Abstract
NMR spectroscopy has distinct advantages for providing insight into protein structures, but faces significant resolution challenges as protein size increases. To alleviate such resonance overlap issues, the ability to produce segmentally labeled proteins is beneficial. Here we show that the S. aureus transpeptidase sortase A can be used to catalyze the ligation of two separately expressed domains of the same protein, MecA (B. subtilis). The yield of purified, segmentally labeled MecA protein conjugate is ∼40%. The resultant HSQC spectrum obtained from this domain-labeled conjugate demonstrates successful application of sortase A for segmental labeling of multi-domain proteins for solution NMR study.
Keywords: Segmental labeling, Sortase A, Protein ligation, MecA, Enzyme-mediated ligation, NMR
The ability to pursue NMR studies of large proteins and biological complexes has steadily improved due to technology development. NMR investigations of multi-domain proteins have specifically benefited from development of segmental labeling techniques that permit selective observation of a single domain out of several. Segmental labeling requires use of a protein ligation method to covalently attach an isotopically labeled protein domain to another, unlabeled one for selective NMR detection. Multiple ligation techniques have been demonstrated in the literature, primarily including native chemical ligation, expressed protein ligation (EPL), protein trans-splicing (PTS) and chemical modification (Dawson et al. 1994; Severinov and Muir 1998; Muralidharan and Muir 2006; Nilsson et al. 2003). Among these, EPL and PTS have been the most successful for segmental labeling of proteins for NMR study (Muona et al. 2010; Skrisovska et al. 2010).
Despite the appeal of using ligases and transpeptidases for segmental labeling, the specific implementation of enzymes for this particular purpose has not been demonstrated yet (Skrisovska et al. 2010; Iwai and Züger 2007; Kobashigawa et al. 2009). A key advantage of enzymatic protein ligation is that the reaction can be conducted using near physiological temperature and pH conditions. Furthermore, enzymatic ligation precludes the use of cysteines on the substrates and concerns regarding redox chemistry, both of which are integral to the more commonly employed EPL and PTS methods. Consequently, enzymatic ligation has potentially greater applicability to a wider range of proteins. In general, the ability to ligate proteins requiring relatively inert amino acid sequences presents greater opportunities for studies of multi-domain proteins by structural biologists.
Sortase A is a transpeptidase found in the Gram-positive bacterium S. aureus. In 2004, it was demonstrated that sortase enzyme can be used to ligate two proteins together in vitro (Mao et al. 2004). Sortase ligations are specific since this enzyme requires the presence of a ‘LPXTG’ sequence motif at the end of the N-terminal chain as well as at least one glycine at the N-terminus of the C-terminal chain (Fig. 1).
Fig. 1.
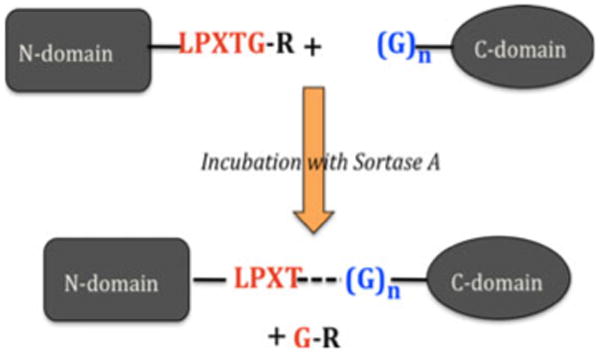
Scheme of protein ligation by sortase A enzyme. The consensus sortase residues required at the end of the N-domain are shown (in red) followed by an ‘R’ group (typically His6, in black). Sortase-specific glycine residue(s) required at the N-terminus of the C-domain are in blue. Ligation of both domains results from formation of the new T-G bond (dashed line) that is catalyzed by sortase
Sortase-mediated ligation was used here to segmentally label MecA, a two-domain B. subtilis protein which functions to proteolytically regulate bacterial competence by targeting a transcription factor for ATP-dependent hydrolysis. Structural studies of intact MecA have focused primarily upon the individual domains as evidenced by the relatively recent X-ray study of the C-terminal domain of this protein (Wang et al. 2009). Detailed NMR analysis of the entire MecA protein would benefit from segmental labeling of specific domains to reduce resonance overlap. Another issue concerns protein dimerization since both the intact as well as the individual domains of this protein form homodimers (Persuh et al. 1999). Line broadening concomitant with dimerization can further contribute to resonance congestion. Clearly, segmental labeling of MecA would enhance NMR studies of this multi-domain protein.
