Abstract
The genome of the halophilic archaeon Halobacterium sp. NRC-1 and predicted proteome have been analyzed by computational methods and reveal characteristics relevant to life in an extreme environment distinguished by hypersalinity and high solar radiation: (1) The proteome is highly acidic, with a median pI of 4.9 and mostly lacking basic proteins. This characteristic correlates with high surface negative charge, determined through homology modeling, as the major adaptive mechanism of halophilic proteins to function in nearly saturating salinity. (2) Codon usage displays the expected GC bias in the wobble position and is consistent with a highly acidic proteome. (3) Distinct genomic domains of NRC-1 with bacterial character are apparent by whole proteome BLAST analysis, including two gene clusters coding for a bacterial-type aerobic respiratory chain. This result indicates that the capacity of halophiles for aerobic respiration may have been acquired through lateral gene transfer. (4) Two regions of the large chromosome were found with relatively lower GC composition and overrepresentation of IS elements, similar to the minichromosomes. These IS-element-rich regions of the genome may serve to exchange DNA between the three replicons and promote genome evolution. (5) GC-skew analysis showed evidence for the existence of two replication origins in the large chromosome. This finding and the occurrence of multiple chromosomes indicate a dynamic genome organization with eukaryotic character.
Halobacterium species are members of the archaeal domain, which includes halophiles, methanogens, thermophiles, and some recently discovered mesophiles. Halobacterium NRC-1, the single halophile to have its genome sequenced thus far, grows optimally in a 4.5 M NaCl medium supplemented with amino acids and other nutrients and is capable of both aerobic and phototropic growth (Ng et al. 1998, 2000). This organism is easy to culture and manipulate in the laboratory, which has made it an excellent model organism among the archaea. It has been extensively studied and shown to contain some of the classic features found in halophilic archaea, for example, an S-layer glycoprotein, ether-linked lipids, and purple membrane. Eukaryotic features of archaea such as DNA replication, transcription, and aspects of translation have also been established in this halophile. Over the last 20 years, many genetic tools have been developed, including efficient DNA-mediated transformation, shuttle plasmids, and a gene replacement system, for Halobacterium NRC-1 and related halophiles (DasSarma et al. 1995; Peck et al. 2000).
The genome of Halobacterium NRC-1 consists of three replicons, a large chromosome of 2 Mb and two smaller replicons, pNRC200 and pNRC100, about 365 kb and 191 kb, respectively, proposed to be minichromosomes because of the presence of essential genes (Ng et al. 1998, 2000). The minichromosomes are relatively AT-rich (pNRC100, 58% G + C; pNRC200, 59% G + C) compared with the large chromosome (∼68% G + C), and account for most of the AT-rich satellite DNA discovered in early genomic studies (Moore and McCarthy 1969; Sapienza and Doolittle 1982). The minichromosomes share a 145-kb region of identity within which exist 2 copies of 33–37-kb inverted repeats. The majority (69 of 91) of IS elements were also localized to the minichromosomes, which, along with the other genomic repeats, accounts for the dynamic nature of the genome (Charlebois and Doolittle 1989; Ng et al. 1991; Hackett et al. 1994). Altogether, 14% of the genome, including the 91 IS elements, 33–37-kb inverted repeats, and 145-kb duplication in pNRC100 and pNRC200, is repeated DNA.
The environment in which Halobacterium NRC-1 grows, which is both extreme and dynamic, presents a significant challenge to its survival. Molar concentrations of salt ions are present and are required for optimal growth, and high intracellular levels of KCl accumulate to maintain osmotic balance (Lanyi 1974). Analysis of some halophilic proteins indicated the presence of high surface negative charge, which is thought to enhance solubility and maintain function at such high salinity (Eisenberg 1995). Halobacterium also encounters intense solar UV irradiation that can cause DNA damage such as the formation of thymine dimers. The high GC composition (65.9%) of Halobacterium reduces the chance of such lesions, and an active photoreactivation pathway exists for repair of thymine dimers (Hescox and Carlberg 1972). Color-specific phototaxis mechanisms are also present that mediate swimming away from high-energy wavelengths of solar radiation (Spudich et al. 2000), and the depth of cells in the water column is regulated by buoyant gas vesicles in response to oxygen and nutrient levels (DasSarma and Arora 1997). Light-driven proton pumping by the purple-membrane proteins permits a period of phototrophic growth (Sumper et al. 1976).
To better understand the basis of adaptation of halophiles to their environment, we have conducted further computational analysis of the Halobacterium NRC-1 genome and proteome. These studies provide a more detailed view of the genome-wide strategies used for survival of this obligate halophile in its hypersaline and high-solar-radiation environment.
RESULTS
Isoelectric Point Prediction and Homology Modeling
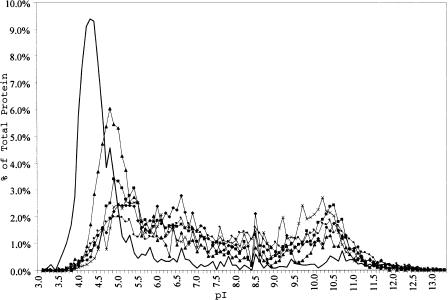
Isoelectric points for the 2630 Halobacterium NRC-1 predicted proteins were calculated, along with predicted proteins from 13 other organisms with completely sequenced genomes, and the data were plotted to show the number of proteins within specific pI ranges (Fig. 1). For all proteomes, except NRC-1, a bimodal distribution of protein pIs is observed with an acidic peak at ∼5.0 and a basic peak at ∼10.3. In contrast, Halobacterium NRC-1 possesses an extremely acidic complement of proteins in its proteome (peak at ∼4.2), and is the only organism that essentially lacks the peak corresponding to basic proteins. Interestingly, the only other organism with a notably acidic proteome is Methanobacterium thermoautotrophicum, also a member of the Euryarchaeota and a distant phylogenetic relative to halophiles. M. thermoautotrophicum, like NRC-1, which accumulates potassium to about 4 M, also contains a relatively high internal concentration of about 1 M potassium ions (Ciulla et al. 1994). These results confirm the expected correlation between high internal salt concentration and protein acidity and show that this is a genome-wide character for Halobacterium NRC-1 (Ng et al. 1998, 2000).
Figure 1.
Predicted percentage of total protein versus isoelectric point distribution for six complete genomes including Methanobacterium thermoautotrophicum (filled triangles), Methanococcus jannaschii (X symbols ), Escherichia coli (filled diamonds), Bacillus subtilis (filled squares), Saccharomyces cerevisiae (filled circles), and Halobacterium NRC-1 (unmarked line).
