Abstract
We present here the first fully integrated, comprehensive map of the canine genome, incorporating detailed cytogenetic, radiation hybrid (RH), and meiotic information. We have mapped a collection of 266 chromosome-specific cosmid clones, each containing a microsatellite marker, to all 38 canine autosomes by fluorescence in situ hybridization (FISH). A 1500-marker RH map, comprising 1078 microsatellites, 320 dog gene markers, and 102 chromosome-specific markers, has been constructed using the RHDF5000-2 whole-genome radiation hybrid panel. Meiotic linkage analysis was performed, with at least one microsatellite marker from each dog autosome on a panel of reference families, allowing one meiotic linkage group to be anchored to all 38 dog autosomes. We present a karyotype in which each chromosome is identified by one meiotic linkage group and one or more RH groups. This updated integrated map, containing a total of 1800 markers, covers >90% of the dog genome. Positional selection of anchor clones enabled us, for the first time, to orientate nearly all of the integrated groups on each chromosome and to evaluate the extent of individual chromosome coverage in the integrated genome map. Finally, the inclusion of 320 dog genes into this integrated map enhances existing comparative mapping data between human and dog, and the 1000 mapped microsatellite markers constitute an invaluable tool with which to perform genome scanning studies on pedigrees of interest.
The modern domestic dog features more than 300 well-defined isolated breeding populations, most of which are readily accessible for mapping genes involved in complex phenotypes (Ostrander and Giniger 1997; Galibert et al. 1998; Ostrander et al. 2000). Indeed, extensive canine pedigrees, coupled with an enormous diversity in morphology and behavior, provides a valuable mechanism for understanding the genetic regulation of mammalian growth and development. In addition, the mating of closely related individuals in order to maximize segregation of desirable traits has led to the propagation of autosomal recessive diseases in the modern dog, many of which are similar or, in some cases, identical to human diseases (e.g., Dodds 1989; Menon et al. 1992; Sharp et al. 1992; Stolzfus et al. 1992; Henthorn et al. 1994; Zheng et al. 1994; Acland et al. 1998, 1999; Lin et al. 1999; Veske et al. 1999). To date, ∼360 genetic diseases have been identified in the dog, representing the largest number reported for any nonhuman mammal (Patterson 2000, 2001). Genetic linkage analysis utilizing domestic dog pedigrees, therefore, should provide a unique and efficient mechanism for understanding the molecular and cellular biology of human health and disease (Patterson et al. 1988; Ostrander et al. 2000).
In the past three years, we and our collaborators have made major advances in the production of a canine meiotic linkage map (Lingaas et al. 1997; Mellersh et al. 1997; Neff et al. 1999; Werner et al. 1999) and a radiation hybrid (RH) map (Priat et al. 1998; Mellersh et al. 2000). The recently published meiotic linkage map was composed of ∼350 markers organized into linkage groups for 37 autosomes and the X chromosome (Werner et al. 1999), whereas the RH map was composed of 600 markers, including 218 genes and 382 microsatellites, organized into 77 RH groups (Priat et al. 1998; Mellersh et al. 2000). By taking advantage of 216 markers mapped on both a set of three-generation reference families (Mellersh et al. 1997) and on a well-characterized canine RH panel (Vignaux et al. 1999), integration of the linkage and RH maps has been accomplished, leading to the production of a 724-marker map (235 genes and 489 microsatellites) with a marker spaced, on average, every 3.7 cM or 24.3 cR5000 (Mellersh et al. 2000). The evolutionary relationship between the canine and human genomes is also now well established through the efforts of two independent groups, both of whom have used bidirectional heterologous chromosome painting to identify evolutionarily conserved regions of human chromosomes (reciprocal Zoo-FISH; Breen et al. 1999c; Yang et al. 1999). Specifically, Breen et al. (1999c) and Yang et al. (1999) propose that 68 and 73 evolutionarily conserved segments, respectively, from the human genome can be visualized by reciprocal Zoo-FISH. Most recently, Sargan et al. (2000) expanded the latter of these reports and assigned all linkage, RH, and syntenic groups from the most recently published canine genome map (Mellersh et al. 2000) to specific dog chromosomes.
Although the accuracy and utility of the map are evidenced by the localization of disease loci for canine renal cancer, narcolepsy, multiple forms of retinitis pigmentosa, hematologic disorders, and several metabolic diseases (e.g., Henthorn et al. 1994; Yuzbasiyan-Gurkan et al. 1997; Acland et al. 1998; Lingaas et al. 1998; Lin et al. 1999; Jónasdóttir et al. 2000), at this juncture those wishing to exploit the dog system for refined mapping and cloning of complex diseases will be met with several significant obstacles. The placement of a marker every 3–4 cM is sufficient to ensure that genome scans of canine families will identify linked markers, but a denser map of more highly informative microsatellites is clearly needed for refining regions defined by linkage, as well as to facilitate the mapping of disease loci within highly inbred families. A more precise understanding of the evolutionary relationship between the canine and human genomes is needed in order that all evolutionary breakpoints may be mapped, and data from the more comprehensively mapped human and murine genomes used for the selection of candidate genes in the study of diseases. The meiotic linkage and RH maps must be fully integrated with the cytogenetic map. The latter represents a particularly challenging task because the similarity in size and banding patterns of many dog chromosomes renders them notoriously difficult to identify by classical cytogenetic methods. Using conventional cytogenetics, the International Committee for the Standardization of the Karyotype of the Dog was able to standardize the karyotype for dog chromosomes 1–21 and the sex chromosomes (Switonski et al. 1996). The Committee then assigned the chromosome-specific paint probes developed by Langford et al. (1996) and recommended that the chromosome numbering of Reimann et al. (1996) should be used for the remaining autosomes (Breen et al. 1998, 1999a). In this report we describe our collective effort to produce an integrated map of the dog genome incorporating cytogenetic, RH, and meiotic data. Building upon a newly developed set of 266 FISH-mapped chromosome-specific cosmid clones, each containing a microsatellite marker that unambiguously identifies one of the 38 autosomes of the dog, we are able for the first time to assign all dog chromosomes to their corresponding meiotic linkage and RH groups. The resulting map features 1078 microsatellites, 320 gene-based markers, and a total of 302 chromosome-specific markers, all of which are assigned by FISH using what is now agreed upon as the standardized chromosome nomenclature. This more than doubles the previously reported density of markers. All known evolutionary breakpoints in the human map are assigned to the canine cytogenetic map and therefore to the RH and linkage maps. Finally, RH mapping of proximal and/or distal markers that were initially mapped by FISH has allowed all but four integrated groups to be orientated. The resulting map, which features robust anchor points on every dog chromosome and a high density of polymorphic markers, provides an invaluable resource for the mapping and cloning of canine genes of interest.
RESULTS
In this paper we present an integrated cytogenetic/RH and meiotic map of the dog. We localized 266 microsatellite-containing cosmid clones on all dog chromosomes through FISH analysis, assigning 1500 markers by RH analysis and 354 markers by meiotic linkage analysis. Through sets of common markers, 251 RH/genetic, 102 RH/cytogenetic, and 52 linkage/cytogenetic, one or more RH groups and one genetic linkage group were assigned to each dog chromosome. This represents an integrated map with 1800 unique markers. In total, 72 RH groups are assigned to all chromosomes of the dog and 39 meiotic linkage groups are assigned to all autosomes plus the X chromosome. The resulting map (poster enclosed with this issue as Fig. 1) covers >90% of the genome.
