Abstract
Sirtuins have emerged in recent years as critical regulators of metabolism, influencing numerous facets of energy and nutrient homeostasis. Here, we review recent advances on the role of this fascinating family of mammalian proteins, and their well orchestrated function in modulating mitochondrial activity.
The discovery of the Sir2 protein as a central regulator of lifespan in yeast more than a decade ago catalyzed an explosion in longevity research and in particularly in studies of the relationship between metabolic control and aging (Kaeberlein et al., 1999; Imai et al., 2010). The subsequent identification of a family of Sir2-related proteins, collectively known as sirtuins, across many species has identified conserved mechanisms of organismal aging which continue to capture the attention of both the academic community and the general public. It is now apparent that sirtuins mediate wide-ranging biological functions, spanning from DNA repair to energy metabolism and oxidative stress responses. Moreover, in addition to aging, alterations in sirtuin function have central roles in neurodegenerative diseases, cancer, circadian rhythms, and much more. Although the closest mammalian homolog of Sir2, SIRT1, has been at center stage for many years, recent work has described distinct and fascinating roles for the other sirtuin family proteins. These studies do not upstage SIRT1, but rather demonstrate that the various members of this protein family act in a carefully orchestrated manner to coordinate metabolic homeostasis. In this Review, we will discuss recent discoveries in the biology of sirtuins with a particular focus on functions in mitochondrial physiology. Our discussion will include both sirtuin family members that directly localize to the mitochondria, as well as to others present elsewhere in the cell yet directly impacting mitochondrial activity by means of influencing the production of metabolic intermediates. Mammalian sirtuins represent essential players in bridging this organelle with the rest of the cell—indeed revealing that “no mitochondrion is an island, entire of itself”.
Sirtuins and mitochondria function
There are seven sirtuins in mammals (SIRT1-7). These proteins share an evolutionarily conserved catalytic core domain but show little similarity in other regions. Sirtuins exhibit distinct subcellular localizations: SIRT1, SIRT6 and SIRT7 are mainly nuclear, SIRT2 is primarily found in the cytosol, and SIRT3, SIRT4 and SIRT5 are mitochondrial proteins. There is some evidence that the mitochondrial targeting of some family members may be dynamic as SIRT1 has been reported to be present in the mitochondria (Aquilano et al., 2010) and SIRT3 can be found in other cellular compartments (Verdin et al., 2010). However, the physiological relevance of this “ectopic” localization remains to be determined.
Sirtuins exert their biological functions primarily through their capacity to modify proteins via catalyzing either deacetylation or ADP-ribosylation. NAD+ is required as a cofactor for this catalytic activity. As NAD+ — and its reduced form NADH — serve as essential electron carriers in a wide range of metabolic processes, sirtuin activity is directly linked to cellular metabolic status and nutrient availability. In turn, sirtuins themselves are important modulators of multiple metabolic processes, including energy production, the urea cycle, fatty acid metabolism, and acetate metabolism.
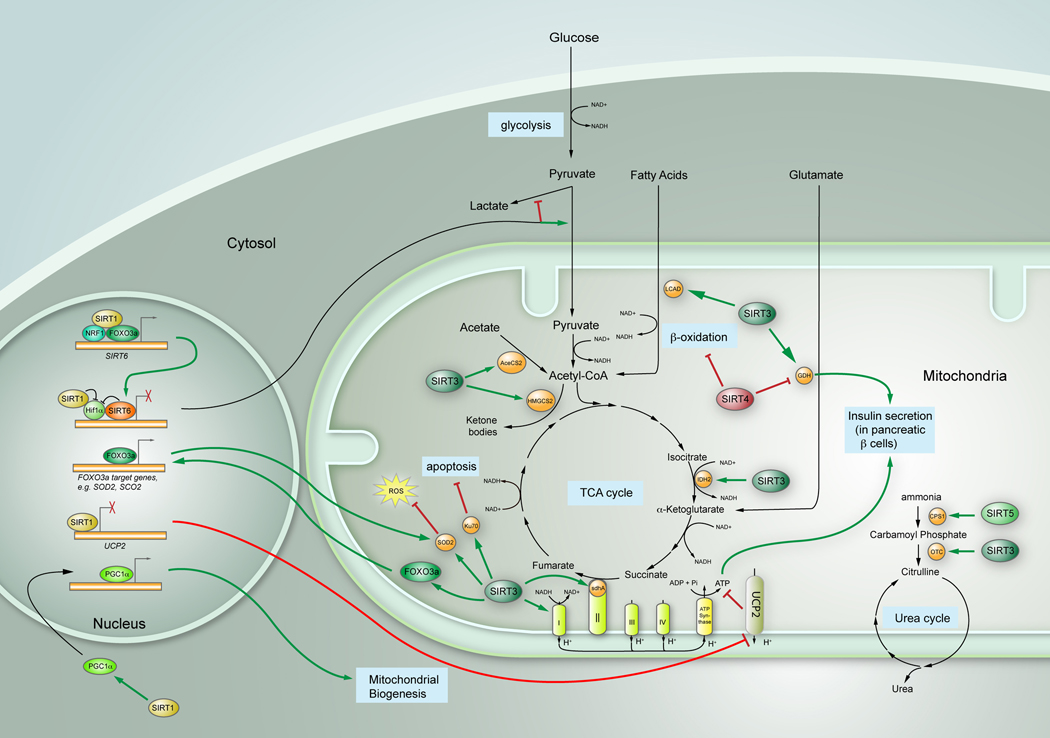