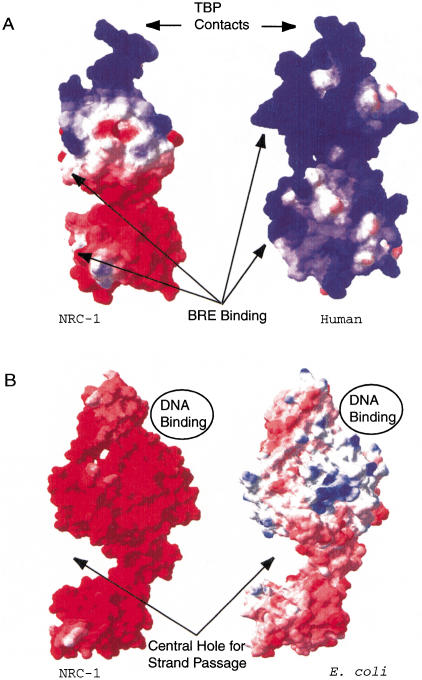
To determine the distribution of negative charges in Halobacterium NRC-1 proteins, we identified candidates appropriate for homology modeling. Modeled structures were generated for the NRC-1 proteins TFBe (one of seven TFB protein homologs in NRC-1; Baliga et al. 2000) and GyrA, based on published crystal structures for each protein (Reece and Maxwell 1991; Berger et al. 1996; Kosa et al. 1997; Littlefield et al. 1999). The best available homolog for TFBe was Pyrococcus woesei TFB (BLAST P value of 6e−24) and for GyrA, Escherichia coli GyrA (BLAST P value of e−119). In the case of TFBe, after confirming a similar structure, subsequent comparisons were made against the human TFIIB protein. Modeled structures of the halobacterial homologs allowed for predictions of amino acids at the protein surface, and for surface charges to then be applied. Using a Coulomb charge calculation, the surface charge was calculated for each protein and a comparison was made for each pair (Fig. 2). Both TFBe and GyrA proteins show a marked increase in surface negative charge when compared to their homologs in nonhalophilic organisms. The increase in surface negative charge is consistent with the lower calculated isoelectric points for the halophile proteins (NRC-1 TFBe has a pI of 6.02, compared with 9.91 for human TFIIB, and NRC-1 GyrA has a pI of 4.31, compared with 5.60 for E. coli GyrA).
Figure 2.
Surface charge comparisons for halophilic and nonhalophilic proteins. Acidic character is indicated by red, basic character is indicated by blue, and neutral areas are indicated by white. (A) NRC-1 TFBe (left) and Human TFIIB (right) are shown with sites for BRE and TBP contacts indicated. (B) NRC-1 (left) and Escherichia coli GyrA (right) are shown with the binding site for the helix that is cleaved as well as the site for strand passage indicated for both molecules.
The data presented in the text and in Figure 2 show the results of surface charge calculations performed using a dielectric constant for the solvent of 80.0. However, in vivo, the dielectric constant decreases from 80.1 to about 48.4 when salinity NaCl concentration is raised from 0 to 5 M (Elcock and McCammon 1998). This reduction in the capacitance of the solvent theoretically results in the reduced deprotonation of acid residues on the protein surface and an overall reduction in charge. The details of amino acid usage in both proteins show an overrepresentation of negatively charged residues and underrepresentation of positively charged residues in the halobacterial homologs that likely compensate for this effect. GyrA from NRC-1 has a total of 163 highly acid residues (Asp and Glu), whereas E. coli GyrA contains 74 such residues. The result is similar in the case of the TFB protein, because the NRC-1 TFBe protein contains 35 negatively charged residues compared with 22 for human TFIIB. The differences in charge also correspond to the surfaces of the proteins, where contact with the solvent and other molecules occurs. Our analysis shows that charged residues near or at the protein surface contribute to an overall charge of −24 for halobacterial GyrA versus −15 for that of E. coli. This result also occurs in the case of TFB, where the halobacterial TFB has a surface charge of −4 versus +14 for the Pyrococcus TFB. This pattern of increased negative charge and consequently lowered pI is observed for the majority of halobacterial proteins when compared with their homologs, with the additional negative charge reflected on the surfaces of the proteins.
Codon Bias
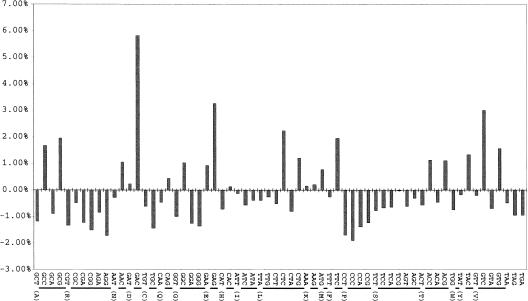
We examined codon usage in Halobacterium NRC-1, beginning with the distribution of GC composition at the three individual codon positions. Our results show an expected result for an organism with high GC content, a third position GC bias. The first, second, and third codon positions of NRC-1 ORFs have GC percentages of 69.50%, 45.97%, and 85.51%, respectively. Additionally, the codon usage in NRC-1 is generally consistent with that expected, when corrected for GC composition (Fig. 3). A few exceptions were noted, corresponding to specific amino acids. The codons GAC and GAG, corresponding to aspartic acid and glutamic acid, respectively, are overrepresented, corresponding to an abundance of acid residues and the preference for a G or C in the third position. Also, the codons GGG and CCC, corresponding to glycine and proline, are underrepresented. This underrepresentation might be explained by the susceptibility of runs of Gs and Cs to site-specific oxidative damage and also to frameshifting during translation (Janel-Bintz et al. 1994; Kawanishi et al. 1999).
Figure 3.
Codon bias plot showing differences in expected triplet frequencies. Each of the 64 triples is plotted along the X axis with one-letter amino acid designations identifying which amino acid a particular group of triplet represents. Results are normalized according to GC composition. The deviation from the expected percentage is plotted on the Y axis.
We also investigated the relations among abundant acidic residues in the proteome, codon frequency, and GC composition. Based on mathematical models of the genetic code, amino acid usage is related to GC content by first-, second-, and third-codon position translations and their correspondence to nonphenotypic mutations (error buffering) in the genetic code (Freeland et al. 2000). Models based on this observation predict an overrepresentation of the amino acids glycine, proline, and arginine in an organism with high GC content (Lobry 1997). However, all of these amino acids are underrepresented in NRC-1. The dominant effect observed instead is clearly the preference for acidic amino acids. Considering the GC composition of NRC-1, aspartic acid and glutamic acid are overrepresented by 41% and 37%, respectively. Therefore, overrepresentation of acidic residues is an adaptation to high salinity that is apparent in the codon usage in this organism.