Cytogenetic Map
Chromosome-Specific FISH Probe Selection
The cytogenetic localization of 266 microsatellite-containing cosmid clones (designated AHT-Kxxx, AHT-Hxxx, or LEI-xxx) was determined by analysis of FISH data. Chromosome assignments were made according to the nomenclature of the International Committee for the Standardization of the Dog Karyotype (Switonski et al. 1996; Breen et al. 1998), and chromosome band assignments were made by reference to the DAPI banded ideogram of Breen et al. (1999a). All physical assignments were conclusively verified by subsequent cohybridization with a differentially labeled whole chromosome paint probe (Langford et al. 1996). The cytogenetic assignment of each of these clones is indicated alongside the corresponding ideogram in Figure 1, demonstrating that the clones are evenly distributed throughout the entire karyotype.
RH Map: Marker Characteristics
Microsatellite Markers
A total of 1078 anonymous markers are now positioned on the canine RH map, of which 786, 20, and 272 are based upon di-, tri- and tetranucleotide repeats, respectively (Table 1). Polymorphism for each marker was evaluated either by heterozygosity (Het) or PIC values; 50% had Het or PIC values >0.5 (indicated by a triple star in Fig. 1) and 20% (mainly the tetranucleotide repeats) had Het or PIC values >0.75. The markers are randomly distributed throughout the autosomes and also the X chromosome. The 561 most polymorphic markers have an average distribution of 1 per 42 cR5000 (4.2 Mb), so indicating that, on average, any point on the map is now within 10 cM of a highly polymorphic marker.
Table 1.
Type and Number of RH Mapped Markers in the Integrated Map
| Markers | No. of markers | Refs. |
|---|---|---|
| Type I markers | 320 | |
| dog gene markers | 252 | 1 |
| cfEST : Dog Expressed Sequenced Tags | 23 | 2 |
| TOAST : Traced Orthologous Amplified Sequence Tags | 31 | 3 |
| huEST : Human Expressed Sequenced Tags | 14 | 4 |
| Type II markers | 1078 | |
| (CA)n dinucleotides mapped in this work | 701 | |
| previously mapped (CA)n microsatellites | 85 | 5 |
| previously mapped trinucleotides | 20 | 6 |
| previously mapped tetranucleotide repeats | 272 | 7 |
| Chromosome-specific markers mapped in this work | 102 |
References correspond to: (1) Priat et al. 1999 and this work; (2) Lin et al. 1997, GenBank accession numbers (Z97…) are listed on the Web site. (3) Jiang et al. 1998; (4) markers provided by G. Gyapay, Généthon, France; (5) Jouquand et al. 2000b and Priat et al. 1998; (6) Jouquand et al. 2000b; (7) Francisco et al. 1996.
All marker characteristics can be found at http://www-recomgen.univ-rennes1.fr/doggy.html.
Gene-Based Markers
The 320 gene-based markers were composed of 252 dog gene markers (Priat et al. 1999; this study), 23 dog EST markers (designed on EST sequences of retinal cDNAs retrieved from GenBank), 14 human EST markers (provided by G. Gyapay, Généthon, France), and 31 orthologous markers (TOASTs; Jiang et al. 1998). The gene-based markers were distributed across all chromosomes with the exception of dog chromosome (CFA) 32 and CFA 36. These data are indicated in Table 1.
Chromosome-Specific Markers
End sequencing of FISH-mapped chromosome-specific cosmid clones was used to derive 68 novel chromosome-specific markers. These markers were added to 35 previously identified FISH mapped markers, 12 of which (connected by green lines in Fig. 1) were reported by Werner et al. (1999). The remaining 23 were previously FISH-mapped at the Animal Health Trust (connected by red lines in Fig. 1) and reported elsewhere as follows: ATB7Bp on CFA 4 (van der Sluis et al. 1999); THY1, CDE3, SLC2A4, and DIO1 on CFA 5 (Thomas et al. 2001b); ALDOA and VCAM1 on CFA 6 (Thomas et al. 2001a); KRT17 on CFA 9 (Miller et al. 1999); CO4107 on CFA 10 (van der Sluis et al. 1999); PROP1 on CFA 11 (Lantiga-van Leeuwen et al. 2000a); IGF1, CPH4, and TRA1 (Ryder 2000); PROC (Leeb et al. 1999); PIT1 (Lantiga-van Leeuwen et al. 2000b); DMD (Schatzberg et al. 1999); SRY (Olivier et al. 1999); or they were mapped at the Animal Health Trust but are currently unpublished (PDEα on CFA 4, DLAA9 on CFA 12, ROM1 on CFA 18, RHO on CFA 20, RB1 on CFA 22, and RTB on CFA 27). Data from all 103 chromosome-specific markers were typed in duplicate.
Radiation Hybrid Mapping
The 900 new markers added here, as compared with the map reported by Mellersh et al. (2000), were typed on the 118-cell-line RHDF5000-2 panel (Vignaux et al. 1999). Data from these markers were added to the previous 600-marker RH map (Mellersh et al. 2000) and computed using the MultiMap software. Pairwise linkage analysis at LOD > 8.0 resulted in 89 RH groups. A total of 1413 out of 1500 markers were incorporated into these RH groups, with the remaining 87 markers remaining unlinked. The presence among RH markers of 102 chromosome-specific markers allowed 72 of the RH groups to be linked to one another resulting in 40 larger groups, each of which was assigned to a specific dog chromosome (Fig. 1). The 17 remaining orphan RH groups, which include 2–12 markers, have a cumulated size of 586 cR5000, representing only 2.5% of the map (data not shown).
The marker order for each RH group was obtained using Multipoint analysis. The order was initially determined using an LOD score of >3.0 (likelihood odds = 1000/1), resulting in a framework map consisting of 579 markers (45%). These markers are underlined in Figure 1. All remaining markers were then integrated relative to the framework markers to give the comprehensive map presented here. For RH groups presenting inconsistent ordering data, the initial pairwise analysis was repeated using a higher LOD score value (LOD > 9) that resulted in the formation of more RH groups. Ordering could then be carried out by multipoint analysis. The number of RH markers assigned to each autosome ranged from 67 markers (CFA 1) to a minimum of 8 (CFA 38), with the smallest dog chromosome, Y, having only 4 markers. The size of RH groups assigned to each chromosome ranged from 1272 cR5000 (CFA 1) to 112 cR5000 (CFA 38), with 17 cR5000 for the Y chromosome. The total size of the RH map is 23,428 cR5000 with 1413 markers mapped to 1354 unique positions. Thus, the average intermarker distance is 17 cR5000. Compared with the previously reported RH map (Mellersh et al. 2000), the present map represents a 2.5-fold increase in marker density, and decreases the average intermarker distance 1.5-fold.
Using maximum likelihood predictions, the canine genome has been estimated to be 26.5 M (Neff et al. 1999). The total length of the RH map presented here is 23,428 cR5000. Taking into account the cR/kb correspondence (see below) and the correspondence of 1 Mb/1 cM, the total size of the RH map thus amounts to 23.5 M. This map, therefore, is estimated to cover 88% of the canine genome. In agreement with this coverage estimate, only 7% of the markers remain unlinked at this point of the RH-map construction.
Meiotic Map: Marker Characteristics
Chromosome-Specific Microsatellite Markers
We selected 38 chromosome-specific cosmid clones (one per autosome) for the isolation of microsatellites. DNA prepared from these clones was shotgun-cloned into M13, and (CA)n-positive subclones were sequenced to identify a microsatellite repeat sequence. In Table 2 we present the primer sequences and optimized PCR conditions for all selected chromosome-specific microsatellite loci.
Table 2.