The overall feasibility of this sortase-mediated segmental labeling approach relies upon several factors. First, both domains of interest must be connected by a linker sequence of suitable length (five or more residues). Then, the sortase consensus five residue sequence, “LPXTG”, must also exist somewhere within this linker. Another requirement is the presence of an N-terminal glycine cap at the beginning of the C-terminal domain. All of these specific residues must then be sterically accessible to sortase (Fig. 1).
To determine whether sortase-mediated ligation would be feasible for segmental labeling of MecA, the amino acid sequence and domain organization of this protein were examined. The two domains are connected via a linker region corresponding to residues 83–93 (Persuh et al. 1999). The last five residues of this linker, LPIPE, were targeted as a potential sortase ligation site since mutation of residues P92 and E93 to T and G, respectively, results in the sortase-compatible sequence, LPITG. The MecA P93T/E94G double mutant represents the starting point for use of sortase-mediated ligation for MecA segmental labeling. Consequently, this particular MecA double mutant (‘MecAmut’), as well as several, slightly modified forms of the individual domains were generated for this study. One domain modification is the addition of a C-terminal hexa-histidine sequence to the MecA N-domain (‘MecAN-His’). A second involves addition of a second glycine at the N-terminus of the MecA C-domain (‘MecAC2G’). Both of these modifications have been documented to improve ligation yields (Mao et al. 2004; Parthasarathy et al. 2007; Clow et al. 2008; Kobashigawa et al. 2009). The amino acid sequences of these various protein constructs are provided in Supplementary Information (Figure S1).
A preliminary, 1 ml test ligation was conducted by incubation of unlabeled protein domain substrates with sortase at room temperature for 24 h. Based upon previous studies (Lu et al. 2007), the final reaction component concentrations were 12.4 μM (MecAN-His), 42.2 μM (MecAC) and 50 μM sortase in a pH 7.5, 50 mM Tris, 150 mM NaCl, 5 mM CaCl2 and 2 mM BME buffer. The reaction was conducted within a 6–8 kDa MWCO dialysis bag with continuous dialysis against the above reaction buffer (Pritz et al. 2007). Reaction samples removed at 0, 2, 4, 6 and 24 h were analyzed by SDSPAGE.
Over time, a new protein band appeared and increased in intensity (Fig. 2). To verify identity of this band, the gel was submitted for peptide sequencing analysis by mass spectrometry. The MecAmut and new gel bands were removed and digested with trypsin. MS/MS sequencing of the resultant tryptic peptide fragments indicated that the sequences of both proteins correlated well with native MecA (Eismann et al. 2008). Since the fragments spanned the N- and C-terminal domains, the unknown band corresponds to a ligated product comprised of both domains (Fig. 2).
Fig. 2.

Confirmation of successful sortase ligation by SDSPAGE and mass spectrometry. The SDSPAGE gel is shown on the left. The left lane corresponds to double mutant MecAmut (band ‘A’) while the remaining lanes correspond to the ligation mixture at the indicated times of incubation. The bands outlined in red were excised from the gel and subjected to tryptic digestion. On the right, the full length MecAmut sequence is shown with the tryptic peptides obtained from product band (‘B’) that were sequenced by MS/MS indicated in red. These sequenced fragments span both domains of full length MecAmut
Upon successful demonstration that sortase ligation of the N- and C-terminal domains results in production of full length MecAmut, conditions for a large scale sortase ligation reaction were optimized. Initial reactions that were performed in batch mode at 37°C resulted in low product yields. Dialysis of the reaction was then employed to favor formation of product at room temperature. At lower temperature, the equilibrium product formation was higher and stable up to 5 days with maximal product observed via SDSPAGE by 72 h. For room temperature and dialysis reaction conditions, the sortase A concentration was varied from 2–50 uM with the optimal concentration determined to be 5 uM. The MecAN-His versus MecAC concentration ratios were subsequently tested and these ranged from 1:1 to 1:5 (MecAN-His:MecAC). In general, saturating the reaction with high concentrations of C-terminal protein relative to N-terminal protein domains results in more efficient ligations with minimal hydrolysis (Mao et al. 2004). Finally, formation of MecA ligation product was further improved when the C-terminal domain was capped with a double rather than a single glycine at its N-terminus (Parthasarathy et al. 2007). This ligation product, ‘MecA-mut2G’, contains an extra glycine within the linker relative to MecAmut (Figure S1).
