those that fall nearby evolutionary breakpoints, can then be prioritized for RH mapping to better define the evolutionary breakpoints and to fill in gaps in the comparative maps.
Figure 2.
Predicted placement of rat Rn.12146 within a conserved rat bin, based on gene order conservation from both human and mouse. Rat backbone in the middle, with comparative map of human on the left and mouse on the right. Red boxes with connecting red lines indicate unique homologous UniGenes that define the conserved segment across rat, human, and mouse. Blue bars indicate the bins into which the rat UniGene is predicted, with the blue lines linking the homologous UniGenes between the species.
Table 3.
Summary of Virtual Map Predictions
| Rat UniGene predictions | |||
| From species | Bin | Stream | Total |
| human | 2153 | 795 | 2948 |
| mouse | 373 | 243 | 616 |
| human, mouse | 78 | 23 | 101 |
| Total | 2604 | 1061 | 3665 |
| Mouse UniGene predictions | |||
| From species | Bin | Stream | Total |
| human | 2055 | 827 | 2882 |
| rat | 1430 | 447 | 1877 |
| human, rat | 245 | 39 | 284 |
| Total | 3730 | 1313 | 5043 |
| Human UniGene Predictions | |||
| From species | Bin | Stream | Total |
| mouse | 61 | 69 | 130 |
| rat | 197 | 110 | 307 |
| mouse, rat | 8 | 4 | 12 |
| Total | 266 | 183 | 449 |
Predictions are separated by species and category. Bin indicates number of virtually mapped unique homologous UniGenes predicted within a conserved segment. Stream indicates number of UniGenes predicted to fall directly upstream or downstream of a unique homologous UniGene anchor currently defining an evolutionary breakpoint prediction. Some predictions are made using conserved segment information from one species and others have been made using both comparative species. The predicted UniGenes have been integrated into the clickable maps found at http://rgd.mcw.edu/VCMAPS and can be queried using the cR distance flanking a region of interest, or by clicking on the rat map backbone.
DISCUSSION
The Future of Comparative Mapping
Given the time to manually generate the maps and the ever-increasing number of genes and ESTs in the UniGene builds and RH maps, we propose that our algorithm and tool set can be used in place of manual builds of the comparative maps for the whole genome. Investigators interested in a given region may wish to conduct a more detailed manual search until the sequences of the human, mouse, and rat genomes are completed. The comparative maps (in clickable format) in Figure 1, as well as those with the mouse and human backbones, have been installed online at the RGD (http://rgd.mcw.edu/VCMAPS), with references from the RGD that allow a visual entry to all of the homology assignments, as well as dbEST and UniGene links to NCBI. Within the next few years, we anticipate that the sequence data from the human, mouse, and rat EST and genome sequencing projects will complete the comparative maps at the sequence level and that sequence-based comparative maps will become the norm. In the interim, there is a need to place more genes on the comparative map to facilitate the discovery of disease genes by linking genomic and phenotypic information between the mouse and rat models with the human. RH mapping is the most powerful interim solution to comparative mapping, as it facilitates higher-resolution maps and has less ambiguity than can be provided by genetic maps. Furthermore, many agricultural and other model organisms will not be sequenced fully, yet sufficient genomic resources (sequenced ESTs, genetic and RH maps) are available to generate virtual comparative maps using our algorithms and tool. Although we acknowledge that there are caveats to using RH maps for local ordering of genes and ESTs, as has been shown when aligning human RH maps with genomic sequence (Agarwala et al. 2000), it certainly is the most powerful and effective approach currently available for global ordering and comparative mapping between species, before genomic sequencing, and for those organisms with genomes that are not likely to be sequenced. Furthermore, the infrastructure we have developed is able to integrate finished sequences of human, mouse, and rat to lead to sequence-based comparative maps as they become available.