Loci and Corresponding PCR Primer Sequences of Chromosome-Specific Markers
| Marker name | Forward primer 5′–3′ | Reverse primer 5′–3′ | PCR size (bp) | Repeat | PCR conditions | Chromosome | Band assignment | Nomenclature Mellersh et al. 2000 |
|---|---|---|---|---|---|---|---|---|
| AHT K338 | ACAAAGCAATTTTTGCTACAAGC | TAATAGGTGTGGGACATTCGTT | 291 | (GT)12 | PCR55/4mM | CFA 1 | q11-q12 | CFA 1 |
| AHT H304Ren | CTCCTTGGTCCCCTCTTTTCTC | TATTCATTTCTCCCTGTGTCCAATC | 249 | – | TD61/2mM | CFA 1 | q37distal-q38 | CFA 1 |
| AHT H72Ren | TTTAGCATTTTCCTCTGGCTTT | ATCTGGTCTGTTTGCCTTCC | 172 | – | TD65/2mM | CFA 2 | q12.3-12.5 | CFA 2 + RH7 |
| LEI 1D6 | GCTCCATCCACATCATTGC | AGCAGCATCGACAATAGCC | 172 | (AC)15 | TD66/4.5mM | CFA 2 | q34distal-q35prox. | CFA 2 + RH7 |
| AHT H255Ren | GTCCCAAAGGCTTTCCAAAT | GATGGCTGGGTCCTGTCTTA | 168 | – | TD67/2mM | CFA 2 | q35prox. | CFA 2 + RH7 |
| AHT H291Ren | CAAATTCTGCCTGAATATACCACAA | TATCAAGCAGTCCTCCTTTGACAGA | 254 | – | TD61/2mM | CFA 3 | q21.1-q21.3prox. | CFA 3 |
| LEI 1E12 | GGAAATGCATGTAGATACATGTG | AGAATGGGAATTGGAAGTCAGG | 152 | (TG)13 | TD61/4.5mM | CFA 3 | q22.3 | CFA 3 |
| AHT H219Ren | CTATGAGTCAAGACTAGGGGCAGTG | AGAGCTGTCTCCCAGAGACGTTAAG | 296 | – | TD61/2mM | CFA 3 | q22.3distal | CFA 3 |
| AHT1.37 | AACTTTTATTTTTTCACTTA | GCTCTAATCAGAACAATCA | 389 | (GT)11 | TD45/3mM | CFA 4 | q22 | Synt. 1 |
| AHT H1.37Ren | ACTGCCCAGAGGAAGGAGAG | TGGTACATTTAGGGGCAACA | 202 | – | TD59/2mM | CFA 4 | q22 | Synt. 1 |
| AHT H142Ren | CATTGGGGTAAAAGCTGCAT | TTTGATGGGGACAGAGCTTC | 163 | – | TD65/2mM | CFA 4 | q33distal-q34.1 | Synt. 1 |
| AHT H68Ren | TGCTTTCTTTGCTTATTCCTATGAT | TGAGAGTCACTCCATTGCTTAGTTT | 248 | – | TD61/2mM | CFA 5 | q11-q12prox. | CFA 5 |
| AHT H248 | GGCCACCCCCCAACAC | TGTAGAGTGGAGCTCAAGAATTTG | 139 | (CA)19 | PCR55/3mM | CFA 5 | q12 | CFA 5 |
| AHT K315 | GCCAAAATCGCTCTCCCA | TTGGAGTGGGCACCAAAA | 260 | (TG)19 | TD46/2mM | CFA 5 | q33distal-q34.1 | CFA 5 |
| AHT H201Ren | CCTCACAGGTTCCCTATATCAGCTT | ATGGAAGGTGGATTAGACAGAGAGG | 248 | – | TD61/2mM | CFA 5 | q35-q36 | CFA 5 |
| AHT H171 | CTCACCAGGCATAGACACTCAG | CTCATTTGTTCACGCACCC | 131 | (GT)19 | TD53/1mM | CFA 6 | q11distal-q12prox. | CFA 6 |
| LEI 2A11 | CATAGCGACATCACATAAGT | GGGCTGATAATTCAGTGAGA | 184 | (AC)17 | TD72/2mM | CFA 6 | q21.3 | CFA 6 |
| AHT H228Ren | GCCTTATTTTTGTCTCTGCTCCAGT | GGGAGGTTCACAGACTTACAGGAAT | 234 | – | TD61/2mM | CFA 6 | q21.3distal-q22 | CFA 6 |
| AHT H289 | GGGACCTGGTAGGCTGAACT | ATGAGCATGATGGTGTTCCA | 153 | – | TD63/4.5mM | CFA 7 | q11 | CFA 7 |
| LEI 1B10 | TTGTCTTTAGACTCAATAGTAGC | CCCAACGATCCGTTCTTTGAC | 119 | (GT)18 | TD63/4.5mM | CFA 7 | q15-q16.2 | CFA 7 |
| AHT H71Ren | CAGGGCAGTGTCCCTAGAAG | GGGTGGTGGACAAAGTCATT | 159 | – | TD69/2mM | CFA 7 | q25distal-q26prox. | CFA 7 |
| AHT H240 | GTCCATGACTGGCTCAGGTTT | TTGAGGCCTAGAACTCTG | 163 | (GT)27 | PCR55/4mM | CFA 8 | q11 | CFA 8 |
| AHT H240Ren | CCCCACTTCCTCTGGTCAGT | GCTCCAATCACCACCATTCT | 141 | – | TD65/2mM | CFA 8 | q11 | CFA 8 |
| LEI 2D2 | CGAGACCACATTGGGCTCC | CCAGACGCTCTCCATGCCTC | 390 | (GAAA)9 | TD63/4.5mM | CFA 9 | q25prox. | CFA 9 |
| AHT H183Ren | TTATCAGGTGAGTCCAGTGTCATCA | TTTCTGATCCTTCAAGCCATAAGGT | 252 | – | TD61/2mM | CFA 9 | q26.1-q26.2prox. | CFA 9 |
| AHT H181Ren | CTAGCATGTTGTAGCCCCCTAAACT | TGAGGTTCAGAGTTTCACACATCTTC | 252 | – | TD61/2mM | CFA 10 | q11prox. | CFA 10 |
| AHT H51 | CTGGGTCTGCGCCTAGCTT | CACAGATTTAACTTTCCAGAACTGC | 407 | (AC)22 | PCT45/3mM | CFA 10 | q14distal-q21prox. | CFA 10 |
| AHT H203Ren | CTTTACCTGCTTATGCTCACTCTCG | TCTTCCTGTGGGAGTTGAGAACAT | 157 | – | TD61/2mM | CFA 10 | q23.2-q24prox. | CFA 10 |
| AHT H279 | GCACATGCTCCATTAACCCC | CACTATCTCCTGGCCTCTTCC | 184 | (GT)19 | PCR55/3mM | CFA 11 | q11distal | Synt. 4 |
| AHT H155Ren | CTGCACACCTACCCCAGAAT | CACGCACGTGCTTAAACATT | 123 | – | TD65/2mM | CFA 11 | q22.3-q23 | Synt. 4 |
| AHT H127 | ATCGACTTGTTCTTTGTCTCAGC | TTTGGATATGATGGGGAATTTA | 178 | – | TD55/2mM | CFA 12 | q22-q24 | CFA 12 |
| AHT H1.47 | GGCAGTGAATCTGTTGTGAGTT | GAGAAATGAGGATAAAACGACACC | 258 | (CA)21 | PCR55/2mM | CFA 12 | q23distal-q24 | CFA 12 |
| AHT H239 | TGTTTCATTGGTCTGCGTGT | TAGGAGGCCGATTCTCTTGA | 263 | – | TD67/2mM | CFA 13 | q21.2 | Synt. 8 |
| AHT H310 | CTTTGCCTGCAGGAGTTTTC | ACGTCAGGCATCCATGCATG | 180 | (GA)12 | PCR55/1.5mM | CFA 13 | q22.2 | Synt. 8 |
| AHT H262Ren | CTCATCCATGCACGTCCTTT | TTAGGGAACAGGAAGCCTGA | 146 | – | TD69/2mM | CFA 14 | q11 | Synt. 9 |
| AHT K207 | CAATCTTCCAACAACCTTGT | GTCCTTGAGGGGTTAATAAT | 94 | (GT)18 | TD56/1.5mM | CFA 14 | q23 | Synt. 9 |
| AHT H257 | CCCACACCTTTGCTGGAC | TGGGAGATCCAAGAGGAGG | 126 | (TG)8C(GT)10 | PCR50/3mM | CFA 15 | q13 | Synt. 6 |
| AHT H17Ren | TAGAGTAAGGGGCTCAAACATCACA | ATGGAACCAGAGAACACTTTGGAA | 237 | – | TD61/2mM | CFA 15 | q24 | Synt. 6 |
| AHT H260 | CGCTATACCCACACCAGGAC | CCACAGAGGAAGGGATGC | 245 | – | TD69/2mM | CFA 16 | q14 | Synt. 12 |