Lateral Gene Transfer
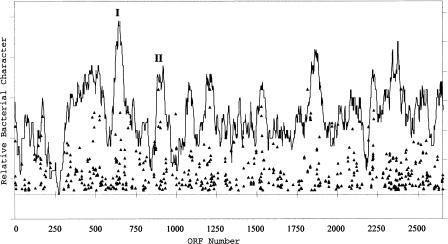
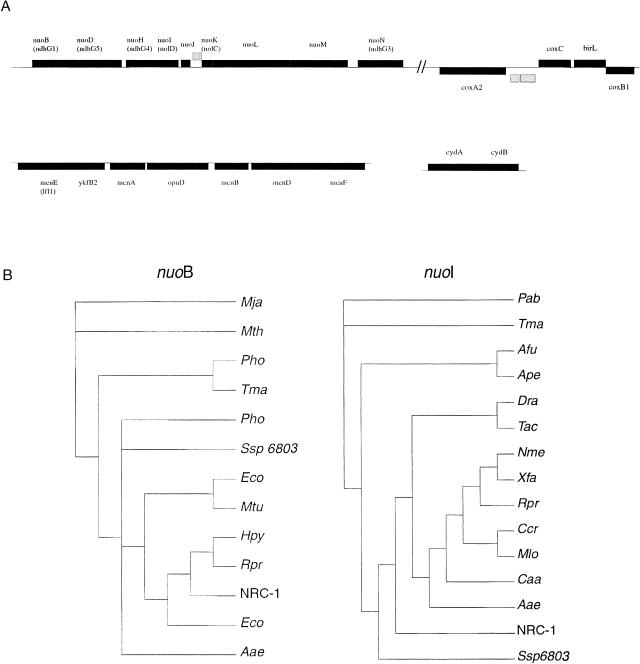
Data from initial examination of halobacterial ORFs on the large chromosome revealed several regions that contained clusters of genes whose closest homologs were bacterial (Fig. 4). Although this evidence per se does not allow for an inference of lateral gene transfer, it indicated that these genes might be worthy of further investigation. Inspection of these regions revealed that they included those encoding electron-transport-chain factors and biosynthetic proteins. Ten nuo genes, encoding subunits of NADH dehydrogenase, along with three of the six NRC-1 cox genes, encoding the three subunits of cytochrome c oxidase, were associated with the larger peak (Fig. 4, peak 1). Six men genes, for menaquinone biosynthesis, were also localized to a second minor peak (Fig. 4, peak 2). Interestingly, gene order for the nuo genes is conserved with respect to E. coli, and for the men genes is conserved compared with both E. coli and Deinococcus radiodurans. GC analysis of these two groups of genes showed them also to be distinguished from the average chromosomal genes in their GC content and variance. The mean GC content of chromosome ORFs is 67.9% compared with 63.9% (P = 1.33E−5) for region 1 and 73.4% (P = 2.28E−3) for region 2. Furthermore, despite their large deviations from the mean GC content of the chromosome, the variance of the data was low (5.72 for 13 genes for region 1; 7.12 for six genes for region 2) when compared against the large chromosome (26.44 for 2059 genes).
Figure 4.
Graphical representation of the relative amount of homology between predicted proteins in the large chromosome of NRC-1 to those encoded by Deinococcus radiodurans and Bacillus subtilis. NRC-1 chromosomal ORFs are plotted on the X axis. Triangles represent NRC-1 ORFs with BLAST P values <e−29, with more significant scores higher on the Y axis. Average bacterial character is shown by the percentage of predicted ORFs in a 40-ORF window that showed BLAST P values of <1 × 10−10. The labeled peaks correspond to regions harboring genes encoding NADH dehydrogenase (I) and menaquinone biosynthesis (II), components of the electron transport chain.
The Halobacterium NRC-1 nuo, men, and cox gene products were subjected to further phylogenetic analysis. An initial search against the NCBI COG database was used to identify appropriate orthologs from which to construct phylogenetic trees (Tatusov et al. 2000). Using the GCG Wisconsin package of programs, phylogenetic trees were constructed and compared for congruity of branching for each subset of gene products (Fig. 5). Our results show two distinct patterns of branching corresponding to the nuo and men genes. Although no archaeal homologs could be identified for the set of men genes, thus preventing detailed phylogenetic analysis, conservation of genetic order and function support the synteny argument of conserved gene order and gene function (Lawrence and Roth 1996). The majority of nuo gene products have closest branching with Synechocystis PCC6803. Interestingly, there does not appear to be gene order conservation between PCC6803 and NRC-1. It is notable that for genes encoding subunits of NADH dehydrogenase, for which archaeal orthologs could be identified (nuoB, nuoD, nouH, nuoI, nuoL), the NRC-1 sequences were most closely related to bacterial orthologs, indicating a deviation from the 16S rRNA tree.
Figure 5.
(A) Genetic maps of aerobic respiration genes. The first row contains the nuo and cox genes in the region from 485,000 to 506,000 bp. The second row contains the men and cyd genes in a chromosomal region from 821,000 to 829,000 bp and pNRC100, respectively. The genetic nomenclature for some nuo and men genes was changed from our earlier work (Ng et al. 2000); changes are indicated parenthetically. (B) Phylogenetic trees of nuoB and nuoI gene products. Branches are labeled for each gene with the corresponding three-letter organism designations except for Ssp 6803 (Synechocystis sp.) and NRC-1 (Halobacterium NRC-1).
Considering the physiology of Halobacterium NRC-1 and its position on 16S ribosomal trees, two alternative possibilities could account for the presence of an intact and functional electron transport chain in this aerobic organism. One possibility is that anaerobic halophiles may have adapted to an oxidizing atmosphere by acquiring the electron transport chain through lateral transfer events from bacterial organisms capable of aerobic respiration. A second possibility is that a common ancestor of the halophiles and methanogens may have harbored an aerobic respiration system similar to that of bacteria that was lost by present-day methanogens. Recent evidence (Salzberg et al. 2001) indicates that with a larger sample of genome sequences from related organisms, evidence for lateral transfer often disappears. This points to the importance of sequencing additional genes from halophiles and related methanogens in order to determine which of these possibilities is more likely.
GC Composition Analysis
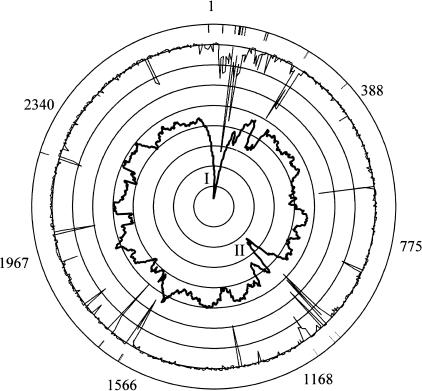
The three replicons of Halobacterium NRC-1 were examined for GC content by calculating the GC percentage of each ORF in the genome and also by χ-squared analysis using 2000-bp windows. The average GC content of chromosomal ORFs and χ-squared data were used to generate a plot on a circular map of the large chromosome (Fig. 6). Two regions of reduced GC content and high significance were observed on the chromosome, where the average GC content is equal to 68%. The first region of lower GC content (I), from 1 to 270,000 bp, has an average GC content of 65.2% and contains 13 of the 22 (59%) observed chromosomal IS elements. A second region of lower GC content (II), from 690,000 to 840,000 bp, has an average GC content of 65.8% and contains an additional 4 (18%) IS elements. The GC percentage of the chromosomal ORFs, calculated without the AT islands, is ∼70%. Also, several minor peaks in the χ-squared analysis correspond to IS elements in the chromosome, indicating an important role for IS elements in mediating rearrangements across all three replicons.