The metabolism of glucose, fatty acid, and amino acids is linked since key metabolic intermediates are shared between the individual pathways. The mitochondria provide the hub that integrates these pathways, serving as a critical site for the production and exchange of metabolic intermediates (Figure 1). Hence, the mitochondria play a critical role in orchestrating complex metabolic networks in order to maintain proper homeostasis. Notably, mitochondrial proteins are subject to posttranslational modifications in response to changes in physiological conditions. For instance, proteomic studies have shown that a large fraction of mitochondrial proteins are acetylated (reviewed in Verdin et al., 2010). Strikingly, acetyl-lysines are found in almost all the enzymes involved in the TCA cycle, the urea cycle and fatty acid metabolism, — key mitochondrial processes — underscoring the significance of this modification in mitochondrial function (reviewed in Verdin et al., 2010). As NAD+-dependent deacetylases, sirtuins are well positioned to control the dynamics of mitochondrial protein acetylation, thus potentially influencing every aspect of mitochondrial function. Indeed, it has been shown that mitochondrial biogenesis itself is regulated by both SIRT1 and SIRT3, through a process involving the transcriptional co-activator PGC1α (Lomb et al., 2010), as it will be discussed below. Furthermore, not only biogenesis but also mitochondrial degradation seems to be regulated by sirtuins, since autophagy of damaged mitochondria in aged kidney is also SIRT1-dependent (Kume et al., 2010 and reviewed in Finkel et al., 2009). Below, we discuss the complex multi-leveled contribution of each member of the sirtuin family to mitochondrial physiology.
Figure 1.
Sirtuins’ regulation of mitochondrial biology. Overview diagram indicating the main metabolic pathways in the mitochondria, and the different roles sirtuins play in those pathways. Green arrows indicate activating effects; red bars denote inhibiting functions.
Energy Production Pathways
The insulin signaling pathway and mitochondrial activity
Insulin—secreted by pancreatic β cells upon nutrient stimulation—is one of the most important regulators of nutrient utilization and metabolic homeostasis. This hormone activates the IRS-PI3K-AKT-mTOR/FOXO1 signaling pathway, promoting protein synthesis, glycolysis, glucose storage, as well as lipid synthesis and storage, while inhibiting gluconeogenesis and ketogenesis (Cheng et al., 2010). Insulin signaling modulates mitochondrial function by altering these pathways, and is in turn regulated by the activity of this organelle (Figure 1). Notably, insulin resistance—a hallmark of metabolic syndrome and Type II diabetes—is accompanied by reduction of mitochondrial OXPHOS activity (Cheng et al., 2010). On the other hand, Reactive Oxygen Species (ROS) produced during mitochondrial OXPHOS promote insulin signaling through oxidation of insulin receptor and inhibition of phosphatases, such as PTP1B and PTEN (Cheng et al., 2010).
Several sirtuins exert their control over metabolic homeostasis by tightly linking insulin secretion to mitochondria activity. In the pancreas, SIRT1 acts as a positive regulator of insulin secretion, through directly binding to and repressing the uncoupling protein 2 (UCP2) gene, which encodes a mitochondrial proton transporter (reviewed in Finkel et al., 2009). UCP2 resides on the inner membrane of mitochondria and promotes proton leakage across the mitochondria membrane, thus uncoupling oxidative respiration from ATP generation. The reduced ATP/ADP ratio in turn blunts insulin secretion. Notably, conditions of short-term fasting reduced the levels of NAD+ in the pancreas, in turn decreasing SIRT1 activity and leading to up-regulation of UCP2 with a concomitant dampening in insulin secretion (Imai et al., 2010). UCP2 plays a critical role in favoring fatty acid metabolism over glucose metabolism, and therefore it will be interesting to determine whether SIRT1, through its role in controlling UCP2, also modulate lipid metabolism beyond its role in insulin secretion. Notably, a recent study reported that SIRT1 mediates resveratrol-induced increase of insulin secretion through a mechanism independent of UCP2 transcriptional repression (Vetterli et al., 2010). In this context, future studies will be required to determine the relevance of the UCP2 pathway in SIRT1-regulated insulin secretion.