| AHT H282Ren | TCCATTAACAAGTGCGTGTCAGATA | CTCAGAGATTCGCTCGACTTAAAGA | 262 | – | TD61/2mM | CFA 17 | q11 | CFA 15 + Synt 16 |
| AHT H265 | GGACCCCTAATACCCAGTAACATT | GTGCAGGCTATAAACCAAAAACTT | 238 | (AC)15 | TD53/4mM | CFA 17 | q15 | CFA 15 + Synt 16 |
| LEI 1E3 | GAGGAGATCCTAGTAACACC | GCTGGTGTTCCTAAAAAGCC | 220 | (TG)14(GA)14 | TD63/4.5mM | CFA 18 | q23-q24prox. | CFA 18 |
| AHT K32 | TACCAAGACCCAACAGCAAG | GAGCCTGCTTCTCCTTAT | 159 | – | TD59/2mM | CFA 18 | q25.1-q25.2 | CFA 18 |
| AHT K292Ren | TTTGTACTCCTAGGACAAGGATTTG | TCCTCCTGTGACAATTTCAGG | 250 | – | TD61/2mM | CFA 18 | q25.3distal | CFA 18 |
| AHT H67 | CAGGGGCTTGTTTTCCTTGA | AGGATGGTTGGTTAAGTG | 282 | (CA)12 | PCR60/3mM | CFA 19 | q21-q22 | CFA 13/19 |
| AHT H216Ren | CGCCCGATTTTCTTTGTTAG | GGCAAAGGAGGACACAGGT | 143 | – | TD69/2mM | CFA 20 | q11prox. | CFA 20 |
| AHT K20 | GTACAGTGGTCACATAGAGG | CTCAAAGGCGTTCTTTCCAG | 169 | – | TD59/2mM | CFA 20 | q15.3prox. | CFA 20 |
| LEI 1B7 | GCAATCCCACCCCAAAGA | CTGTTGCTATGATCATGGGTG | 198 | (GAAA)4(GA) 18(GAAA)13 | TD63/4.5mM | CFA 20 | q15.3distal | CFA 20 |
| AHT H284Ren | CTCTGAGAACAGGAACAGCAAATTC | CTCAGTGAACTCACCCCAAGACATT | 243 | – | TD61/2mM | CFA 20 | q16-q17 | CFA 20 |
| AHT H298 | CCAGGCATTCGAGGGTG | TCCCAGAACTTGAGGAACCATAG | 103 | (GT)12 | PRC45/3mM | CFA 21 | q13.3distal-q21prox. | Synt. 2 |
| AHT H211 | AGCTTCAGGCACAGGGGT | CCAGGGAGGTGAAAAATGACT | 348 | (GT)19 | TD53/2mM | CFA 22 | q11.2 | Synt. 7 |
| AHT K253 | CTCAAAGGCGTTCTTTCCAG | GCACATGGAGGACAAGCAC | 146 | (TG)20 | TD56/2mM | CFA 23 | q13 | CFA 22 |
| AHT H180 | TCTGTGCTTTGATTAGCAGCAT | GGAGCCTGACAGTGAGTGGA | 250 | – | TD59/2mM | CFA 23 | q23 | CFA 22 |
| AHT H138 | ACTGGGTGTCCTATTGATGTTTT | TATACACAGCGCCCAACACAC | 204 | (GT)19 | TD53/2mM | CFA 24 | q11 | Synt. 5 |
| AHT H9bRen | AGTTCAGTCCAGGGCACCTA | CGTGCAGGATTCCCTCTTTA | 154 | – | TD69/2mM | CFA 24 | q24 | Synt. 5 |
| AHT K120 | GGGTCTCAGACACTGTGTGC | TTGCAGGCTAGGCTTTTCTC | 115 | (CA)17 | TD61/2mM | CFA 25 | q21.3-q22 | Synt. 10 |
| AHT H226 | GGGCAAGCTACCTTTCCACTCT | CTTTTTAATGAGCCCTTCATGGAAT | 210 | – | TD61/2mM | CFA 25 | q24distal | Synt. 10 |
| AHT K211 | TTAGCAGCCGAGAAATACGC | ATTCGCCCGACTTTGGCA | 92 | (CA)13 | TD63/4mM | CFA 26 | q13distal-q21 | CFA 26 |
| AHT K200 | GATTTATCAGTTTGGAGGGTG | TGTAGAAATCCTGAACATTACA | 65 | (GT)14 | TD61/2mM | CFA 26 | q23-q24prox. | CFA 26 |
| AHT K18 | CAGATGAGGAAGTGTGCGTG | CTGCCTCCTCTAGCCTTCTG | 89 | (GT)15 | TD59/2mM | CFA 27 | q11 | CFA 16 |
| AHT H197Ren | CGAGGGGCCTAAAATCTAGC | GGCAGGACCCCAAATTAGAT | 156 | – | TD69/2mM | CFA 28 | q11 | Synt. 14 |
| AHT K135 | GCAGCTCTCAGCACCCACAC | GTGAGTCCACTTGGGGCG | 223 | (GT)12 | TD60/2mM | CFA 28 | q18 | Synt. 14 |
| AHT K40 | TAACGTGCTGGTTTTAGACTGATC | CACATGGAGCCTGCTTCTC | 221 | (CA)18 | TD56/2mM | CFA 29 | q13-q15 | Synt. 3 |
| AHT H235Ren | GCCTGCTTCACAAAATGACA | GGCAAGTTTCCTGAGCTGAC | 172 | – | TD69/2mM | CFA 29 | q22-q23.1 | Synt. 3 |
| AHT H134Ren | CCCGTCCTTAACCACATGAT | AGCTGGTGACCCAGGTAGTG | 167 | – | TD69/2mM | CFA 30 | q13-q14prox. | CFA 30 |
| LEI 1F11 | TTTCAGAGTTCTGATGCTCC | GTCAGTTCAATTAGCCAGAG | 175 | (GT)19 | TD54/4.5mM | CFA 30 | q15.1distal-q15.2 | CFA 30 |
| AHT H246 | ATTTTCATTCCGAGGTTCTAACTG | ATTCACCATCTCGTAGCCTTTATC | 260 | (GT)16 | TD53/3mM | CFA 31 | q15.3 | Synt. 13 |
| AHT H327 | TCAGCTTATGTCCTTACATTTG | TCCCGGTGCATGGAG | 301 | (CA)21 | TD57/4mM | CFA 32 | q11.2distal-q13 | Synt. 15 |
| AHT H327Ren | TGCCAGATTCCTCAACTTCC | CTGCACACAAGAGGTGATAAGC | 139 | – | TD69/2mM | CFA 32 | q11.2distal-q13 | Synt. 15 |
| AHT K336 | GGTGAAGATGGGGTGATTTG | TCTTCAGATTCCACCGGTTT | 200 | – | TD69/2mM | CFA 33 | q15.1distal-q15.2 | CFA 29-35 |
| AHT H131Ren | CCCACCCCAAGTATGTCCTA | CCTCCTGCTCTTGAAGCAAC | 145 | – | TD63/2mM | CFA 34 | q12-q13 | RH1-b |
| AHT H163 | AATATAATGGAAAGGAAGATTAGGC | CAATCCAACAGTACCTTGAAAATG | 223 | (CA)22 | PCR45/1.5mM | CFA 34 | q14 | RH1-b |
| AHT H53b | TGGTGTGTGGAAAAGGGACT | AGCAAGCTGGTCAAGAACTCC | 167 | (AC)10 | PCR55/2mM | CFA 35 | q14 | RH5-b |
| AHT H53Ren | CACTGGACTGTGGGCAGAGAT | TCTGAACTGGGCATGGATC | 309 | – | TD69/2mM | CFA 35 | q14 | RH5-b |
| AHT H1261D | AGCTGCTTCTCCCTCCTTTAGTCTG | AGCTGAAAGGTTTCCGGAGG | 100 | (AC)12 | PVR58/1.5mM | CFA 35 | q14 | RH5-b |
| AHT H31Ren | CAGGTTCCCTCTCCACCTTT | TTTCAGGCAAGCCTCTCATT | 150 | – | TD65/2mM | CFA 36 | q13 | Linkage-gp 1 |
| AHT H130 | GTTTCTCTCCCTTCGGGTTC | GACGTGTGTTCACGCCAG | 130 | (GT)22 | TD59/3mM | CFA 36 | q14 | Linkage-gp 1 |
| AHT H128Ren | AGTCCCAGGCAGGAGAGAAT | TGATCCCCGATTGTGTTGTA | 148 | – | TD67/2mM | CFA 37 | q11-12prox. | Synt. 11 |
| AHT H174 | TCAACGGAGTCTTTGAGGAT | AAGGAGTACATTTTCAAGAA | 121 | (TG)16 | PCR45/3mM | CFA 37 | q16 | Synt. 11 |
| AHT H91 | CCCAGTCTTTGTTCTCCTG | AGGACTCTTTCAGGTACTCGGTC | 206 | (GT)17 | PCR45/3mM | CFA 38 | q15.2 | Synt. 17 |
| AHT H91Ren | GGACCGAGTACCTGAAAGAGTC | TTCTTTAACCAGTCAGCCACAC | 200 | – | TD67/2mM | CFA 38 | q15.2 | Synt. 17 |