In order to generate a segmentally labeled form of MecA consisting of 15N-labeled N-terminal domain only, 15N-labeled MecAN-His and unlabeled MecAC2G substrates were mixed and incubated with sortase in a pH 7.5, 50 mM Tris, 150 mM NaCl, 10 mM CaCl2 and 2 mM BME buffer. The final concentrations of reaction components were 5 μM sortase, 15uM 15N MecAN-His and 51 uM unlabeled MecAC2G in 26 ml. The reaction was conducted within a 6–8 kDA MWCO dialysis bag that was incubated for 72–96 h at 20–22°C with daily reaction buffer exchange and monitoring via SDSPAGE (Figure S2).
Once product formation reached steady state (approximately 72 h), the reaction was quenched by addition of EDTA (20 mM final concentration, Parthasarathy et al. 2007), followed by overnight dialysis at 8°C against a 50 mM Tris, 150 mM NaCl and 5 mM EDTA, pH 7.5 buffer. The mixture was concentrated via ultrafiltration (Millipore Ultra-free 3 kDa) and purified by SEC FPLC chromatography (Superdex 75). Fractions containing MecAmut were verified by SDSPAGE (Figure S3) before they were pooled and concentrated via ultrafiltration. The crude yield of conjugate, based upon SDSPAGE and FPLC peak integration (Figure S3) is estimated to be 60%. The NMR sample corresponded to 0.5 mM protein (quantitated with Bradford reagent, Bio-Rad) in a 90%:10% H2O:D2O, 25 mM sodium phosphate, 5 mM BME, 5 mM EDTA, pH 7.5 buffer. The final quantity of purified, segmentally 15N-labeled MecAN-MecAC conjugate obtained was 4 milligrams. This corresponds to a final yield calculated relative to the limiting reagent, 15N-MecAN-His, of 40%.
Sortase-mediated ligations of protein-peptide or peptide-peptide systems have maximal efficiencies approaching 70% (Pritz et al. 2007; Mao et al. 2004). Reported protein–protein ligation efficiencies have varied from low (Mao et al. 2004) to 90% (Kobashigawa et al. 2009). For the latter, 90% represents an apparent ligation efficiency that is based upon qualitative disappearance of the SDSPAGE N-terminal domain substrate band (Kobashigawa et al. 2009). For our study, we chose to report traditional product yields because gel band intensities can decrease for various reasons including ligation progress as well as degradation. Based upon the amount of conjugate protein obtained relative to the amount of limiting protein substrate, the crude and final purified yield are 60 and 40%, respectively, for our system. It is also anticipated that these yields may be affected by the dimeric properties of both MecA protein domains.
In general, the final recovery of sortase-mediated ligation product will depend upon linker accessibility as well as efficiency of product purification. Based upon these results, we believe that sortase A represents a viable alternative for segmental labeling of multi-domain proteins.
One-dimensional proton NMR spectra were recorded from the segmentally labeled MecA protein using various edit/filter options. In Figure S4, effects of segmental labeling are evident from observation of only a subset of 15N-coupled amide proton resonances in the 15N-edited spectrum, relative to those recorded in a reference, unedited/unfiltered spectrum. The number and pattern of observable 15N-edited proton resonances are also distinctive from those of the 14N-coupled amide proton resonances observed in the 15N-filtered spectrum (Figure S4).
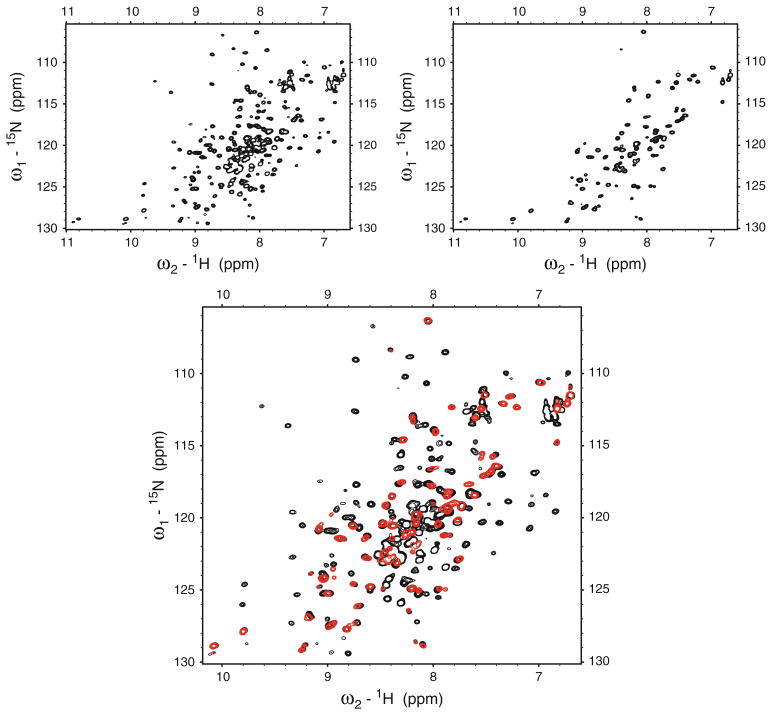
Comparison of 2D 1H-15N HSQC spectra recorded from the native and double mutant forms of two-domain MecA indicate that both proteins adopt the same global fold with only a very small number of observed resonance shifts (less than 5%, Figure S5). Comparison of the 2D 1H-15N HSQC spectra of segmentally labeled 15N-MecAN-MecAC (MecAmut2G) conjugate versus fully 15N-labeled MecA-mut indicates that the conjugate yields far fewer resonance peaks, with minimal differences between the N-terminal domain resonance patterns of both (Fig. 3).