Figure 6.
Circular representation of the Halobacterium NRC-1 chromosome showing the GC composition of ORFs, χ-squared analysis, and location of IS elements. The outer scale refers to ORF identification numbers. Bars associated with the outmost circle denote the position of the chromosomal IS elements. χ-squared analysis is plotted as a thin black line, and average GC content of ORF is the innermost plot in darker black, each against their relative position on the circular map. The Roman numerals I and II inside the plot indicate relatively AT-rich islands.
Based on our GC composition analysis, these two regions of the chromosome look similar to the minichromosomes pNRC100 and pNRC200 in both reduced GC content and number of IS elements. These regions further resemble the minichromosomes in that they possess a lower fraction of named genes with recognizable homologs in the database. Like pNRC100 and pNRC200, which have an average of 30% named genes, the two regions of the chromosome contain 35% named genes, compared with the large chromosome, which contains 42% named genes. A reversal of the situation described above is observed in the genome, where a 15-kb region present on the inverted repeat of both smaller replicons is higher in GC content (64%) than the average GC composition of the replicon (58%; Ng et al. 1998). These results indicate that genomic regions with diverse GC composition occur in all three replicons.
GC Skew
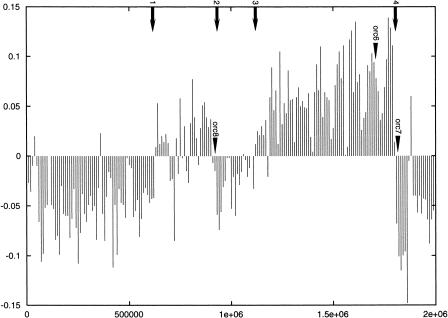
GC skew is a statistical method to measure the observation of a strand-specific overrepresentation of guanine. In organisms that have a measurable GC skew, the point where the data plot changes from positive to negative or from negative to positive (inflection point) is sometimes an indication of the origin or terminus for DNA replication (McLean et al. 1998). For the large chromosome of Halobacterium NRC-1, the GC-skew plot shows four such inflection points (point 1 at 633,000 bp; point 2 at 909,000 bp; point 3 at 1,121,000 bp; point 4 at 1,805,000 bp), indicating the possibility of two distinct replicons, each with an origin of replication and corresponding termination site (Fig. 7).
Figure 7.
Plot of the GC skew of the Halobacterium NRC-1 large chromosome. The GC ratio (Y axis) is plotted over the length of the chromosome (X axis). The numbered arrows show inflection points, where the plot crosses the zero mark. The positions of the three chromosomal orc1/cdc6 genes are indicated.
Genes characteristically located near origins of replication that are used to confirm the results of GC skew include orc1/cdc6 in some archaea, and dnaA, dnaN, gyrA, and gyrB in bacteria. Homologs of the origin recognition protein and replication initiation protein (orc1/cdc6) in yeast (Bell and Stillman 1992) have been found located near the origins of M. thermoautotrophicum and three Pyrococcus species (Myllykallio et al. 2000). For Halobacterium NRC-1, we found three homologs of orc1/cdc6 genes on the large chromosome and 9 or 10 total copies in the genome. Two of these genes are located near the GC-skew inflection points, 909,000 bp and 1,805,000 bp, which correspond to the predicted termination regions. Interestingly, gyrB and gyrA occur in tandem in the region spanning positions 672,176–676,486 bp near the first inflection point. These two genes occur in the same order (gyrB followed by gyrA) in close proximity to the origin in the genomes of Borrelia burgdorferi, Mycoplasma genitalium, Mycoplasma pneumoniae, Bacillus subtilis, and Mycobacterium tuberculosis.
Similar results to those described above were obtained for the large chromosome using an oligomer-skew method, which measures the skews of short sequences compared to their complements (data not shown). Neither the GC-skew nor oligomer-skew analyses identify any clear replication origins on pNRC100 and pNRC200. Altogether, the results indicate that multiple mechanisms of replication are probably operating in Halobacterium NRC-1, including an eukaryote-like system with multiple origins of replication on the large chromosome of NRC-1.
DISCUSSION
We have conducted detailed computational analysis of the complete genome sequence of Halobacterium NRC-1, which has revealed some interesting and unique traits of halophilic archaea. A general characteristic of halophilic proteins is acidity. Homology modeling indicates that significant negative charge occurs at the protein surface. The halophile genome is a mosaic of diverse GC composition and regions studded with IS elements. This novel genome structure may reflect the adaptation of halophiles to a dynamic environment and may have led to the acquisition of new genes and metabolic capabilities, for example, aerobic respiration, through lateral gene transfer during the evolution of halophiles. The dynamic genome is manifested in the occurrence of multiple and variable replicons, including two putative replicons comprising the large chromosome.
One of our most significant findings is that the halophile genome is essentially unique in coding for an acidic proteome, as suggested by early studies (Lanyi 1974). As shown in Figure 1, protein acidity is a general proteome-wide characteristic of this organism, and most proteins have pIs within the range of 3.5 to 5.0. Such a dramatic skew indicates that these proteins have adapted to the high salt environment in a manner directly related to their acidic isoelectric points. Because Halobacterium NRC-1 has a neutral internal pH (pH 7.2; Tsujimoto et al. 1988) the added hydration effect of charged residues and the mutual repulsion provided by a high concentration of surface negative charges have been suggested as reasons for the acidity of many halophilic proteins. Indeed, models employing a salt-dependent dielectric constant to solve the Poisson–Boltzmann equation for electrostatics on various proteins accurately account for experimental evidence showing reduced stability and activity of halophilic proteins below their optimum salt concentrations (Elcock and McCammon 1998). This earlier work has showed halophilic proteins to have, in addition to an abundance of acidic residues on the protein surface, structures that harness the high ionic strength of the solvent to form additional salt bridges that aid in monomer and multimer stability. Moreover, comparisons between crystal structures of malate dehydrogenase from a halophile and the nonhalophilic lactate dehydrogenase homolog have also indicated that an increase in the number of negatively charged residues on the surface of the protein helps to maintain activity at high salt concentrations (Dym et al. 1995).