In contrast to SIRT1, SIRT4 appears to repress insulin secretion, by modulating the activity of Glutamate Dehydrogenase (GDH), a mitochondrial enzyme that converts glutamate into the TCA-cycle intermediate α-ketoglutarate (Verdin et al., 2010). GDH is known to promote amino acid-stimulated insulin secretion. Under normal diet conditions, GDH is ADP-ribosylated and repressed by SIRT4, allowing cells to use glucose as the primary energy source. During Calorie Restriction (CR) or low glucose conditions, however, the repressive control on GDH is relaxed in order to increase the amount of α-ketoglutarate, which fuels the TCA cycle for ATP production to meet the energy demands of cells (Verdin et al., 2010). Since SIRT4 protein levels do not seem to change during CR, it will be interesting to investigate how SIRT4 activity is regulated under different nutrient conditions. Interestingly, GDH can be deacetylated by another mitochondrial sirtuin, SIRT3 (Verdin et al., 2010). At least in vitro, acetylation of GDH reduces its activity while SIRT3 mediated deacetylation of GDH activates it (Verdin et al., 2010). Since no significant changes of GDH activity can be detected in SIRT3 deficient animals, the physiological significance of the SIRT3-GDH regulatory pathway remains to be determined (Nakagawa et al., 2009).
It is striking to note that SIRT1, SIRT4, and possibly SIRT3, act in an opposite albeit coordinated manner, to regulate insulin secretion in response to a range of dietary conditions, including ad libitum feeding, short–term fasting and chronic calorie restriction. Although our mechanistic understanding of this coordination is incomplete, it is clear that the crosstalk between sirtuins relates to distinct and critical functions in maintaining metabolic homeostasis, a reoccurring theme that will be mentioned throughout this Review.
Glycolysis
Glycolysis is the first step in a metabolic pathway to obtain high-energy compounds (e.g. ATP) from glucose. Glucose, the primary carbon source of energy in most cells, is imported by glucose transporters (GLUTs) and metabolized through a series of steps to generate pyruvate (Figure 1). Pyruvate can be further oxidized into Acetyl-CoA, to enter the Tricarboxylic Acid (TCA) cycle in the mitochondria. Under conditions of nutrient excess, most of the glucose is converted into pyruvate to enter mitochondrial respiration, whereas, under low glucose or low oxygen conditions, pyruvate is diverted to produce lactate through anaerobic glycolysis. A key regulator of this glycolytic switch is Hypoxia Induced Factor 1α (Hif1α), a transcription factor that is activated during hypoglycemia or hypoxia, in turn inducing expression of multiple genes that promote anaerobic glycolysis while inhibiting the TCA cycle (Figure 1). Although anaerobic glycolysis is far less energy-efficient (2 mol ATP/mol of glucose, against 36 mol ATP/mol glucose in the TCA cycle), it provides critical intermediate metabolites necessary to maintain macromolecular biosynthesis. Such a switch provides an important advantage to rapidly dividing cells, as is the case in tumor cells (i.e. the well recognized “Warburg effect”) (Warburg, 1956).
SIRT3 has recently been found to indirectly inhibit Hif1α activity through ROS reduction, thereby repressing glycolysis (Finley et al., 2011). A more direct connection between Sirtuins and Hif1α comes from SIRT1 and SIRT6: SIRT1 represses glycolysis by inhibiting Hif1α activity through direct interaction and deacetylation (Lim et al., 2010). Interestingly, SIRT6 also suppresses glycolysis under normal conditions through co-repressing Hif1α target genes, via functioning as a histone H3K9 deacetylase at the promoters of these glycolytic genes (Zhong et al., 2010)(Figure 1). As Hif1α is largely, although not completely inactivated by proteasomal degradation under high glucose or high oxygen conditions, this extra layer of regulation imposed by SIRT1 and SIRT6 may maximize ATP production, inhibiting unnecessary lactate production, while at the same time allowing rapid activation of this glycolytic switch under conditions of nutrient stress.