Accuracy of Virtual Mapping
Two tests were executed to examine the accuracy of the predictions. In the first test, 243 rat UniGenes, predicted in a previous iteration of the comparative map (bin predictions), were subsequently RH mapped in the wet-lab and tested directly using the next successive comparative map. Of the 243 rat UniGenes tested (representing a total of 2713 ESTs), the location of 143 of 243 (59%) were confirmed, using a 50-cR or <10-cM bin interval, whereas 100 were wet-lab mapped to locations outside the bin prediction. If the criteria were relaxed so that the predicted and tested placement must be on the same chromosome, the accuracy of prediction increased to 71%, indicating that inaccuracies in the RH placement may impact the predictions because of the low density of the initial comparative map from which these predictions were made. Because of a lower density of anchors between the species, the minor intrachromosomal rearrangements that often occur within conserved segments may not have been evident. To evaluate this possibility, the Whitehead Institute/Massachusetts Institute of Technology (MIT) public Mouse EST RH Mapping Project release 8 (7606 mapped ESTs; http://www-genome.wi.mit.edu/mouse_rh/index.html) was used to build comparative maps (data not shown). One hundred eleven predicted mouse UniGenes from release 8 were tested against MIT release 9 RH maps (8413 ESTs mapped), using the approach as described above in second test. Ninety-five of 111 (86%) predicted locations were confirmed to map within the predicted bin. With respect to mapping to the correct chromosome, 99 of 111 (89%) met these criteria. Therefore, it appears that as map density increases, the predictive ability of this method concurrently increases. It is also possible that because of intrachromosomal rearrangements, we may not be able to increase the accuracy of the virtual mapping greater than this level. Nonetheless, the virtual mapping described here provides a valuable starting point for an investigator interested in testing an EST with a specific homology or wanting to follow up on ESTs shown to have different expression via microarrays, SAGE, or other techniques. We also anticipate that, as was shown here, with consecutive iterations of the comparative map, the accuracy of prediction will increase as the density of mapped ESTs (and thus UniGenes) increases. The algorithm and tools, coupled with the emerging databases, continued RH mapping of rat and mouse ESTs, and genomic sequencing, will result in increased accuracy of the detailed comparative maps.
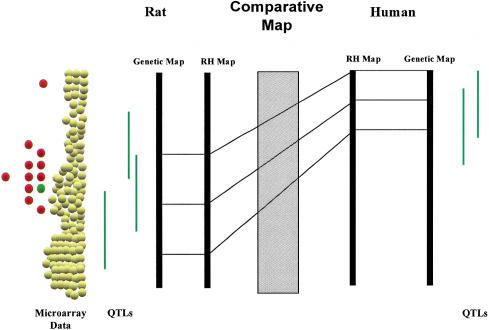
The comparative maps are a very powerful means to integrate data attached to the genome in rat, human, and mouse. For example, quantitative trait loci (QTLs) mapped for hypertension-related phenotypes in the rat, combined with comparative map data, have been used to predict regions of the human genome to be investigated at a higher resolution (e.g., by an association study using single nucleotide polymorphisms), and several of these regions have been independently identified in human and mouse (Stoll et al. 2000). The gene(s) associated with the disease could then be validated using mouse knockout or other transgenic strategies, establishing a mammalian genome platform to facilitate gene discovery. The generation of this platform could be taken a step further to, for example, integrate data generated by microarray studies (Fig. 3). On a larger scale, the algorithm used here to generate the comparative maps between rat, human, and mouse can be applied to other species with similar resources to create a mammalian genome platform that can be used not only for functional genomics but also for better understanding of the evolution of mammalian genomes.
Figure 3.
Functional genomics integrated with comparative mapping. Quantitative trait loci (QTLs) in green are similar traits found to be in conserved regions between rat and human using the comparative maps based on RH backbones. Microarray studies in animal models identify differentially expressed UniGenes in tissue between disease inbred and normal inbred rat strain. Genes found to be differentially expressed (red circles and green circles) and mapping in the QTL region common between rat and human can be further studied in human populations for their potential contribution to the disease.