Loci and corresponding PCR primer sequences used to anchor RH and meiotic linkage groups to their corresponding dog chromosome. Where a microsatellite marker was isolated from the clone, the repeat motif is indicated in Column 5. For the PCR conditions (Column 6) the annealing temperature, as either as a single temperature (PCRxx) or a standard touchdown program (TDxx), is presented followed by the magnesium chloride concentration. The chromosome to which each marker was assigned by FISH is indicated in Column 7. The band assignment for each marker is based on the DAPI banded ideogram reported in Breen et al. (1999b). The nomenclature of the RH groups used by Mellersh et al. (2000) are presented in the final column.
Meiotic Linkage Mapping
Microsatellites from each of the 38 dog autosomes were genotyped on dog reference families that have been described previously (Mellersh et al. 1997), and the data were merged with those from the previous meiotic linkage map described by Werner et al. (1999). We identified 39 linkage groups, containing a total of 350 markers. Of these 38 groups could be assigned to a chromosome via a chromosome-specific microsatellite marker. An additional linkage group did not contain a chromosome-specific microsatellite and could not, therefore, be conclusively assigned to a chromosome (data not shown).
The chromosome-specific microsatellites could be assigned to their respective linkage groups with varying levels of significance. For 32 autosomes, CFA 1–12, 14–27, 29, 31, 33, 35–37, the chromosome-specific microsatellites were linked to other microsatellites in the corresponding group with LOD scores >5.0, indicating significant evidence of linkage. Meiotic linkage data for the six remaining chromosomes were as follows: the chromosome-specific microsatellite for CFA 13 (AHTH310) was linked to one other marker (FH2394) in the group with a LOD score >4.0, and to multiple other markers in the same group with LOD scores >3.0. The markers representing CFA 28 (AHTK135), CFA 32 (AHTH327), and CFA 34 (AHTH163) were each linked to a single marker (LEI006, CPH2, and FH2377, respectively) in their corresponding groups with a LOD score >3.0. For CFA 30 (LEI-1F11) and CFA 38 (AHTH91), the best two-point LOD scores were 2.709 (to PEZ7) and 2.912 (to FH2244), respectively. However, RH data for these two markers did enable them to be assigned to their respective groups with significant LOD scores (>8.0), thus validating the assignment of the whole group.
The Integrated Map: Integration of the Three Maps
The use of FISH analysis to integrate the RH and meiotic maps with the cytogenetic map is illustrated in Figure 2. A comparison of the RH, meiotic, and cytogenetic maps shows that they are highly concordant. The only observed discrepancies in the colinearity between the maps is the inversion of a segment containing four markers on CFA 4, and 20 inverted pairs of markers distributed on other autosomes. Chromosome-specific FISH-mapped markers were RH-mapped on all chromosomes as follows: 8 dog chromosomes harbor 3–6 probes, 20 have 2 probes, and 12 chromosomes have 1 probe. RH/meiotic groups could be readily orientated on 33 chromosomes owing to the localization of at least one FISH probe at one end of the chromosome. Another chromosome, CFA 19, could be oriented because of syntenic conservation between human and dog chromosomes. The maps for the remaining four autosomes (i.e., CFA 21, CFA 32, CFA 35, and CFA 36), each harboring one midchromosome probe, and for the two sex chromosomes cannot as yet be orientated (Fig. 1).
Figure 2.
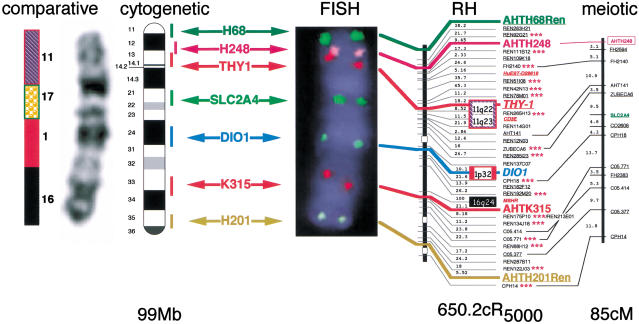
Multicolor fluorescence in situ hybridization of canine clones to dog chromosome 5 (CFA 5). The integrated RH and meiotic maps for CFA 5 are shown on the right hand side with their corresponding sizes noted below each map. Six clones were selected on the basis of their positions along the length of the RH map and are indicated in colored text. A seventh clone, SLC2A4, was selected on the basis of its position in the meiotic map. Each of the seven clones was labeled with one of the following five fluorochromes: Spectrum Green dUTP (green signal), Spectrum Orange dUTP (gold signal), Spectrum Red dUTP (red signal), DEAC (aqua signal), and Cy5 (pink signal). All seven clones were cohybridized by FISH to elongated dog chromosomes. The resulting FISH image of a DAPI-counterstained CFA 5 is shown in the middle of the figure, illustrating the hybridization signals of all seven clones, along with the assignment of each clone to the DAPI-banded ideogram of CFA 5 (Breen et al 1999a). On the far left is the enhanced DAPI-banded image of the same CFA 5 alongside the corresponding conserved human chromosome segments identified by Breen et al. (1999c).