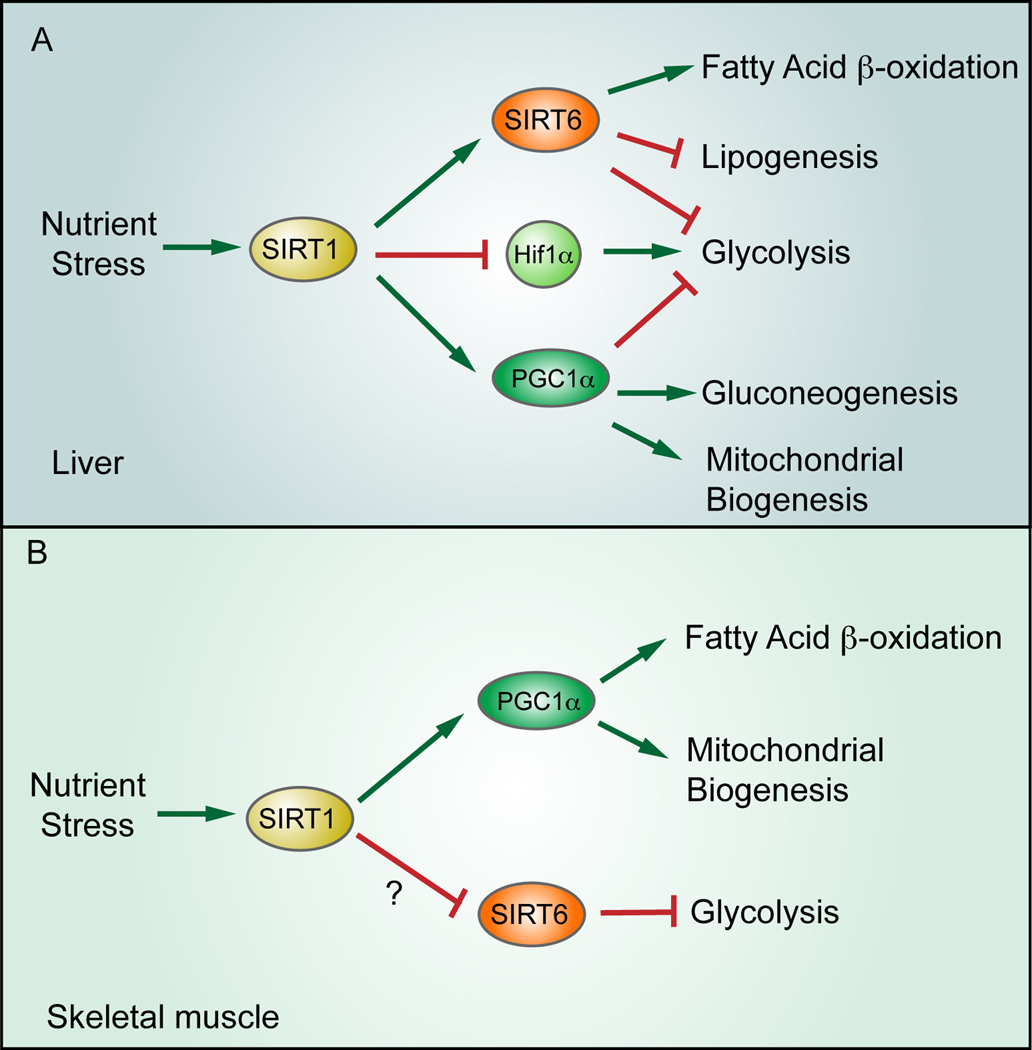
Recent studies further illustrate the potential crosstalk between SIRT1 and SIRT6. Kim and colleagues found that SIRT1 enhances SIRT6 expression by interacting with FOXO3a and NRF1 at the SIRT6 promoter (Kim et al., 2010)(Fig. 1). SIRT1 has previously been shown to increase under conditions of nutrient scarcity, causing in liver increased mitochondrial biogenesis, increased gluconeogenesis and reduced glycolysis; in muscle, SIRT1 activation also leads to increased mitochondrial biogenesis, in turn inducing in this tissue fatty acid oxidation. These effects have been attributed to increased SIRT1 activity following AMP-activated protein kinase (AMPK)-mediated increase in cellular NAD+ levels. In turn, SIRT1 mediates the activation of the PGC1-α transcriptional co-activator, a known modulator of this metabolic adaptation to nutrient stress (reviewed in Verdin et al., 2010)(Fig. 2). Kim et al. propose another layer of regulation in studies that employed liver-specific SIRT6 deficient mice. They show that SIRT1 also activates SIRT6 in the liver, which further supports the switch from glycolysis toward fatty acid oxidation. Of note, initial studies found that germline SIRT6 KO mice die from a lethal hypoglycemia due to increased glycolysis and concomitant uncontrolled glucose uptake in skeletal muscle and fat (Fig. 2; Zhong et al., 2010). As opposed to liver, activation of SIRT1 in muscle during nutrient stress is expected to decrease SIRT6 activity, favoring a switch towards glycolysis, an hypothesis that remain to be tested.
Figure 2.
Differential effect for sirtuins in liver and muscle metabolism. Under nutrient stress, SIRT1 is activated in liver, in turn modulating the activity of multiple downstream pathways to promote gluconeogenesis and fatty acid oxidation, while inhibiting lipogenesis and glycolysis. In contrast, nutrient stress in muscle appears to enhance glycolysis, possibly through SIRT1-mediated downregulation of SIRT6.