METHODS
Establishment of Sequence Alignment Criteria
The alignment criteria for testing DNA sequence similarities were derived by a sophisticated test of UniGene sequences from 1000 UniGenes (per organism) using the gapped BLAST program. For the three species, 100 common orthologs (between R-M, M-H, and R-H) were selected from the ortholog data curated and assigned by The MGI group at The Jackson Laboratories (http://www.informatics.jax.org/). The test data sets were based on curated homologous genes and excluded those homologous genes based solely on the similarities in DNA or protein sequences. To take into consideration the potential confounding issue of paralogous genes, we included 10 putative paralogous genes, each corresponding to one of the remaining 90 orthologous genes, in each of the three data sets. Three test data sets were created (R-M, M-H, and R-H) of 1000 UniGenes, each composed of 90 curated orthologs to the other organisms (as chosen from the MGI data) and 10 curated paralogs plus an additional 900 randomly chosen UniGenes not found in the MGI data sets. For each pair, sequences corresponding to the genes of the first organism were used as BLAST probes to the target collection of sequences of the second organism. To determine the optimal BLAST threshold, a series of processes were executed using each combination of (minimal base pair aligned length, % alignment) for base pair length ranging from 50 to 150 bp in 5-bp intervals and percent alignment ranging from 65% to 100% in 5% intervals. After compression and scoring (see Compression and Scoring Algorithm), the predicted homologous UniGene one-to-one objects were compared with the curated orthologous pairs. Sensitivity, specificity and ACP (average conditional probability, an overall statistical evaluation for both specificity and sensitivity) of predicting the correct homologous UniGenes under each aligned length and percentage combination were calculated. The optimal BLAST threshold for positive prediction of homology for R-H was 100 bp, (95%); for R-M, 100 bp (85%); and for M-H, 95 bp (85%). On comparison with other determinations (Makalowski and Boguski 1998: 100 bp, 85%; HomoloGene algorithm, NCBI: 100 bp, 85%), we determined that the optimal parameters for virtual comparative mapping were 100 bp (85%).
Construction of Comparative Maps between Rat, Mouse, and Human
All rat, mouse, and human ESTs represented in the UniGene database (NCBI; http://www.ncbi.nlm.nih.gov/UniGene/index.html) were downloaded to a local database and screened for sequence identity using the methodology described above. A compression algorithm, described below, collects and parses the following data into an anchor file: (1) the GenBank accession IDs of the probe ESTs, showing alignment with the target species; (2) RH map location (if available); (3) the associated UniGene ID, with all other mapped ESTs in that UniGene; and (4) the UniGene IDs of the homologous ESTs and related RH map information, any available gene symbols and descriptions, and location (cytogenetic, genetic, and/or RH) data. This file is then compressed into homologous UniGene objects by parsing and reorganizing all data by UniGene ID (see Compression and Scoring Algorithm, below). This compression results in the identification of many-to-many UniGene objects (it may be that ESTs from multiple UniGenes in one species align with ESTs from multiple UniGenes in another species, see Fig. 4). All many-to-many associations are then scored based on the quality and quantity of the gene and EST sequence alignments, consistent map information, and the consistency of assembled aligned sequences. The best one-to-one assignments are then predicted, and results are sorted accordingly. The scoring algorithm proved to be 91% accurate in predicting known orthologs (based on the 1000 gene test set); therefore, most of the homologies we determine using this algorithm are likely orthologs. After scoring and sorting, all one-to-one homologous UniGene objects are located in an anchor file, which is used to construct the comparative maps.
Figure 4.
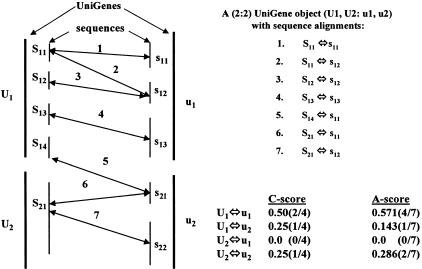
Diagram illustrating the compression and scoring algorithm. The object on the left represents a relatively simple (2:2) UniGene object represented by U1, U2:u1, u2. UniGene U1, defined by sequences S11, S12, S13, S14 and U2, by sequence S21, have potential similarity to u1 (s11, s12, s13) and u2 (s21, s22). Sequence alignments of the various constituent sequences represented by the two-ended arrows between sequence vertical bars. Alignment relationships are summarized on the upper right, and calculated scores for the four possible clustered links are shown on the lower right. The C-score calculates the ratio of the number of observed clustered links among aligned sequences. The A-score is calculated using all possible links between any aligned sequences within a UniGene object.