By comparing the size in megabases of a chromosome (deduced by the bivariate flow-sorting measurements of Langford et al. 1996) to the size of the RH group harboring FISH markers located at the two ends of the chromosome (this was possible for CFA 1, CFA 10, CFA 12, CFA 14, and CFA 20), a correspondence of 1 cR5000 for 100 kb was calculated. This result compares well with those reported for human and murine genomes at comparable radiation doses (McCarthy et al. 1997; Stewart et al. 1997).
Coverage of the Integrated Map
Our data indicate that RH groups span the length of all but three autosomes, CFA 32, CFA 36, and CFA 38, for which we observed a clear difference between the size of the chromosome and that of the corresponding RH group. Specifically, CFA 32 and CFA 36 are each composed of one RH group, covering approximately half of the chromosome, whereas CFA 38 is covered by one RH group representing approximately one-third of the chromosome. The X chromosome is more poorly covered by two RH groups and four single markers. Regions not covered by the RH map can be estimated to 200 Mb, or ∼8% of the total genome size. However, unlinked markers and orphan RH groups are likely to fill the gaps.
The size of meiotic linkage groups in Figure 1 approximates to their chromosome length for all but eight autosomes: CFA 6, CFA 17, CFA 19, CFA 21, CFA 24, CFA 32, CFA 34, and CFA 36. Coverage of chromosomes by their corresponding meiotic linkage group is 60% for CFA 19 and CFA 21, 50% for CFA 6 and CFA 24, 30% for CFA 17 and CFA 32, 20% for CFA 36, and 10% for CFA 34. The X chromosome is covered at 50%. However, by combining data from the RH and meiotic linkage maps, only half of both CFA 32 and CFA X and two-thirds of CFA 36 are not represented in the integrated map. A summary of the relevant statistics of the integrated map is shown in Table 3.
Table 3.
Integrated Map Statistics by Chromosome
| CFA | Cytogenetic map no. of markers | RH map no. of markers | Linkage map no. of markers | cyto/RH common markers | RH/linkage common markers | cyto/linkage common markers | Integrated map(1) unique markers | Gene(2) content | ZOO-FISH(3) segments | RH(4) segments |
|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 12 | 67 | 17 | 2 | 15 | 1 | 78 | 14 | 4 | 3 |
| 2 | 11 | 59 | 17 | 3 | 11 | 2 | 71 | 14 | 4 | 3 |
| 3 | 11 | 43 | 21 | 3 | 11 | 1 | 60 | 5 | 3 | 3 |
| 4 | 13 | 51 | 11 | 4 | 8 | 1 | 62 | 12 | 2 | 3 |
| 5 | 29 | 37 | 13 | 7 | 9 | 2 | 61 | 5 | 4 | 3 |
| 6 | 15 | 36 | 14 | 6 | 10 | 2 | 47 | 10 | 3 | 3 |
| 7 | 13 | 53 | 15 | 3 | 8 | 1 | 69 | 10 | 2 | 2 |
| 8 | 9 | 37 | 12 | 2 | 8 | 2 | 46 | 11 | 1 | 1 |
| 9 | 30 | 56 | 14 | 7 | 8 | 4 | 81 | 26 | 2 | 2 |
| 10 | 9 | 41 | 16 | 3 | 10 | 2 | 51 | 4 | 3 | 2 |
| 11 | 4 | 45 | 13 | 3 | 9 | 1 | 49 | 6 | 2 | 2 |
| 12 | 3 | 49 | 11 | 2 | 7 | 1 | 53 | 13 | 1 | 1 |
| 13 | 4 | 33 | 7 | 1 | 5 | 1 | 37 | 5 | 2 | 2 |
| 14 | 3 | 32 | 8 | 2 | 4 | 1 | 36 | 8 | 1 | 1 |
| 15 | 8 | 35 | 11 | 5 | 10 | 2 | 37 | 5 | 3 | 2 |
| 16 | 7 | 23 | 6 | 1 | 4 | 1 | 30 | 4 | 3 | 2 |
| 17 | 6 | 40 | 4 | 3 | 4 | 2 | 41 | 10 | 2 | 2 |
| 18 | 17 | 40 | 11 | 4 | 7 | 2 | 55 | 11 | 1 | 2 |
| 19 | 3 | 35 | 7 | 2 | 6 | 1 | 36 | 2 | 2 | 2 |
| 20 | 17 | 53 | 13 | 5 | 7 | 2 | 69 | 12 | 2 | 2 |
| 21 | 2 | 49 | 8 | 1 | 6 | 1 | 51 | 13 | 1 | 1 |
| 22 | 3 | 47 | 7 | 2 | 7 | 1 | 47 | 6 | 1 | 1 |
| 23 | 4 | 23 | 9 | 3 | 6 | 1 | 26 | 2 | 1 | 1 |
| 24 | 9 | 27 | 9 | 2 | 9 | 1 | 33 | 4 | 1 | 1 |
| 25 | 11 | 23 | 9 | 2 | 6 | 1 | 34 | 5 | 2 | 3 |
| 26 | 7 | 32 | 7 | 2 | 5 | 3 | 36 | 7 | 2 | 2 |
| 27 | 4 | 45 | 11 | 2 | 10 | 1 | 47 | 14 | 1 | 1 |
| 28 | 6 | 26 | 6 | 2 | 6 | 1 | 29 | 6 | 1 | 1 |
| 29 | 2 | 29 | 8 | 2 | 6 | 1 | 30 | 2 | 1 | 1 |
| 30 | 7 | 20 | 5 | 2 | 3 | 1 | 26 | 4 | 1 | 1 |
| 31 | 6 | 20 | 6 | 2 | 3 | 1 | 26 | 3 | 1 | 2 |
| 32 | 1 | 10 | 4 | 1 | 3 | 1 | 10 | 0 | 1 | 0 |
| 33 | 1 | 16 | 4 | 1 | 3 | 1 | 16 | 3 | 1 | 1 |
| 34 | 2 | 27 | 2 | 2 | 2 | 1 | 26 | 2 | 1 | 1 |
| 35 | 2 | 17 | 2 | 1 | 1 | 1 | 18 | 2 | 1 | 1 |
| 36 | 2 | 9 | 2 | 2 | 2 | 1 | 8 | 0 | 1 | 0 |
| 37 | 4 | 22 | 7 | 2 | 4 | 1 | 26 | 3 | 1 | 1 |
| 38 | 3 | 8 | 3 | 1 | 2 | 1 | 10 | 1 | 1 | 1 |
| X | 1 | 20 | 0 | 1 | 6 | 0 | 14 | 8 | 0 | 1 |
| Y | 1 | 4 | 0 | 1 | 0 | 0 | 4 | 1 | 0 | 1 |
| Total assigned | 302 | 1339 | 350 | 102 | 251 | 52 | 1586 | 273 | 67 | 65 |
| No. of orphan mrks | 0 | 74 | 4 | 0 | 0 | 0 | 78 | 17 | 0 | 0 |
| Sub-total | 302 | 1413 | 354 | 102 | 251 | 52 | 1664 | 290 | 67 | 65 |
| Unlinked | 0 | 87 | 0 | 0 | 0 | 0 | 91 | 30 | 0 | 0 |
| Total | 302 | 1500 | 354 | 102 | 251 | 52 | 1755 | 320 | 67 | 65 |
This table displays a summary of relevant data of the three maps composing the integrated map. The number of markers per dog chromosome is presented.
(1) Total number of unique markers reported (i.e., total of the three maps minus common mapped markers).
(2) The gene content column reports the number of genes mapped on the RH map.
(3) Human chromosome segments identified by Zoo-FISH data reported in Breen et al. (1999a). An additional four conserved regions were subsequently identified by Yang et al. (1999) and are denoted by ‘+1’ beside CFA 4,18,25 and 31.
(4) Human orthologous segments identified from the RH map.