Therefore, both experimental work and computational analysis have implicated increased surface charge as a means of counteracting the decrease in dielectric constant that occurs at high salinity and thus providing enhanced solubility. It is notable that even DNA-binding proteins, such as TFBs, that are generally known to be basic, owing to their requirement to make close contacts with the negatively charged DNA, are found to be either neutral or acidic in NRC-1. A question that arises from these observations is how protein–protein and protein–DNA interactions are affected in halobacterial species. The result of homology modeling has revealed, in a number of cases, proteins without any distinct regions of neutral or basic charge associated with the putative binding site. These data showing a highly acidic proteome as a genomic-wide adaptation to a hypersaline environment highlight the importance and value of future global structural genomics projects to understanding organisms at the level of their proteome. Structural conservation of halobacterial proteins with their nonhalophilic homologs and the high solubility of halophilic proteins make Halobacterium NRC-1 attractive for large-scale crystallographic studies. This halophile should serve as an ideal model for the emerging and exciting field of structural genomics.
Wobble rules allow maximal flexibility in the third-codon position GC content, followed by weaker effects from the first and second codon positions (Deschavanne and Filipski 1995; Lobry 1997). A study of 12 bacterial genomes showed the third codon position to be most affected by higher or lower GC composition (Majumdar et al. 1999). Our results with NRC-1 are consistent with previous reports and show a strong GC bias (86%) in the third codon position. We also examined the effects of GC content and the acidic proteome on amino acid usage. Statistical models report a predicted overrepresentation of glycine, proline, and arginine in high-GC organisms based on codon usage and codon positions available to nonphenotypic mutation. We have reported on the extreme acidity of the NRC-1 proteome and investigated whether selective pressure for charged residues is evident. In our study, we found that the acidic proteome seems to be the dominant force in codon and amino acid selection. Overrepresentation of aspartic acid and glutamic acid codons were seen instead of codons for glycine, proline, and a number of other amino acids that are uncharged at physiological pH.
The recent availability of many completely sequenced microbial genomes (Nelson et al. 2000) has facilitated studies aimed at discovery of lateral gene transfer (LGT). Although whole-genome comparisons have supported the overall structure of the 16s rRNA tree (Fitz-Gibbon and House 1999; Snel et al. 1999), the importance of LGT as a major force in evolution has also come to light as a driving force in prokaryotic evolution (Doolittle 1999; Nelson et al. 1999, 2000; Campbell 2000). Aerobic respiration, which is common among halophiles yet absent from phylogenetically related archaea, is an observation now amenable to further inquiry. In our studies, we find all of the components of the electron transport chain, including NADH dehydrogenase (nuo), menaquinone (men), and cytochrome oxidase (cox), with high bacterial character and clustered in several regions of the genome. A second cluster of cox genes is also found in a separate locus on the large chromosome. Furthermore, nuo, men, and cox genes show both distinct GC percentages and low GC variance when compared with the chromosome on which they are located. Two other genes (cydA and B) encoding subunits of an alternate cytochrome d oxidase, putatively used under oxygen-limiting growth conditions, were previously found on the large inverted repeats of pNRC100 and pNRC200 in dynamic AT- and IS-rich regions (Ng et al. 1998). Many of the genes involved in the electron transport chain are found in conserved syntenic regions of E. coli and D. radiodurans. Phylogenetic analysis of nuo, cox, and cyd gene products indicated close bacterial homology in the trees generated, distinct from NRC-1 as a whole, but conserved with respect to gene cluster and genomic locus. Additionally, the presence of men genes with conserved gene order is consistent with our findings. These data provide evidence for the coordinated transfer of functional sets of aerobic respiration genes in Halobacterium NRC-1 through a number of genetic transfer events.
Prior to our examination of Halobacterium NRC-1, early studies had shown that halophile genomes consist of two fractions with differing GC content (Moore and McCarthy 1969; Pfeifer and Betlach 1985). Our analysis of the NRC-1 genome shows, further, that the great majority of IS elements, and thus by association most genomic rearrangements, occur in the relatively AT-rich satellite fraction that is part of all three replicons. The preferential integration of IS elements into relatively AT-rich regions, and their subsequent proliferation at nearby sites, could have contributed to the observed genomic structure. The clustering of IS elements within certain genomic regions, especially pNRC100 and pNRC200, could account for scrambling of the genome via extensive rearrangements, recombinations, and DNA exchanges mediated by IS elements. The possibility also exists that a fraction of the NRC-1 genome (e.g., the AT-rich satellite) was introduced by a large lateral transfer event, although evidence for such a mechanism is lacking.
An interesting possibility is that the IS element-rich regions of the genome might function as a source for the generation of genetic diversity, allowing the organism to better respond to a wide range of environmental variables such as salinity, light, oxygen, and nutrients. For example, the single ISH1 element in the genome preferentially inactivates the bop gene, and may serve to maintain a purple membrane-deficient fraction of cells in the population (DasSarma 1989). Similarly, the generation of partially and completely gas-vesicle-deficient strains of NRC-1 by DNA rearrangements associated with the gvp gene cluster of pNRC100 and pNRC200 may provide a mechanism for stratification of the population (DasSarma 1993). In addition to specific mutational mechanisms, IS elements could also play a role in mediating wholesale DNA rearrangements of genetic material in NRC-1, including exchanges of genes on the three replicons. The correspondence of IS elements and genes with unusual composition, as shown by peaks in the χ-squared plot (Fig. 6), is further evidence of genetic transactions and rearrangements involving these elements. A comparative genomic analysis of halophiles generally, and Halobacterium species specifically, would provide a better understanding of the role of IS elements and AT-rich satellite DNA in biology and evolution.
The factors affecting a particular organism's tendency to have higher or lower GC content have been the object of much study (Deschavanne and Filipski 1995). In some instances mutational tendencies driving base composition amelioration toward a certain GC percentage involving transitions and transversion have been correlated with the presence or absence of particular repair pathways, and transcriptional efficiency (Deschavanne and Filipski 1995; Xia 1996). To this end, the genome of Halobacterium NRC-1 represents an interesting case in which two distinct GC fractions are present and seemingly maintained in the genome. In our investigation into codon usage and GC composition of the chromosome, pNRC100, and pNRC200, we have found that although there are distinct regions corresponding to relatively GC-rich or AT-rich DNA, there is a gradient of base composition that is distributed among all three replicons. It is difficult to envision circumstances that would result in this observation, aside from some advantageous function for the organism. Roles for the two fractions are presumably separated so that the major GC-rich fraction of the genome, more stable and representing the majority of DNA on the large chromosome, serves to encode for NRC-1's housekeeping and critical function genes, whereas the AT-rich and IS-element-enriched fraction perhaps functions as a dynamo for the generation of metabolic and regulatory diversity through the uptake, shuffling, and possible integration of duplicated genes and foreign DNA. The novel structure of the NRC-1 genome and its high rate of rearrangements make this halophile an excellent model system for studies of plasmid/chromosome dynamics and prokaryotic evolution.