TCA cycle
The TCA cycle is at the core of a network of cellular metabolic processes. It links glycolysis and pyruvate oxidation with oxidative phosphorylation (Figure 1). In addition, amino acids and fatty acids can be converted to acetyl-CoA and enter the TCA cycle. The TCA cycle itself also provides various metabolic intermediates for amino acid synthesis. SIRT3 may potentially affect the TCA cycle by deacetylating isocitrate dehydrogenase 2 (Idh2)(Someya et al., 2010) and succinate dehydrogenase complex subunit A (SdhA) (Cimen et al., 2010), thereby increasing their enzymatic activity. Idh2 is responsible for catalyzing the conversion of isocitrate to α-ketoglutarate and CO2, in this way reducing NAD+ to NADH, while SdhA converts succinate to fumarate. Whereas SIRT3-mediated deacetylation of Idh2 has been linked to protection against age-associated hearing loss (see below), it remains as yet unclear whether SIRT3 deacetylation of these TCA components affects the TCA cycle per se. In this context, it will be interesting to determine whether point-mutants mimicking constitutively acetylated proteins exhibit TCA cycle defects, an experiment which will validate the role of SIRT3 in modulating the Krebs cycle.
Electron Transport Chain, Oxidative Stress and Mitochondrial Dependent Apoptosis
Sirtuins play important roles in regulating the electron transport chain (ETC). First, SIRT3 augments the activity of Complex I through direct interaction and deacetylation of several of its subunits. In line with this observation, ATP levels are reduced over 50% in the heart, liver and kidney in SIRT3 deficient mice (Finkel et al., 2009), highlighting the importance of SIRT3 in maintaining basal levels of ATP. Second, the above-mentioned SIRT3 substrate and TCA cycle enzyme, SdhA, is also a component of Complex II. The activation of SdhA caused by SIRT3 deacetylation stimulates Complex II and help cells cope with the increased levels of NAD+ that follows nutrient deprivation (Cimen et al., 2010). Third, ATP synthase α and cytochrome C are deacetylated by SIRT3 and SIRT5, respectively, although the physiological significance of these interactions remains to be explored (reviewed in Verdin et al., 2010).
A common theme emerges from these studies: lack of nutrients increases the levels of NAD+, activating sirtuins (mainly SIRT1 and SIRT3 at present), which in turn enhance ATP production in order to meet energy demands. At the same time, sirtuins keep this machinery from overdrive, protecting cells from stress-induced apoptosis, as discussed below.
ETC is a major site for the production of ROS, which can serve as signaling molecules vital for normal cellular functions. However, abnormal ROS production can lead to serious consequences, such as oxidation of protein, fatty acids and lipids, as well as severe DNA damage, which can impair cell viability. Cells protect themselves against ROS by expressing a group of antioxidant enzymes, such as SOD2/MnSOD, which convert superoxide to hydrogen peroxide.
Sirtuins appear to be at the center of ROS metabolic conversion, as well influencing the direct apoptotic response to ROS. Notably, SIRT3-deficient cells experience increased ROS levels and genomic instability when exposed to stress, and these factors lead to tumorigenesis in vivo (Verdin et al., 2010). As mentioned above, Finley et al. extended this notion one step further by demonstrating that increased ROS level stabilizes Hif1α, contributing to a metabolic reprogramming in cancer cells (Finley et al., 2011). How does SIRT3 reduce ROS? Two studies have independently demonstrated that SIRT3 directly deacetylates and activates SOD2/MnSOD in response to stress (Tao et al., 2010; Qiu et al., 2010). Intriguingly, the lysine residues deacetylated by SIRT3 identified by the two groups are different (K122 in one and K53/K89 in the other), raising the question of how and whether these two events are coordinated. In addition to direct deacetylation of SOD2/MnSOD, SIRT3 also interacts with the FOXO3a transcription factor in the mitochondria and increases its binding to the promoters of mitochondrial genes. FOXO3a in turn increases expression of a cohort of genes that encode superoxide scavengers, such as SOD2/MnSOD, SCO2, and Catalase (Verdin et al., 2010). Interestingly, nuclear localization of SIRT1 in failing hearts has been shown to induce MnSOD expression as a protective measure to promote cardiomyocyte survival (Tanno et al., 2010), although the molecular mechanism behind this induction has not been fully explored.
In addition to their roles in ROS metabolism, sirtuins appear to exert a protective function against mitochondria-dependent apoptosis. SIRT3 and SIRT4 are required for maintaining mitochondrial NAD+ pool mediated by NAD+ biosynthesis enzyme NAMPT to protect cells from apoptosis induced by genotoxic reagents (reviewed in Finkel et al., 2009). although the mechanisms underline this protective role is unclear. The authors proposed that mitochondrial permeability transition pore (mPTP) might be involved, which indeed turned out to be the case. Cyclophilin-D, a component of mPTP, is found to be deactylated by SIRT3. Consequently, mPTP opening is inhibited, and apoptosis prevented (Shulga et al., 2010; Hafner et al., 2010). Several other targets of SIRT3 have been identified to promote SIRT3-dependent protection under stress: genotoxic and oxidative stress up-regulates SIRT3 protein levels, protecting mouse cardiomyocytes from stress-induced apoptosis through deacetylation of Ku70 (Sundaresan et al., 2008). As mentioned above, SIRT3 mediated the protective effects of calorie restriction on age-associated hearing loss by deacetylating and activating Idh2, in turn reducing ROS and protecting against apoptosis of spiral ganglion neurons and sensory hair cells (Someya et al., 2010). Overall, SIRT3 seems to work as multi-layer coordinators to ensure that cells are properly protected against oxidative damage, whereas the function of SIRT4 in this process, although essential, remains to be explored.