Compression and Scoring Algorithm
The UniGene-to-UniGene homology prediction in this work is based on the complete collection of the data and information that is consistent with the goal of both identifying unique homologs and mapping UniGenes (as opposed to mapping ESTs). No other homology prediction algorithms (published or available on the Web) incorporate map information into their predictions. In addition, we compute a weighted score of all the alignment information to test which of all possible UniGene-to-UniGene combinations are the most likely orthologs, given the available data. For the goal of this work, it is imperative that potentially irreconcilable information between sequence alignment, mapped ESTs, and EST assemblies be resolved before comparative maps can be constructed. A compression and scoring algorithm was developed that would allow the systematic prediction of unique, mapped, homologous, UniGene anchors. The algorithm is best shown by example (Fig. 4); here we denote the UniGenes (EST and cDNA sequence assemblies) of two organisms (U and u) by UI and uJ, and their constituent sequences by SIk and sJl, respectively. UniGene objects are denoted by an (M:N)-tuple, representing the number and identity of the UniGenes of each organism that have alignment association by their respective sequence constituents. The object in Figure 4 represents a relatively simple (2:2) UniGene object represented by U1, U2:u1, u2.
In this figure, UniGene U1, defined by sequences S11, S12, S13, S14 and U2, by sequence S21, have potential homology with u1 (s11, s12, s13, s14) and u2 (s21, s22). The potential for identity of the homolog is defined by the sequence alignments of the various constituent sequences and represented by the two-ended arrows between sequence vertical bars. We grouped the related alignments together as the UniGene 2:2 object U1, U2:u1, u2. This single UniGene object consists of four potential unique homologous UniGene anchors ([U1, u1], [U1, u2], [U2, u1], and [U2, u2]). Other alignments can result in more complicated UniGene N:M objects, giving weight to other combinations of objects and potential anchors. UniGene objects fall into four natural categories: category I, one-to-one (1:1); category II, one-to-many (1:M); category III, many-to-one (N:1); and category IV, many-to-many (N:M). One-to-one objects are the basis of the comparative maps, although there are examples of 1:M, N:1, and N:M objects theoretically useful in building maps. For the purpose of these comparison maps, we developed a scoring algorithm to reduce (compress) the three more complex categories of objects into the 1:1 category. The 1:1 object with the highest score is extracted and used as the unique homologous UniGene anchor.
A hierarchy of scores was developed to test the hypothesis that each potential 1:1 object is the most likely unique homologous UniGene anchor, given the available data. The likelihood is defined by three scores, C, A, and P. The C-score calculates the ratio of the number of observed clustered links among aligned sequences between all UniGene pair combinations to the total number of possible links. Clustered links are defined as groups of sequences that are networked together by cross-species alignment and clustered by residing in common UniGenes. In this case, we assume that multiple alignments are most likely the consequence of oversampling the original coding sequence, and thus, they provide false positive weight to the underlying homology. Returning to the representative example (Fig. 4), we have four clustered links: S11 aligns to s11 and s12, and S12 aligns to s12; we say that the alignment links 1, 2, and 3 are clustered together as it may be that S11 and S12 and s11 and s12 are simply resampled EST sequences and thus are providing redundant alignment information. Thus, these links are only counted once. Link 4 does not cluster with any other links. Links 6 and 7 are clustered together, but link 5 aligns U2 and u1, whereas links 6 and 7 align U2 and u2. Therefore, we count link 5 as a separated cluster. As a result, in this UniGene object, there are a total of 4 clusters of links. Of all possible clustered links, U1:u1 accounts for 2, U1:u2 for 1, U2:u1 for 0, and U2:u2 for 1. The C-scores are calculated in the panel to the right. The advantage of the C-score is that it eliminates the effect of redundant ESTs. The A-score is calculated using all possible links between any aligned sequences within a UniGene object. For U1, U2:u1, u2 there are a total of seven links, four define U1:u1, one in U1:u2; U2:u1 has none, and U2:u2 has one. The A-score counts all evidence of homology but is biased to oversampled data sets. Finally, a P-score is a qualitative measurement of the certainty in map information of a mapped UniGene; it is the sum of the map information value for a pair of UniGene homologs, each mapped to one position on a chromosome. A map information value of 0.5 is assigned to any UniGene with ESTs that are all mapped to one position (sequences RH mapped to within a 5-cR interval to their mean position are considered mapped to the same position) on one chromosome. UniGenes mapped to m (>1) positions on n (>1) chromosomes are assigned a value of (0.5/m)n. A P-score between 0.5 and 1.0 indicates one of the two UniGene homologs is mapped to one position on a chromosome. P-scores <0.5 indicate that both UniGene homologs are mapped to multiple positions on one chromosome or more.