GC skew has been used for numerous bacterial genomes to locate the replication origin (Lobry 1996), although, for most archaeal organisms and extremophile bacteria (e.g., Thermotoga maritima), GC skew gives no clear indication of any origin of replication. The exceptions thus far are M. thermoautotrophicum and the three sequenced Pyrococcus species, which appear to have a single origin based on a combination of GC skew, skewed distribution of short oligomers, and experimental results (Salzberg et al. 1998; Myllykallio et al. 2000). Based on GC-skew analysis, Halobacterium NRC-1 also appears to have two origins in the large chromosome. Interestingly, we also found multiple Cdc6/Orc1 (9 or 10 in total) protein-coding homologs, two of which are located at GC-skew inflection points, consistent with these regions serving as origins of replication. This situation may have arisen as a result of a recent replicon fusion, although the similarity to the Halobacterium GRB genome is not consistent with this idea. Similarly, the possibility of a recent inversion causing a GC-skew artifact appears to be unlikely. If the function of two distinct origins can be demonstrated for replication of the NRC-1 large chromosome, it would establish an interesting model for studies of replication among archaeal and eukaryotic systems.
The diverse topics covered in this paper highlight, at once, the amount that has been learned from the sequence of Halobacterium NRC-1 and the number of questions still remaining to be answered. Recent success in solving the structure of the large ribosomal subunit from a related halophile, Haloarcula marismortui (Ban et al. 2000), genomic and genetic dissection of the archaeal regulon for purple membrane synthesis in Halobacterium NRC-1 (Baliga et al. 2001), and reverse genetic analysis of carotenoid and retinal biosynthesis (Peck et al. 2001) further solidify halophiles as viable and indispensable model systems in both prokaryotic and eukaryotic fields. Undoubtedly, on-going work using genetic arrays, gene knockouts, and further computational analysis will yield insights into life in extreme environments generally and adaptation to hypersaline environments specifically. The advantages in halobacterial systems such as facile growth and developed genetic tools, augmented by the complete genome sequence, will only accelerate advances in an already exciting field.
METHODS
Isoelectric Point Prediction
Each of the 2630 predicted proteins of NRC-1 was submitted, using a Perl script, to the GCG Wisconsin Package program PEPTIDESORT. A second Perl script was then used to extract the isoelectric data from the output file and organize it into a file that could be opened in Excel. An Excel spreadsheet was used for calculating the average pI and for plotting the data. For comparisons against the 13 other genomes, FASTA format proteins sequences were retrieved from the NCBI web site and converted into GCG format files. Excel was again used to tabulate the 14 sets of data into one spreadsheet. Six were selected for graphing.
Homology Modeling
The program Swiss PDB viewer (version 3.7 beta 2) (http://expasy.ch/spdbv/mainpage.html) was used to load NRC-1 protein sequences and locate appropriate crystal structure on which to base a model. Alignment of primary sequence and structural information resulted in predictions of protein folding and surface charge. Coulomb charge calculations were then mapped onto the predicted surface for a visual representation of surface charge. Calculations of surface accessibility were made to determine which residues were located at the surface of the proteins.
Codon Bias
The GCG program CODONFREQUENCY was run recursively to determine the number and percentages of all codon triplets of predicted NRC-1 proteins. These data were collected and sorted using Perl scripts and Excel. Averages for each triplet were calculated for each of the three replicons. Comparisons against expected values to determine over- or underrepresentation were corrected based on the 65.9% GC content of NRC-1. Predictions of amino acid usage were performed by summing the codon usage data for each amino acid.
Lateral Gene Transfer
BLASTable databases were constructed using the total set of predicted proteins from representative organisms. Deinococcus radiodurans and Bacillus subtilis were used to represent the bacterial domain, given their relatively high degree of homology (PAM values) with NRC-1 (Ng et al. 2000). Saccharomyces cerevisiae was used to represent the eukaryotic branch, and a composite database of Archaeoglobus fulgidus, Aeropyrum pernix, Pyrococcus abyssi, and Methanococcus jannaschii was used for the archaea. Recursive searches using all NRC-1 proteins as query sequences against the constructed databases resulted in >10,000 individual output files, which were then screened using Perl scripts, with the output being further sorted in Excel.
BLAST P values were used in a Boolean manner with values >e−10 assigned a value of 1. A total of the Boolean interpretation was plotted in a sliding 40 ORF window. Peaks of significant bacterial character to the exclusion of archaeal homology were further analyzed by visual inspection of the genes present in those peaks. Strength of homologies, conservation of functional operons, and preserved genetic order were important aspects that were considered.
Phylogenetic Analysis
To construct phylogenetic trees for the proteins involved in electron transport, an initial search was done against the COG database using NRC-1 sequences as queries. Identified orthologs were then aligned using the GCG program PILEUP. Neighbor-joining tree phylograms were constructed, bootstrapped, and displayed using PAUPSEARCH and PAUPDISPLAY.
GC Analysis
The GC content of the 2-Mb chromosome of NRC-1 was analyzed by calculating the GC composition of each predicted ORF on the replicon and graphing. Using the Perl scripting language, a program was written to read each nucleotide sequence (FASTA format) and tabulate the nucleotide percentages for each ORF. A spreadsheet was created using Microsoft Excel software for data tabulation and graph construction. Specific sets of ORFs were determined to have statistically significant differences in mean GC percentage and standard deviation by T-test for two samples assuming unequal variances.
GC- and Oligomer-Skew Analysis
The three replicons of Halobacterium NRC-1 were examined for GC skew. The ratio of guanine to cytosine along with the skews of all 8-mers was calculated for NRC-1 using the method described previously (Salzberg et al. 1998). Eight GC-rich oligomers (CCGCCGCC, CCACCGCC, CGCCGCCC, GGCGGCGA, CGCCGCCA, CCGCCCGC, CGCCCGCC, and CCACCACC) that were found to have significant skews were used in calculating the origin of replication.
Acknowledgments
This work was supported by collaborative research grants from the National Science Foundation to S.D. (DEB-9812330) and L.H. (DEB-9900497).
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL dassarma@umbi.umd.edu; FAX (410) 234-8896.
Article and publication are at http://www.genome.org/cgi/doi/10.1101/gr.190201.