In this context, SIRT5 can deacetylate cytochrome C, a protein essential for apoptosis, suggesting a role for this sirtuin in stress-induced, mitochondrial-dependent apoptosis (Verdin et al., 2010). Despite this biochemical observation, it remains unclear whether SIRT5-dependent deacetylation of cytochrome C inhibits or induces apoptosis, and therefore it physiological relevance remains unknown.
Urea Cycle
During periods of fasting, amino acids in muscle are catabolized as a carbon source for gluconeogenesis, and the newly formed glucose is metabolized for energy. However, ammonia generated in this process is highly toxic and must be converted to urea for proper excretion, a process that occurs through the urea cycle. The rate limiting first step of the urea cycle converts ammonia to carbamoyl phosphate and is catalyzed by carbamoyl phosphate synthetase 1 (CPS1). Recently, SIRT5 was found to interact with and deacetylate CPS1, thus enhancing its enzymatic activity (Nakagawa et al., 2009) (Fig.1). SIRT5 deficient mice exhibited increased CPS1 acetylation and decreased CPS1 activity. Concomitantly, these animals suffer from hyper-ammonia during starvation. In addition, starvation activates SIRT5, suggesting that SIRT5 positively regulates the urea cycle through CPS1 deacetylation during fasting. Notably, an earlier report showed increased CPS1 acetylation levels during CR (Schwer et al., 2009). These seemingly contradictory results suggest that different levels of nutrient limitation might affect the requirement for amino acid catabolism and the urea cycle. The second step in the urea cycle, where carbamoyl phosphate is converted to citrulline by ornithine transcarbamoylase (OTC), is regulated by another mitochondrial sirtuin, SIRT3. SIRT3 directly deacetylates OTC and stimulates its enzymatic activity during CR (Hallows et al., 2011)(Fig. 1). Although unexplored, it is tempting to speculate that increased ammonia levels might coordinately stimulate activity of both SIRT3 and SIRT5, which in turn will act upon CPS1 and OTC, respectively, to robustly activate the urea cycle. Future studies would be required to test this possibility.
Lipid and acetate metabolism
Lipid homeostasis depends on a well-orchestrated balance between fatty acid synthesis, lipid oxidation and lipid storage. This important metabolic pathway is also tightly intertwined with carbohydrate and protein metabolism. Therefore, control of the latter two pathways by sirtuins discussed above also likely causes indirect effects on lipid metabolism. In addition, several studies have defined direct roles for sirtuins in lipid regulation as well (Lomb et al., 2010). For example, SIRT1 and SIRT2 negatively regulate adipocyte differentiation and triglyceride storage through PPARγ repression and FOXO1 deacetylation, respectively (Lomb et al., 2010). Liver-specific deletion of SIRT1 in mice impairs the fatty acid β-oxidation and leads to hepatic steatosis and inflammation under high fat diet, possibly through direct interaction between SIRT1, PPARα and PGC-1α (reviewed in Lomb et al., 2010). Interestingly, another study using the same animals, when challenged with western style diet at an older age, produced contradictory results (reviewed in Lomb et al., 2010). These results suggest that there might be different regulatory mechanisms for SIRT1 on liver physiology at different stages in life. Recently, the significance of mitochondrial sirtuins on lipid metabolism has been put forward by several reports. SIRT3 deacetylates long-chain acyl co-A dehydrogenase (LCAD) and promotes mitochondrial fatty acid β-oxidation (FAO) upon fasting (reviewed in Verdin et al., 2010). Mitochondrial 3-hydroxy-3-methylglutaryl CoA synthase 2 (HMGCS2) is an additional novel substrate of SIRT3 that contributes to lipid utilization (Shimazu et al., 2010). Upon fasting, HMGCS2 is deacetylated and activated by SIRT3 in order to generate ketone bodies from acetyl-CoA, which is produced by FAO in the liver. Ketone bodies can then be used by extrahepatic tissues — such as brain — as an alternative energy source. In addition, a recent proteomics study revealed several other potential SIRT3 substrates in FAO (Smith et al., 2010). On the other hand, SIRT4 seems to have a negative role on FAO, since knocking down SIRT4 results in increased FAO gene expression. Curiously, this effect is SIRT1 dependent, and SIRT4 knockdown is accompanied by increased expression of SIRT1 and SIRT3 (Nasrin et al., 2010), revealing another intriguing example of crosstalk among sirtuins (Figure 1).