Potential UniGene objects, unique homologous UniGene anchors, are scored and ranked based on their C-, A-, and P-scores, in that order. In the U1, U2:u1, u2 example, the unique homologous UniGene anchor is U1, u1 based on the C-score. If needed, the A-score would be used to rank the four options (and U1, u1 would again score highest). In our experience, the P-score is not generally used in ranking (uniqueness is determined by the first two scores generally); however, in every case we have tested, the P-score has ranked a unique 1:1. As data become more abundant, the ranked scoring system will take into account all available data and can be used to incorporate other more refined information while still being used to predict 1:1 anchors. In addition, extensions to the compression algorithm and minor revisions of the scoring systems can be developed to compress and score category II, III, and IV objects (all potential paralog relationships.)
Manual Validation of Maps
For comparison of the maps presented here to those described by Watanabe et al. (1999), we directly compared the displayed R-H comparative maps, using the rat as a backbone species. For each chromosome, we identified which conserved segments were in common, which were identified only in our maps, and which were identified only in the Watanabe maps. We based consensus on presence of the conserved segment but did not consider the chromosomal location because the map information for the human was based on different mapping methods (human RH versus cytogenetic mapping).
For manual annotation and validation of the current maps, 142 homologous UniGenes on the comparative maps of rat chromosome 3 were checked for their predicted orthologous relationship to human using the HomoloGene database at NCBI (http://www.ncbi.nlm.nih.gov/HomoloGene/) and the protein similarities found in the UniGene database at NCBI (http://www.ncbi.nlm.nih.gov/UniGene/), as well as position data using various databases at NCBI, including the UniGene Web page and its links to LocusLink (http://www.ncbi.nlm.nih.gov/LocusLink/), the human GeneMap99 (http://www.ncbi.nlm.nih.gov/genemap99/), and RATMAP (http://ratmap.gen.gu.se/). If the rat UniGene corresponded to a described gene, protein similarities were checked in the UniGene page. If protein similarities were determined in human, their corresponding nucleotide sequence was identified using Entrée at NCBI, and its corresponding UniGene was determined using the LinkOut option. The maps were then annotated with information including the UniGene ID(s) of the homologous human and/or mouse ESTs, any available gene symbols and descriptions, and location (cytogenetic, genetic, and/or RH) data. The determination of orthologous relationships and associated map information was then compared between the manually annotated map and the map generated using our algorithm.
Virtual Mapping of Additional Genes and ESTs
The computer tools were developed to build and display the virtual comparative maps, using publicly available rat RH maps (RGD, http://rgd.mcw.edu), human GB4 RH maps (GeneMap99, http://www.ncbi.nlm.nih.gov/genemap99/), and mouse RH maps (Mouse Genome Center, http://www.mgc.har.mrc.ac.uk/). Virtual mapping was performed from these comparative maps, using the rat, mouse, and human backbones. Using the rat as a backbone, conserved segments of human and mouse were identified, based on our algorithm. If two UniGenes lie within an uninterrupted conserved segment in one species, additional one-to-one homologous UniGenes between those flanking markers are virtually mapped, based on the map position of the homolog in the other species. If a UniGene defines a potential evolutionary breakpoint, additional one-to-one homologous UniGenes are predicted upstream and/or downstream of that marker. In this case, homologous UniGenes directly upstream or downstream (depending on which end of the conserved segment is being considered) of the UniGene flanking the breakpoint are identified and prioritized for wet-lab mapping to either confirm a segment defined by a single anchor or to extend and better define the evolutionary breakpoint. Predictions were made for all three species' backbones as described above for rat.