REFERENCES
- Baliga NS, Goo YA, Ng WV, Hood L, Daniels CJ, DasSarma S. Is gene expression in Halobacterium NRC-1 regulated by multiple TBP and TFB transcription factors? Mol Microbiol. 2000;36:1184–1185. doi: 10.1046/j.1365-2958.2000.01916.x. [DOI] [PubMed] [Google Scholar]
- Baliga NS, Kennedy SP, Ng WV, Hood L, DasSarma S. Genomic and genetic dissection of an archaeal regulon. Proc Natl Acad Sci. 2001;98:2521–2525. doi: 10.1073/pnas.051632498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ban N, Nissen P, Hansen J, Moore PB, Steitz TA. The complete atomic structure of the large ribosomal subunit at 2.4 Å resolution. Science. 2000;289:905–920. doi: 10.1126/science.289.5481.905. [DOI] [PubMed] [Google Scholar]
- Bell SP, Stillman B. ATP-dependent recognition of eukaryotic origins of DNA replication by a multiprotein complex. Nature. 1992;357:128–134. doi: 10.1038/357128a0. [DOI] [PubMed] [Google Scholar]
- Berger JM, Gamblin SJ, Harrison SC, Wang JC. Structure and mechanism of DNA topoisomerase II. Nature. 1996;379:225–232. doi: 10.1038/379225a0. ; erratum 380: 179. [DOI] [PubMed] [Google Scholar]
- Campbell AM. Lateral gene transfer in prokaryotes. Theor Popul Biol. 2000;57:71–77. doi: 10.1006/tpbi.2000.1454. [DOI] [PubMed] [Google Scholar]
- Charlebois RL, Doolittle WF. Transposable elements and genome structure in Halobacteria. In: Berg DE, Howe MM, editors. Mobil DNA. Washington, DC.: American Society of Microbiology Press; 1989. pp. 297–307. [Google Scholar]
- Ciulla R, Clougherty C, Belay N, Krishnan S, Zhou C, Byrd D, Roberts MF. Halotolerance of Methanobacterium thermoautotrophicum delta H and Marburg. J Bacteriol. 1994;176:3177–3187. doi: 10.1128/jb.176.11.3177-3187.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DasSarma S. Mechanisms of genetic variability in Halobacterium halobium: The purple membrane and gas vesicle mutations. Can J Microbiol. 1989;35:65–72. doi: 10.1139/m89-010. [DOI] [PubMed] [Google Scholar]
- ————— Identification and analysis of the gas vesicle gene cluster on an unstable plasmid of Halobacterium halobium. Experientia. 1993;49:482–486. doi: 10.1007/BF01955149. [DOI] [PubMed] [Google Scholar]
- DasSarma S, Arora P. Genetic analysis of the gas vesicle cluster in haloarchaea. FEMS Microbiology Letters. 1997;153:1–10. [Google Scholar]
- DasSarma S, Arora P, Lin F, Molinari E, Yin LR. Wild-type gas vesicle formation requires at least ten genes in the gvp gene cluster of Halobacterium halobium plasmid pNRC100. J Bacteriol. 1995;176:7646–7652. doi: 10.1128/jb.176.24.7646-7652.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deschavanne P, Filipski J. Correlation of GC content with replication timing and repair mechanisms in weakly expressed E. coli genes. Nucleic Acids Res. 1995;23:1350–1353. doi: 10.1093/nar/23.8.1350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doolittle WF. Phylogenetic classification and the universal tree. Science. 1999;284:2124–2129. doi: 10.1126/science.284.5423.2124. [DOI] [PubMed] [Google Scholar]
- Dym O, Mevarech JL, Sussman L. Structural features that stabilize halophilic malate dehydrogenase from an archaebacterium. Science. 1995;267:1334–1346. doi: 10.1126/science.267.5202.1344. [DOI] [PubMed] [Google Scholar]
- Eisenberg H. Life in unusual environments: Progress in understanding the structure and function of enzymes from extreme halophilic bacteria. Arch Biochem Biophys. 1995;318:1–5. doi: 10.1006/abbi.1995.1196. [DOI] [PubMed] [Google Scholar]
- Elcock AH, McCammon JA. Electrostatic contributions to the stability of halophilic proteins. J Mol Biol. 1998;280:731–748. doi: 10.1006/jmbi.1998.1904. [DOI] [PubMed] [Google Scholar]
- Fitz-Gibbon ST, House CH. Whole genome-based phylogenetic analysis of free-living microorganisms. Nucleic Acids Res. 1999;27:4218–4222. doi: 10.1093/nar/27.21.4218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freeland SJ, Knight RD, Landweber LF, Hurst LD. Early fixation of an optimal genetic code. Mol Biol Evol. 2000;17:511–518. doi: 10.1093/oxfordjournals.molbev.a026331. [DOI] [PubMed] [Google Scholar]
- Hackett NR, Bobovnikova Y, Heyrovska N. Conservation of chromosomal arrangement among three strains of the genetically unstable archaeon Halobacterium salinarium. J Bacteriol. 1994;176:7711–7718. doi: 10.1128/jb.176.24.7711-7718.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hescox MA, Carlberg DM. Photoreactivation in Halobacterium cutirubrum. Can J Microbiol. 1972;18:981–985. doi: 10.1139/m72-152. [DOI] [PubMed] [Google Scholar]
- Janel-Bintz R, Maenhaut-Michel G, Fuchs RP. MucAB but not UmuDC proteins enhance −2 frameshift mutagenesis induced by N-2-acetylaminofluorene at alternating GC sequences. Mol Gen Genet. 1994;245:279–285. doi: 10.1007/BF00290107. [DOI] [PubMed] [Google Scholar]
- Kawanishi S, Oikawa S, Murata M, Tsukitome H, Saito I. Site-specific oxidation at GG and GGG sequences in double-stranded DNA by benzoyl peroxide as a tumor promoter. Biochemistry. 1999;38:16733–16739. doi: 10.1021/bi990890z. [DOI] [PubMed] [Google Scholar]
- Kosa PF, Ghosh G, DeDecker BS, Sigler PB. The 2.1-Å crystal structure of an archaeal preinitiation complex: TATA-box-binding protein/transcription factor (II)B core/TATA-box. Proc Natl Acad Sci. 1997;94:6042–6047. doi: 10.1073/pnas.94.12.6042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanyi JK. Salt-dependent properties of proteins from extremely halophilic bacteria. Bacteriol Rev. 1974;38:272–290. doi: 10.1128/br.38.3.272-290.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrence JG, Roth JR. Selfish operons: Horizontal transfer may drive the evolution of gene clusters. Genetics. 1996;143:1843–1860. doi: 10.1093/genetics/143.4.1843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Littlefield O, Korkhin Y, Sigler PB. The structural basis for the oriented assembly of a TBP/TFB/promoter complex. Proc Natl Acad Sci. 1999;96:13668–13673. doi: 10.1073/pnas.96.24.13668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lobry JR. Asymmetric substitution patterns in the two DNA strands of bacteria. Mol Biol Evol. 1996;13:660–665. doi: 10.1093/oxfordjournals.molbev.a025626. [DOI] [PubMed] [Google Scholar]