Comparative Mapping
For each dog chromosome presented in Figure 1, we have illustrated the regions of evolutionarily conserved chromosome segments that were observed in the Zoo-FISH studies of Breen et al. (1999c). Although Yang et al. (1999) used a different chromosome nomenclature from that recently endorsed (International Society for Animal Genetics 2000, G. Dolf, DogMap Chairman, pers. comm.) and in use here, correspondence between the two chromosome numbering systems was tentatively identified by Sargan et al. (2000). Our localization of 320 gene markers on the RH map has now verified this correspondence. This has allowed us to confirm that these two studies were grossly comparable in their assessment of the distribution of human conserved chromosome segments throughout the dog karyotype. We observed differences in five cases. Four differences were at the proximal end of CFA 4, CFA 18, and CFA 31 and the telomeric end of CFA 25. This was because of the lack of visible signal at the centromeric and telomeric ends of the dog chromosomes reported by Breen et al. (1999c), a phenomenon that has also been reported for other species (Jauch et al. 1992; Wienberg et al. 1992; Nash et al. 1998). However, hybridization signals were observed in these regions by Yang et al. (1999), and we have now confirmed the presence of these regions by identification of corresponding type I markers in the RH map. The additional information for these four regions provided by Yang et al. (1999) is therefore included in Figure 1, highlighted with (Y) beside the data. A fifth difference was observed toward the distal end of CFA 10, where Yang et al. (1999) reported a small block of HSA 12 between blocks of HSA 22 and HSA 2 reported by Breen et al. (1999c). This finding has not yet been confirmed by RH mapping. Zoo-FISH data between dog and human identified no fewer than 67 (Breen et al. 1999c) or 75 conserved chromosome segments (Yang et al. 1999). RH mapping of dog genes whose human locations are known has thus far allowed the detection of 65 conserved segments.
For 229 RH-mapped dog genes, the human localization of the putative ortholog was known. All but two dog chromosomes (CFA 32 and CFA 36) harbor at least one (and up to 22) gene marker for which a physically mapped human ortholog is known. The absence of any mapped gene on CFA 32 and CFA 36 precluded the use of the RH data for comparative assessment of these two autosomes. Conserved syntenic fragments between dog and human are indicated in Figure 1, with colored boxes to the immediate left of the RH markers. Apparently 18 dog chromosomes correspond to only one human fragment each, that is, CFA 8/HSA 14q, CFA 12/HSA 6pq, CFA 14/HSA 7pq, CFA 21/HSA 11pq, CFA 22/HSA 13q, CFA 23/HSA 3pq, CFA 24/HSA 20pq, CFA 27/HSA 12pq, CFA 28/HSA 10q, CFA 29/HSA 8q, CFA 30/HSA 15q, CFA 33/HSA 3q, CFA 34/HSA 3q, CFA 35/HSA 6p, CFA 37/HSA 2q, CFA 38/HSA 1q, X/X, and Y/Y. Another 13 dog chromosomes (CFA 7, CFA 9, CFA 10, CFA 11, CFA 13, CFA 15, CFA 16, CFA 17, CFA 18, CFA 19, CFA 20, CFA 26, and CFA 31) correspond to two human fragments each, and the remaining 9 dog chromosomes correspond to three or four different human chromosomal fragments each. In contrast, only four human chromosomes share exclusive conserved synteny with a dog chromosome (HSA 14/CFA 8, HSA 20/CFA 24, HSA 21/CFA 31, and chromosome X). The remainder are split into two to eight chromosomal segments (e.g., HSA 1 is split into eight fragments in the dog, corresponding to regions of CFA 2, CFA 4, CFA 5, CFA 6, CFA 7, CFA 15, CFA 17, and CFA 38; Breen et al. 1999c; Yang et al. 1999).
DISCUSSION
Great strides have been made over the past few years toward the development of a canine genome map. Prior to this study, however, there were a number of limitations associated with the canine map. Chromosomal assignment of meiotic linkage groups had been made for only 19 of the 38 dog autosomes (Mellersh et al. 2000), with the remainder being referred to as anonymous linkage groups. In addition, the chromosomal orientation of the meiotic linkage and RH groups was largely unknown. Furthermore, estimates of genome coverage had been entirely theoretical, and little was known about the extent to which the RH and meiotic linkage maps extended along the length of their respective chromosomes. In this study we have addressed these issues. We have conclusively assigned 266 cosmid clones to their specific dog chromosomes, by FISH, using the chromosome numbering system endorsed by the ISAG 2000 DogMap workshop (G. Dolf, DogMap Chairman, pers. comm.). Because all the cosmid clones had been prescreened to contain a microsatellite repeat, they represent not only additional chromosome-specific genetic markers but also the key resource that enabled us, for the first time, to integrate the canine meiotic linkage and RH maps with the cytogenetic map.
Meiotic linkage data from 38 chromosome-specific microsatellite markers, derived from these clones, were used to anchor meiotic linkage groups to each of the 38 autosomes. RH mapping of 102 chromosome-specific markers similarly allowed us to assign 72 RH groups to all chromosomes comprising the dog karyotype, and provide an integrated cytogenetic, meiotic linkage, and RH map of the dog. Moreover, FISH mapping of other previously RH-mapped markers strengthen the chromosomal assignments made in this study, for example, GALK1, RARA, THRA1, NF1, and KRT17 on CFA 9 (Werner et al. 1997; Miller et al. 1999), C04107 on CFA10 (Van der Sluis et al. 1999), PROP1 on CFA11 (Lantinga-van Leuween et al. 2000a), and PIT1 on CFA31 (Lantinga-van Leuween et al. 2000b). Sixteen of the 19 integrated assignments reported by Werner et al. (1999) and Mellersh et al. (2000) are in agreement with the nomenclature used in this study. However, there are three discrepancies in chromosome nomenclature. In this report the chromosomes we refer to as CFA 17, CFA 23, and CFA 27 were described by Werner et al. (1999) and Mellersh et al. (2000) as CFA 15, CFA 22, and CFA 16, respectively. Reciprocal exchange of probes from both laboratories now confirms that the nomenclature used here is correct (P. Werner and M. Breen, data not shown). Also in this study, when possible, we used RH and meiotic linkage mapping to position chromosome-specific markers that had been FISH-mapped to both the proximal and distal ends of each chromosome. This approach allowed us to orientate all but four of the integrated RH and meiotic groups on the chromosomes to which they have been assigned. The exceptions are those groups assigned to CFA 21, CFA 32, CFA 35, and CFA 36, which will require additional markers to reveal their true orientations. In the previous meiotic (Werner et al. 1999) and RH (Mellersh et al. 2000) maps, calculation of the coverage was based on theoretical considerations. Integration of the RH, meiotic, and cytogenetic maps allowed a more accurate assessment of chromosome coverage, as is demonstrated clearly in Figure 1. Good coverage is evident for all autosomes, with the exception of half of CFA 32 and X as well as two-thirds of CFA 36. The integrated RH/cytogenetic map would, therefore, confirm genome coverage >90%. Chromosomes for which the map coverage is less than complete can now be targeted by specific efforts to provide full coverage. For instance, the meiotic linkage maps for three chromosomes in particular, CFA 6, CFA 19, and CFA 24, appear to be short (to varying extents) at the proximal and/or distal ends, and the map for CFA 17 appears not to represent the distal half of the chromosome. In all these cases we will aim to select the markers at the ends of both the RH and meiotic maps, in order to isolate BAC clones that may then be FISH-mapped to determine the true extent of the chromosome coverage of the existing maps.