Acetate, through its reversible conversion to Acetyl-CoA, provides an alternative carbon source for various metabolic pathways, such as amino acid synthesis, fatty acid metabolism and the TCA cycle (Figure 1). AceCS2 catalyzes this reaction and plays important roles in thermogenesis during fasting conditions. Notably, SIRT3 can deacetylate AceCS2 and promote its activity (reviewed in Finkel et al., 2009). Acetyl-CoA generated in this process can be used in the TCA cycle to promote aerobic respiration and subsequent oxidative phosphorylation, or diverted for fatty acid synthesis (FAS). Therefore, through activation of AceCS2, SIRT3 also indirectly modulates both the TCA cycle and FAS.
Closing Remarks and Perspective
Over the last decade, studies on the biology of sirtuins have led to a remarkable growth in our knowledge of metabolic processes involving mitochondrial function. A number of sirtuins have emerged as “master regulators” of mitochondrial metabolism, acting as sensors of NAD/NADH levels, in turn to convert information about nutrient status into changes in cell physiology, by way of deacetylating or ADP-ribosylating key enzymes in multiple pathways (Figure 1). In the next few years we will certainly witness the expansion of this list of sirtuin substrates and the deciphering of the crosstalk among the different sirtuins. In addition to providing detailed understanding of mitochondrial function, this work is likely to open up new opportunities for therapeutic intervention in a broad range of metabolic and degenerative diseases.
Acknowledgments
We apologize to those authors whose original work could not be referenced due to space constraints. We would like to thank Nabeel Bardeesy and all the members of the Mostoslavsky's lab for helpful comments. R.M. is a Sidney Kimmel Scholar, the recipient of the Massachusetts Life Science Center New Investigator Award, a Howard Goodman Awardee and the recipient of a Young Investigator AFAR Grant. Work in the Mostoslavsky's lab is funded in part by NIH grants RO1DK088190-01A1 and RO1GM093072-01.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Aquilano K, Vigilanza P, Baldelli S, Pagliei B, Rotilio G, Ciriolo MR. Peroxisome proliferator-activated receptor gamma co-activator 1 alpha (PGC-1{alpha}) and sirtuin 1 (SIRT1) reside in mitochondria: possible direct function in mitochondrial biogenesis. J. Biol. Chem. 2010;1:1–19. doi: 10.1074/jbc.M109.070169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng Z, Tseng Y, White MF. Insulin signaling meets mitochondria in metabolism. Trends Endocrinol. Metab. 2010;21:589–598. doi: 10.1016/j.tem.2010.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cimen H, Han M-J, Yang Yongjie, Tong Q, Koc H, Koc EC. Regulation of succinate dehydrogenase activity by SIRT3 in mammalian mitochondria. Biochemistry. 2010;49:304–311. doi: 10.1021/bi901627u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finkel T, Deng C-X, Mostoslavsky R. Recent progress in the biology and physiology of sirtuins. Nature. 2009;460:587–591. doi: 10.1038/nature08197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finley LW, et al. SIRT3 Opposes Reprogramming of Cancer Cell Metabolism through HIF1α Destabilization. Cancer Cell. 2011;19:416–428. doi: 10.1016/j.ccr.2011.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hafner AV, Dai J, Gomes AP, Xiao C-Y, Palmeira CM, Rosenzweig A, Sinclair Da. Regulation of the mPTP by SIRT3-mediated deacetylation of CypD at lysine 166 suppresses age-related cardiac hypertrophy. Aging. 2010;2:914–923. doi: 10.18632/aging.100252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hallows WC, et al. Sirt3 Promotes the Urea Cycle and Fatty Acid Oxidation during Dietary Restriction. Mol. Cell. 2011;41:139–149. doi: 10.1016/j.molcel.2011.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imai S-ichiro, Guarente L. Ten years of NAD-dependent SIR2 family deacetylases: implications for metabolic diseases. Trends Pharmacol. Sci. 2010;31:212–220. doi: 10.1016/j.tips.2010.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaeberlein M, McVey M, Guarente L. The SIR2/3/4 complex and SIR2 alone promote longevity in Saccharomyces cerevisiae by two different mechanisms. Genes & Dev. 1999;13:2570–2580. doi: 10.1101/gad.13.19.2570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim HS, et al. Hepatic-Specific Disruption of SIRT6 in Mice Results in Fatty Liver Formation Due to Enhanced Glycolysis and Triglyceride Synthesis. Cell Metab. 2010;12:224–236. doi: 10.1016/j.cmet.2010.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kume S, et al. Calorie restriction enhances cell adaptation to hypoxia through Sirt1-dependent mitochondrial autophagy in mouse aged kidney. J. Clin. Inv. 2010;120:1043–1055. doi: 10.1172/JCI41376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim J-H, Lee Y-M, Chun Y-S, Chen J, Kim J-E, Park J-W. Sirtuin 1 Modulates Cellular Responses to Hypoxia by Deacetylating Hypoxia-Inducible Factor 1α. Mol. Cell. 2010;38:864–878. doi: 10.1016/j.molcel.2010.05.023. [DOI] [PubMed] [Google Scholar]