Fig. 3.
2D 1H-15N HSQC spectra of fully 15N-labeled MecA double mutant (MecAmut) (top left) and N-terminally labeled 15N-MecAN-MecAC (MecAmut2G) protein conjugate (top right). Overlay of the two spectra is shown at bottom (black—fully labeled protein, red—N-terminal domain labeled). Spectra were acquired on a 600 MHz Varian Inova spectrometer at 303 K
These results represent the first application of sortase-mediated ligation for segmental labeling of a multi-domain protein. This enzymatic approach involves micromolar protein domain concentrations and near physiological conditions. Although the fully native versus the ligated MecA proteins differ by three amino acids within the linker region, the detected NMR differences indicate that this does not significantly affect the segmentally labeled protein in terms of the global structure of MecA. Truncation mutants at the N-terminus of the C-terminal domain of MecA that encompass the LPIPE target site suggest that this linker region is quite flexible, as judged by strong resonance intensities and predominantly random-coil chemical shifts in the corresponding 2D 1H-15N HSQC spectra (Cavanagh et al. unpublished data). The NMR results provide no evidence for any significant changes in inter-domain contacts in MecA arising from the ligation process.
Segmental isotopic labeling via sortase-mediated ligation has significant implications for improving the spectral quality and simplifying analyses of multi-domain proteins for structural biology studies by NMR or other methods. This methodology can be employed for example to support NMR experiments designed to detect inter-domain NOEs from proteins with different domain isotope labeling patterns, and to facilitate characterization of individual domain dynamics via NMR spin relaxation studies.
Supplementary Material
Acknowledgments
The authors thank H. Mao for early discussions regarding sortase. S. Dave, P. Kaur, E. Lao and M. Zandi are thanked for assistance at various stages of this project. Dr. M. Howell is gratefully acknowledged for many helpful discussions. Dr. K. Greis (University of Cincinnati Proteomics Facility) is thanked for assistance with in-gel tryptic digestion, MS/MS experiments. Funding from NIH (GM55769 and GM65156 to JC, GM063855 to MR, and RR19077 and RR027755 to the UC College of Medicine NMR facility) is acknowledged.
Abbreviations
- HSQC
Heteronuclear single quantum coherence
- SDS-PAGE
Sodium dodecyl sulfate polyacrylamide gel electrophoresis
- SEC
Size-exclusion chromatography
- NOE
Nuclear Overhauser effect
- MS
Mass spectrometry
Footnotes
Electronic supplementary material The online version of this article (doi:10.1007/s10858-010-9464-2) contains supplementary material, which is available to authorized users.
Contributor Information
Mary Anne Refaei, Department of Chemistry, University of Cincinnati, Cincinnati 45221-0172, USA.
Al Combs, Department of Molecular Genetics, Biochemistry and Microbiology, University of Cincinnati, Cincinnati 45267-0524, USA.
Douglas J. Kojetin, Department of Molecular Genetics, Biochemistry and Microbiology, University of Cincinnati, Cincinnati 45267-0524, USA
John Cavanagh, Department of Molecular and Structural Biochemistry, North Carolina State University, Raleigh, NC 27695, USA.
Carol Caperelli, College of Pharmacy, University of Cincinnati, Cincinnati 45267-0004, USA.
Mark Rance, Department of Molecular Genetics, Biochemistry and Microbiology, University of Cincinnati, Cincinnati 45267-0524, USA.
Jennifer Sapitro, Department of Chemistry, University of Cincinnati, Cincinnati 45221-0172, USA.
Pearl Tsang, Email: pearl.tsang@gmail.com, Department of Chemistry, University of Cincinnati, Cincinnati 45221-0172, USA.