Acknowledgments
This work has been supported by RO1s HL9826–03 (H.J.J.) and HL59789–03 (V.C.S.) and was accomplished by a large group of people. Here we cite them and their contributions as suggested by Rennie et al. (1997) for manuscripts with large author lists. Overall project leadership was by H.J.J. and V.C.S. For the Medical College of Wisconsin, the project leader was A.E.K. For RH mapping at the Medical College of Wisconsin were J.G.-H. (team leader), Kim Orlebeke, Jeff Eckert, Angela Lemke, Rebekah Kopec, Tim Mull, Stephanie Brown, Mary Granados, Rebecca Majewski, M. Stoll, M. Shiozawa, M.N., Michelle Runte, Nicole Johnson, and Uli Broeckel; for Bioinformatics, P.J.T. (project leader), D.C., Jian Lu, Y.S.C., S.T., Zhitao Wang, Hui Zhu, and Wei Wang. For the University of Iowa, the project leaders were V.C.S. (RH mapping), M.B.S. (Gene Discovery), and T.L.C. (Bioinformatics). For RH mapping at the University of Iowa were Michael Raymond (team leader), Jane Zhang, Nichole Butters, and Christine Sun; for Gene Discovery: Tammy Kucaba (team leader), N. Altman, J. Assouline, N. Bedford, B. Berger, R. Brown, K. Crouch, M. Donohue, G. Doonan, B. Johnson, R. Kinkaid, S. Mackerly, E. Mallet, V. Miljkovic, B. Rhoads, C. Smith, and H. Young; for Bioinformatics, Judy Barkal (team leader), Hakeem Abdulkawy, Clay Birkett, Allen Gavin, Kang Liu, Kevin Pedretti, Chad Roberts, Natalie Robinson, and Todd E. Scheetz.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
NOTE ADDED IN PROOF
The orientation of rat chromosomes 11 and 18 are inverted on the poster map accompanying this paper. The on-line version of these maps link to the updated VCMaps, where these chromosomes have been corrected to reflect pter to qter orientation.
Footnotes
E-MAIL ablack@mcw.edu; FAX (414) 456-6516.
Article and publication are at http://www.genome.org/cgi/doi/10.1101/gr.173701.
REFERENCES
- Adams MD, Celniker SE, Holt RA, Evans CA, Gocayne JD, Amanatides PG, Scherer SE, Li PW, Hoskins RA, Galle RF, et al. The genome sequence of Drosophila melanogaster. Science. 2000;287:2185–2195. doi: 10.1126/science.287.5461.2185. [DOI] [PubMed] [Google Scholar]
- Agarwala R, Applegate D, Maglott D, Schuler G, Schaffer A. A fast and scalable radiation hybrid construction and integration strategy. Genome Res. 2000;10:350–364. doi: 10.1101/gr.10.3.350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersson L, Archibald A, Ashburner M, Audun S, Barendse W, Bitgood J, Bottema C, Broad T, Brown S, Burt D, et al. Comparative genome organization of vertebrates: The first international workshop on comparative genome organization. Mamm Genome. 1996;7:717–734. doi: 10.1007/s003359900222. [DOI] [PubMed] [Google Scholar]
- Blake J, Eppig J, Richardson J, Davisson M. The Mouse Genome Database (MGD): Expanding genetic and genomic resources for the laboratory mouse. Nucleic Acids Res. 2000;28:108–111. doi: 10.1093/nar/28.1.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carver EA, Stubbs L. Zooming in on the human-mouse comparative map: Genome conservation re-examined on a high-resolution scale. Genome Res. 1997;7:1123–1137. doi: 10.1101/gr.7.12.1123. [DOI] [PubMed] [Google Scholar]
- Castle W, Wachter W. Variations of linkage in rats and mice. Genetics. 1924;9:1–12. doi: 10.1093/genetics/9.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark M. Comparative genomics: The key to understanding the Human Genome Project. BioEssays. 1999;21:121–130. doi: 10.1002/(SICI)1521-1878(199902)21:2<121::AID-BIES6>3.0.CO;2-O. [DOI] [PubMed] [Google Scholar]