- Lomb DJ, Laurent G, Haigis MC. Sirtuins regulate key aspects of lipid metabolism. Biochim. Biophys. Acta. 2010;1804:1652–1657. doi: 10.1016/j.bbapap.2009.11.021. [DOI] [PubMed] [Google Scholar]
- Nakagawa T, Lomb DJ, Haigis MC, Guarente L. SIRT5 Deacetylates carbamoyl phosphate synthetase 1 and regulates the urea cycle. Cell. 2009;137:560–570. doi: 10.1016/j.cell.2009.02.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nasrin N, et al. SIRT4 regulates fatty acid oxidation and mitochondrial gene expression in liver and muscle cells. J. Biol. Chem. 2010;285:31995–32002. doi: 10.1074/jbc.M110.124164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu X, Brown K, Hirschey MD, Verdin E, Chen D. Calorie Restriction Reduces Oxidative Stress by SIRT3-Mediated SOD2 Activation. Cell Metab. 2010;12:662–667. doi: 10.1016/j.cmet.2010.11.015. [DOI] [PubMed] [Google Scholar]
- Schwer B, et al. Calorie restriction alters mitochondrial protein acetylation. Aging Cell. 2009;8:604–606. doi: 10.1111/j.1474-9726.2009.00503.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimazu T, et al. SIRT3 Deacetylates Mitochondrial 3-Hydroxy-3-Methylglutaryl CoA Synthase 2 and Regulates Ketone Body Production. Cell Metab. 2010;12:654–661. doi: 10.1016/j.cmet.2010.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shulga N, Pastorino JG. Ethanol sensitizes mitochondria to the permeability transition by inhibiting deacetylation of cyclophilin-D mediated by sirtuin-3. J. Cell Sci. 2010;123:4117–4127. doi: 10.1242/jcs.073502. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Smith BC, Settles B, Hallows WC, Craven MW, Denu JM. SIRT3 Substrate Specificity Determined by Peptide Arrays and Machine Learning. ACS Chem. Biol. 2010;6:146–157. doi: 10.1021/cb100218d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Someya S, Yu W, Hallows WC, Xu J, Vann JM, Leeuwenburgh C, Tanokura M, Denu JM, Prolla Ta. Sirt3 Mediates Reduction of Oxidative Damage and Prevention of Age-Related Hearing Loss under Caloric Restriction. Cell. 2010;143:802–812. doi: 10.1016/j.cell.2010.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sundaresan NR, Samant Sa, Pillai VB, Rajamohan SB, Gupta MP. SIRT3 is a stress-responsive deacetylase in cardiomyocytes that protects cells from stress-mediated cell death by deacetylation of Ku70. Mol. Cell. Biol. 2008;28:6384–6401. doi: 10.1128/MCB.00426-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanno M, Kuno A, Yano T, Miura T, Hisahara S, Ishikawa S, Shimamoto K, Horio Y. Induction of manganese superoxide dismutase by nuclear translocation and activation of SIRT1 promotes cell survival in chronic heart failure. J. Biol. Chem. 2010;285:8375–8382. doi: 10.1074/jbc.M109.090266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tao R, et al. Sirt3-Mediated Deacetylation of Evolutionarily Conserved Lysine 122 Regulates MnSOD Activity in Response to Stress. Mol. Cell. 2010;40:893–904. doi: 10.1016/j.molcel.2010.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verdin E, Hirschey MD, Finley LWS, Haigis MC. Sirtuin regulation of mitochondria: energy production, apoptosis, and signaling. Trends Biochem. Sci. 2010;35:669–675. doi: 10.1016/j.tibs.2010.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vetterli L, Brun T, Giovannoni L, Bosco D, Maechler P. Resveratrol potentiates glucose-stimulated insulin secretion in INS-1E beta-cells and human islets through Sirt1 dependent mechanism. J. Biol. Chem. 2010;286:6049–6060. doi: 10.1074/jbc.M110.176842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warburg O. On the origin of cancer cells. Science (New York, N.Y.) 1956;123:309–314. doi: 10.1126/science.123.3191.309. [DOI] [PubMed] [Google Scholar]
- Zhong L, et al. The histone deacetylase Sirt6 regulates glucose homeostasis via Hif1alpha. Cell. 2010;140:280–293. doi: 10.1016/j.cell.2009.12.041. [DOI] [PMC free article] [PubMed] [Google Scholar]