- Lobry JR. Influence of genomic G + C content on average amino-acid composition of proteins from 59 bacterial species. Gene. 1997;205:309–316. doi: 10.1016/s0378-1119(97)00403-4. [DOI] [PubMed] [Google Scholar]
- Majumdar S, Gupta SK, Sundararajan VS, Ghosh TC. Compositional correlation studies among the three different codon positions in 12 bacterial genomes. Biochem Biophys Res Commun. 1999;266:66–71. doi: 10.1006/bbrc.1999.1774. [DOI] [PubMed] [Google Scholar]
- McLean MJ, Wolfe KH, Devine KM. Base composition skews, replication orientation, and gene orientation in 12 prokaryote genomes. J Mol Evol. 1998;47:691–696. doi: 10.1007/pl00006428. [DOI] [PubMed] [Google Scholar]
- Moore RL, McCarthy BJ. Base sequence homology and renaturation studies of the deoxyribonucleic acid of extremely halophilic bacteria. J Bacteriol. 1969;99:255–262. doi: 10.1128/jb.99.1.255-262.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myllykallio H, Lopez P, Lopez-Garcia P, Heilig R, Saurin W, Zivanovic Y, Philippe H, Forterre P. Bacterial mode of replication with eukaryotic-like machinery in a hyperthermophilic archaeon. Science. 2000;288:2212–2215. doi: 10.1126/science.288.5474.2212. [DOI] [PubMed] [Google Scholar]
- Nelson KE, Clayton RA, Gill SR, Gwinn ML, Dodson RJ, Haft DH, Hickey EK, Peterson JD, Nelson WC, Ketchum KA, et al. Evidence for lateral gene transfer between archaea and bacteria from genome sequence of Thermotoga maritima. Nature. 1999;399:323–329. doi: 10.1038/20601. [DOI] [PubMed] [Google Scholar]
- Nelson KE, Paulsen IT, Heidelberg JF, Fraser CM. Status of genome projects for nonpathogenic bacteria and archaea. Nat Biotechnol. 2000;18:1049–1054. doi: 10.1038/80235. [DOI] [PubMed] [Google Scholar]
- Ng WL, Kothakota S, DasSarma S. Structure of the gas vesicle plasmid in Halobacterium halobium inversion isomers, inverted repeats, and insertion sequences. J Bacteriol. 1991;173:3933. doi: 10.1128/jb.173.12.3933b.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng WV, Ciufo SA, Smith TM, Bumgarner RE, Baskin D, Faust J, Hall B, Loretz C, Seto J, Slagel J, et al. Snapshot of a large dynamic replicon in a halophilic archaeon: Megaplasmid or minichromosome? Genome Res. 1998;8:1131–1141. doi: 10.1101/gr.8.11.1131. [DOI] [PubMed] [Google Scholar]
- Ng WV, Kennedy SP, Mahairas GG, Berquist B, Pan M, Shukla HD, Lasky SR, Baliga NS, Thorsson V, Sbrogna J, et al. Genome sequence of Halobacterium species NRC-1. Proc Natl Acad Sci. 2000;97:12176–12181. doi: 10.1073/pnas.190337797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peck RF, DasSarma S, Krebs MP. Homologous gene knockout in the archaeon Halobacterium salinarum with ura3 as a counterselectable marker. Mol Microbiol. 2000;35:667–676. doi: 10.1046/j.1365-2958.2000.01739.x. [DOI] [PubMed] [Google Scholar]
- Peck RF, Echavarri-Erasun C, Johnson EA, Ng WV, Kennedy SP, Hood L, DasSarma S, Krebs MP. brp and blh are required for synthesis of the retinal cofactor of bacteriorhodopsin in Halobacterium salinarum. J Biol Chem. 2001;276:5739–5744. doi: 10.1074/jbc.M009492200. [DOI] [PubMed] [Google Scholar]
- Pfeifer F, Betlach M. Genome organization in Halobacterium halobium: A 70 kb island of more (AT) rich DNA in the chromosome. Mol Gen Genet. 1985;198:449–455. doi: 10.1007/BF00332938. [DOI] [PubMed] [Google Scholar]
- Reece RJ, Maxwell A. DNA gyrase: Structure and function. Crit Rev Biochem Mol Biol. 1991;26:335–375. doi: 10.3109/10409239109114072. [DOI] [PubMed] [Google Scholar]
- Salzberg SL, Salzberg AJ, Kerlavage AR, Tomb JF. Skewed oligomers and origins of replication. Gene. 1998;217:57–67. doi: 10.1016/s0378-1119(98)00374-6. [DOI] [PubMed] [Google Scholar]
- Salzberg SL, White O, Peterson J, Eisen JA. Microbial genes in the human genome: Lateral transfer or gene loss? Science. 2001;292:1903–1906. doi: 10.1126/science.1061036. [DOI] [PubMed] [Google Scholar]
- Sapienza C, Doolittle WF. Unusual physical organization of the Halobacterium genome. Nature. 1982;295:384–389. doi: 10.1038/295384a0. [DOI] [PubMed] [Google Scholar]
- Snel B, Bork P, Huynen MA. Genome phylogeny based on gene content. Nat Genet. 1999;21:108–110. doi: 10.1038/5052. [DOI] [PubMed] [Google Scholar]
- Spudich JL, Yang CS, Jung KH, Spudich EN. Retinylidene proteins: Structures and functions from archaea to humans. Annu Rev Cell Dev Biol. 2000;16:365–392. doi: 10.1146/annurev.cellbio.16.1.365. [DOI] [PubMed] [Google Scholar]
- Sumper M, Reitmeier H, Oesterhelt D. Biosynthesis of the purple membrane of halobacteria. Angew Chem Int Ed Engl. 1976;15:187–194. doi: 10.1002/anie.197601871. [DOI] [PubMed] [Google Scholar]
- Tatusov RL, Galperin MY, Natale DA, Koonin EV. The COG database: A tool for genome-scale analysis of protein functions and evolution. Nucleic Acids Res. 2000;28:33–36. doi: 10.1093/nar/28.1.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsujimoto K, Semadeni M, Huflejt M, Packer L. Intracellular pH of halobacteria can be determined by the fluorescent dye 2‘,7‘-bis(carboxyethyl)-5(6)-carboxyfluorescein. Biochem Biophys Res Commun. 1988;155:123–129. doi: 10.1016/s0006-291x(88)81058-1. [DOI] [PubMed] [Google Scholar]
- Xia X. Maximizing transcription efficiency causes codon usage bias. Genetics. 1996;144:1309–1320. doi: 10.1093/genetics/144.3.1309. [DOI] [PMC free article] [PubMed] [Google Scholar]