- DeBry R, Seldin M. Human/mouse homology relationships. Genomics. 1996;33:337–351. doi: 10.1006/geno.1996.0209. [DOI] [PubMed] [Google Scholar]
- Deloukas, P., Schuler, G.D., Gyapay, G., Beasley, E.M., Soderlund, C., Rodriguez-Tome, P., Hui, L., Matise, T.C., McKusick, K.B., Beckmann, J.S., et al. A physical map of 30,000 human genes. Science 282: 744–746. [DOI] [PubMed]
- Desnick R, Patterson D, Scarpelli D. Animal models of inborn errors of metabolism. NY: Alan R. Liss; 1982. [Google Scholar]
- Fitch W. Homology: A personal view on some of the problems. Trends Genet. 2000;16:227–231. doi: 10.1016/s0168-9525(00)02005-9. [DOI] [PubMed] [Google Scholar]
- Grutzner F, Himmelbauer H, Paulsen M, Ropers H, Haaf T. Comparative mapping of mouse and rat chromosomes by fluorescence in situ hybridization. Genomics. 1999;55:306–313. doi: 10.1006/geno.1998.5658. [DOI] [PubMed] [Google Scholar]
- Gyapay G, Schmitt K, Fizames C, Jones H, Vega-Czarny N, Spillett D, Muselet D, Prud'Homme JF, Dib C, Auffray C, et al. A radiation hybrid map of the human genome. Hum Mol Genet. 1996;5:339–346. doi: 10.1093/hmg/5.3.339. [DOI] [PubMed] [Google Scholar]
- Haldane J. The comparative genetics of color in rodents and carnivora. Biol Rev Camb Philos Soc. 1927;2:199–212. [Google Scholar]
- Lander ES, Linton LM, Birren B, Nusbaum C, Zody MC, Baldwin J, Devon K, Dewar K, Doyle M, FitzHugh W. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- Levan, G., Szpirer, J., Szpirer, C., Klinga, K., C. Hanson, C., and Islam, M.Q. 1991. The gene map of the norway rat (Rattus norvegicus) and comparative mapping with mouse and man. Genomics 10: 699–718. [DOI] [PubMed]
- Lyons L, Laughlin TF, Copeland NG, Jenkins NA, Womack JE, Obrien SJ. Comparative anchor tagged sequences (CATS) for intergrative mapping of mammalian genomes. Nat Genet. 1997;15:47–56. doi: 10.1038/ng0197-47. [DOI] [PubMed] [Google Scholar]
- Makalowski W, Boguski MS. Evolutionary parameters of the transcribed mammalian genome: An anlysis of 2,820 orthologous rodent and human sequences. Proc Natl Acad Sci. 1998;4:9407–9412. doi: 10.1073/pnas.95.16.9407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall E. Public-private project to deliver mouse genome in 6 months. Science. 2000;290:242–243a. doi: 10.1126/science.290.5490.242a. [DOI] [PubMed] [Google Scholar]
- Nadeau JH. Maps of linkage and synteny homologies between mouse and man. Trends Genet. 1989;5:82. doi: 10.1016/0168-9525(89)90031-0. [DOI] [PubMed] [Google Scholar]
- Nadeau JH, Taylor BA. Lengths of chromosomal segments conserved since divergence of man and mouse. Proc Natl Acad Sci. 1984;81:814–818. doi: 10.1073/pnas.81.3.814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nash W, O'Brien S. Conserved regions of homologous G-banded chromosomes between orders in mammalian evolution: Carnivores and primates. Proc Natl Acad Sci. 1982;79:6631–6635. doi: 10.1073/pnas.79.21.6631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oakey RJ, Watson ML, Seldin MF. Construction of a physical map on mouse and human crhomosome 1: Comparison of 13 Mb of mouse and 11 Mb of human DNA. Hum Mol Genet. 1992;1:613–620. doi: 10.1093/hmg/1.8.613. [DOI] [PubMed] [Google Scholar]
- O'Brien S, Menotti-Raymond M, Murphy W, Nash W, Wienberg J, Stanyon R, Copeland N, Jenkins N, Womack J, Graves J. The promise of comparative genomics in mammals. Science. 1999;286:458–481. doi: 10.1126/science.286.5439.458. [DOI] [PubMed] [Google Scholar]
- Pennisi E. Finally, the book of life and instructions for navigating it. Science. 2000a;288:2304–2307. doi: 10.1126/science.288.5475.2304. [DOI] [PubMed] [Google Scholar]