References
- Clow F, Fraser JD, Proft T. Immobilization of proteins to biacore sensor chips using Staphylococcus aureus sortase A. Biotechnol Lett. 2008;30:1603–1647. doi: 10.1007/s10529-008-9718-1. [DOI] [PubMed] [Google Scholar]
- Dawson PE, Muir TW, Clark-Lewis I, Kent SB. Synthesis of proteins by native chemical ligation. Science. 1994;266:776–779. doi: 10.1126/science.7973629. [DOI] [PubMed] [Google Scholar]
- Eismann T, Huber N, Shin T, Kuboki S, Galloway E, Wyder M, Edwards MJ, Greis KD, Shertzer HG, Fisher AB, Lentsch AB. Peroxiredoxin-6 protects against mitochondrial dysfunction and liver injury during ischemia/reperfusion in mice. Am J Physiol Gastrointest Liver Physiol. 2008;296:G266–G274. doi: 10.1152/ajpgi.90583.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwai H, Züger S. Protein ligation: application in NMR studies of proteins. Biotechnol Genet Eng Rev. 2007;24:129–146. doi: 10.1080/02648725.2007.10648096. [DOI] [PubMed] [Google Scholar]
- Kobashigawa Y, Kumeta H, Ogura K, Inagaki F. Attachment of an NMR-invisible solubility enhancement tag using a sortase-mediated protein ligation method. J Biomol NMR. 2009;43:145–150. doi: 10.1007/s10858-008-9296-5. [DOI] [PubMed] [Google Scholar]
- Lu C, Zhu J, Wang Y, Umeda A, Cowmeadow RB, Lai E, Moreno GN, Person MD, Zhang Z. Staphylococcus aureus sortase A exists as a dimeric protein in vitro. Biochemistry. 2007;46:9346–9354. doi: 10.1021/bi700519w. [DOI] [PubMed] [Google Scholar]
- Mao H, Hart SA, Schink A, Pollok BA. Sortase-mediated ligation: a new method for protein engineering. J Am Chem Soc. 2004;126:2670–2671. doi: 10.1021/ja039915e. [DOI] [PubMed] [Google Scholar]
- Muona M, Aranko AS, Raulinaitis V, Iwai H. Segmental isotopic labeling of multi-domain and fusion proteins by protein trans-splicing in vivo and in vitro. Nat Protoc. 2010;5:574–587. doi: 10.1038/nprot.2009.240. [DOI] [PubMed] [Google Scholar]
- Muralidharan V, Muir TW. Protein ligation: an enabling technology for the biophysical studies of proteins. Nat Methods. 2006;3:429–438. doi: 10.1038/nmeth886. [DOI] [PubMed] [Google Scholar]
- Nilsson BL, Hondal RJ, Soellner MB, Raines RT. Protein assembly by orthogonal chemical ligation methods. J Am Chem Soc. 2003;125:5268–5269. doi: 10.1021/ja029752e. [DOI] [PubMed] [Google Scholar]
- Parthasarathy R, Subramanian S, Boder ET. Sortase A as a novel molecular “stapler” for sequence-specific protein conjugation. Bioconjug Chem. 2007;18:469–476. doi: 10.1021/bc060339w. [DOI] [PubMed] [Google Scholar]
- Persuh M, Turgay K, Mandic-Mulec I, Dubnau D. The N- and C-terminal domains of MecA recognize different partners in the competence molecular switch. Mol Microbiol. 1999;33:886–894. doi: 10.1046/j.1365-2958.1999.01544.x. [DOI] [PubMed] [Google Scholar]
- Pritz S, Wolf Y, Kraetke O, Klose J, Bienert M, Beyermann M. Synthesis of biologically active peptide nucleic acid-peptide conjugates by sortase-mediated ligation. J Org Chem. 2007;72:3909–3912. doi: 10.1021/jo062331l. [DOI] [PubMed] [Google Scholar]
- Severinov K, Muir TW. Expressed protein ligation, a novel method for studying protein-protein interactions in transcription. J Biol Chem. 1998;273:16205–16209. doi: 10.1074/jbc.273.26.16205. [DOI] [PubMed] [Google Scholar]
- Skrisovska L, Schubert M, Allain FHT. Recent advances in segmental isotope labeling of proteins: NMR applications to large proteins and glycopeptides. J Biomol NMR. 2010;46:51–65. doi: 10.1007/s10858-009-9362-7. [DOI] [PubMed] [Google Scholar]
- Wang F, Mei Z, Qi Y, Yan C, Xiang S, Zhou Z, Hu Q, Wang J, Shi Y. Crystal structure of the MecA degradation tag. J Biol Chem. 2009;384:34376–34381. doi: 10.1074/jbc.M109.053033. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.