- Pennisi E. Rat genome off to an early start. Science. 2000b;289:1267–1269. b. [PubMed] [Google Scholar]
- Rennie D, Yank V, Emanuel L. When authorship fails: A proposal to make contributors accountable. JAMA. 1997;279:579–585. doi: 10.1001/jama.278.7.579. [DOI] [PubMed] [Google Scholar]
- Sawyer J, Hozier JC. High resolution of mouse chromosomes: Banding conservation between man and mouse. Science. 1986;232:1632–1635. doi: 10.1126/science.3715469. [DOI] [PubMed] [Google Scholar]
- Scalzi J, Hozier JC. Comparative genome mapping: Mouse and rat homologies revealed by fluorescence in situ hybridization. Genomics. 1998;47:44–51. doi: 10.1006/geno.1997.5090. [DOI] [PubMed] [Google Scholar]
- Scherthan H, Cremer T, Arnason U, Weier HU, Lima-de-Faria A, Fronicke L. Comparative chromosome painting discloses homologous segments in distantly related mammals. Nat Genet. 1994;6:342–347. doi: 10.1038/ng0494-342. [DOI] [PubMed] [Google Scholar]
- Sellar GC, Jordan SA, Bickmore WA, Fantas JA, vanHeyningen V, Whitehead AS. Ther human serum amyloid A protein (SAA) superfamily gene cluster: Mapping to chromosome 11p15.1 by physical and genetic linkage anlaysis. Genomics. 1994;19:221–227. doi: 10.1006/geno.1994.1051. [DOI] [PubMed] [Google Scholar]
- Sheetz T, Raymond M, Nishimura D, McClain A, Roberts C, Birdett C, Gardiner J, Zhang J, Butters N, Sun C, et al. Generation of a high-density rat EST map. Genome Res. 2001;11:497–502. doi: 10.1101/gr.151601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steen RG, Kwitek-Black AE, Glenn C, Gullings-Handley J, Van Etten W, Atkinson OS, Appel D, Twigger S, Muir M, Mull T, et al. A high-density intergrated genetic linkage and radiation hybrid map of the laboratory rat. Genome Res. 1999;9:AP1–AP8. [PubMed] [Google Scholar]
- Stoll M, Kwitek-Black AE, Cowley AW, Jr, Harris EL, Harrap SB, Krieger JE, Printz MP, Provoost AP, Sassard J, Jacob HJ. New target regions for human hypertension via comparative genomics. Genome Res. 2000;10:473–482. doi: 10.1101/gr.10.4.473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stubbs L, Rinchik EM, Goldberg E, Rudy B, Handel MA, Johnson D. Clustering of six human 11p15 gene homologs within a 500 kb interval of proximal mouse chromosome 7. Genomics. 1994;24:324–332. doi: 10.1006/geno.1994.1623. [DOI] [PubMed] [Google Scholar]
- Thomas JW, Summers TJ, Lee-Lin SQ, Braden Maduro VV, Idol JR, Mastrian SD, Ryan JF, Jamison DJ, Green ED. Comparative genome mapping in the sequence-based era: Early experience with human chromosome 7. Genome Res. 2000;10:624–633. doi: 10.1101/gr.10.5.624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- VanEtten W, Steen R, Nguyen H, Castle A, Slonim D, Ge B, Nusbaum C, Schuler G, Lander E, Hudson T. Radiation hybrid map of the mouse genome. Nat Genet. 1999;22:384–387. doi: 10.1038/11962. [DOI] [PubMed] [Google Scholar]
- Watanabe TK, Bihoreau MT, McCarthy LC, Kiguwa SL, Hishigaki H, Tsuju A, Browne J, Yamasaki Y, Mizoguchi-Miyakita A, Oga K, et al. A radiation hybrid map of the rat genome containing 5,255 markers. Nat Genet. 1999;22:27–36. doi: 10.1038/8737. [DOI] [PubMed] [Google Scholar]
- Wienberg J, Stanyon R. Chromosome painting in mammals as an approach to comparative genomics. Curr Opin Genet Dev. 1995;5:792–797. doi: 10.1016/0959-437x(95)80013-u. [DOI] [PubMed] [Google Scholar]
- Zhang Z, Schwartz S, Wagner L, Miller W. A greedy algorithm for aligning DNA sequences. J Comput Biol. 2000;7:203–214. doi: 10.1089/10665270050081478. [DOI] [PubMed] [Google Scholar]