The meiotic linkage groups assigned to seven chromosomes (CFA 21, CFA 32, CFA 33, CFA 34, CFA 35, CFA 36, and CFA 38) do not yet contain a sufficient number of appropriately placed markers to accurately determine the extent of chromosome coverage. We are therefore pursuing these chromosomes from a comparative angle by targeting type I markers that have been mapped in the corresponding evolutionarily conserved chromosome segments as identified by Breen et al. (1999c) and Yang et al. (1999).
The present study provides a significant advance in the development of an effective genome map of the dog. The genome-wide integration of the meiotic and RH maps with the cytogenetic map, together with the increase in the number and density of RH-mapped markers, will improve our ability to use the canine genome to identify important genes in major ways. It will now be possible to assign accurately markers linked to canine disease genes to their corresponding position in the human genome, using the set of conserved genes presented in this study in combination with previously established gross levels of conserved synteny, reported by Breen et al. (1999c) and Yang et al. (1999). This, in turn, will provide a mechanism for identifying appropriate positional candidate genes whose disease-causing roles can subsequently be investigated. In addition, the significant increase in the number of informative microsatellites on the RH map will facilitate the direct identification of a disease-associated gene from that of a linked marker. The 561 polymorphic di- and tetranucleotide microsatellites in the integrated map will provide complete coverage of the dog genome because any position in the genome is within an average of 10 cM of at least one of these microsatellites. Finally, this integrated map will facilitate the cloning of desired genes, because it allows us to exploit the dog as a powerful new genetic system for mapping complex traits that have been difficult to identify through corresponding studies of human families.
METHODS
Generation of Chromosome-Specific Data
A dog cosmid genomic library in pWE15 (Stratagene) was gridded manually and screened for the presence of CA/GT repeats, using high stringency, as described previously (Holmes et al. 1993). A total of 2304 cosmids were screened, and 328 (26%) were found to be positive. DNA from all positive clones was prepared using either a QIAprep (QIAGEN) or Wizard Plus SV (Promega).
Fluorescence in Situ Hybridization (FISH)
Metaphase chromosomes of the dog and all probes were prepared for FISH as described by Breen et al. (1999b). Posthybridization stringency washes were performed in 50% formamide: 2× SSC for 3 min at 42°C (three washes), followed by 2× SSC for 3 min at 42°C (three washes). Immunological detection of the probes was performed with Texas Red conjugated avidin DCS (Vector Labs; 1:500) and FITC conjugated anti-digoxin (Sigma; 1:500) for biotin-labeled and digoxigenin-labeled probes, respectively. Chromosomes were counterstained in 80 ng/mL 4‘,6-diaminidino-2-phenylindole (DAPI) and mounted in antifade solution (Vectashield, Vector Laboratories).
Image Acquisition and Chromosome Assignment
Images were acquired and processed using a FISH workstation comprising a fluorescence microscope (Axiophot, Zeiss) equipped with an FITC/Texas Red/DAPI excitation filter set and a cooled CCD camera (KAF 1400, Photometrics), both driven by dedicated software (SmartCapture, Vysis Inc.). The digital image of each DAPI-stained metaphase spread was processed using a high-pass spatial filter to reveal enhanced DAPI bands. Each clone was assigned to a chromosome band according to the DAPI-banded nomenclature of Breen et al. (1999a). Verification of the chromosomal assignment of all clones was made by subsequent cohybridization in the presence of a differentially labeled chromosome paint probe (Langford et al. 1996). In this way a panel of 266 chromosome-specific cosmid clones was developed from which a subset was selected for subsequent analysis.
RH Map Data
Generation of RH Markers
Accession numbers and marker characteristics appear at http://www-recomgen.univ-rennes1.fr/doggy.html.
Microsatellite markers with the motif (CA)n were isolated from a small-insert dog genomic library (Jouquand et al. 2000b). Tri- and tetranucleotides were described in Mellersh et al. (2000) and Jouquand et al. (2000b). The degree of polymorphism was determined using five mongrel dogs as described previously (Jouquand et al. 1999, 2000b). The heterozygosity value was calculated as follows: Het = 1 − Σ (Pi)2, with Pi corresponding to allele frequencies. Dog gene markers and dog ESTs from a retinal cDNA library were retrieved from public databases. Primers were designed utilizing the Primer3 program.
Chromosome-Specific Markers
Sequence information was obtained from at least one chromosome-specific cosmid clone per chromosome and, where possible, from at least two clones that had been FISH-mapped to proximal and distal ends of the corresponding chromosome. For each chromosome-specific marker, sequence data were generated from the corresponding cosmid clone, in the flanking regions of each microsatellite locus. For all these markers, primers were designed to amplify fragments of 200–500 bp under unique PCR conditions, as described previously (Priat et al. 1999). Primer pairs were initially screened using 50 ng of dog template DNA, 50 ng of hamster template, and a 1:3 mix of dog and hamster DNA. Examination of the resulting PCR products by agarose gel electrophoresis allowed for selection of markers that gave the correct size PCR products, and that produced canine-specific products that were easily distinguished from any hamster product.
RH Typing and RH Map Construction
Markers were typed on the RHDF5000-2 dog/hamster radiation hybrid panel. This panel, consisting of 118 hybrids, was selected from the original RHDF5000 panel (Vignaux et al. 1999). Amplification reactions from all markers were performed as previously described (Priat et al. 1998).
One-third of the (CA)n microsatellite markers were typed by agarose gel electrophoresis (Priat et al. 1998; Mellersh et al. 2000), and the remainder by the fluorescent detection method previously described (Jouquand et al. 2000a). Markers were scored and data were recorded using suitable data acquisition software (G. Brenterch and N. Soriano, pers. comm.). Map construction was performed using the MultiMap software (Matise et al. 1994). RH linkage groups were constructed using the find-all-linkage-groups function of MultiMap. Within each RH group, markers were then ordered using Multipoint analysis, and distances were calculated as follows: D = −ln(1 − θ), where θ is the breakage frequency. Distances are referred to as centirays (cR5000), in reference to the 5000-rad value used to construct the panel.
Meiotic Linkage Data
A subset of 38 chromosome-specific cosmid clones was processed to characterize their respective microsatellite markers.
Subcloning and Identification of Microsatellite Markers
DNA from at least one clone per chromosome was subjected to restriction digestion with Sau3A1, HaeIII, or Alu1, and the resulting fragments were subcloned into either the BamH1 site of M13mp18 or the HincII site of M13mp19 (Oncor Appligene). Single-strand preparations or PCR products were sequenced from either end of the insert, with M13 primers using an ABI-377 sequencer (PE Biosystems). For clones in which the CA/GT repeat lay outside the region of readable sequence, a set of six primers [(GT)10N, where N = A, C, or T; (TG)10N, where N = A, G, or C] was used to sequence outward from the repeat to identify flanking sequences suitable for the design of primers to amplify the microsatellite. Microsatellites characterized at the Animal Health Trust are denoted by the prefix AHT, and those characterized at the University of Leicester are denoted by the prefix LEI.
Design of Suitable Locus-Specific PCR Primers
PCR primers were designed either manually or with the aid of the program PRIMER v3. All primers were synthesized commercially (Genset; Amersham Pharmacia Biotech). PCR reactions were carried out on an MJ-Tetrad thermal cycler. Microsatellites were typed either by incorporation of fluorescently labeled dUTP during the PCR and analyzed on an ABI-377 sequencer with GENESCAN and GENOTYPER software (PE Biosystems) or by end-labeling of one primer with [γ33P]dATP using T4 polynucleotide kinase.
Genotyping of Chromosome-Specific Microsatellites
Microsatellites were genotyped on a panel of canine reference pedigrees described previously (Mellersh et al. 1997; Neff et al. 1999). The reference panel comprises 16 interrelated three-generation pedigrees and contains 212 individuals, including 163 F2 offspring. Several distinct breeds of dog are represented in the panel including Miniature and Toy Poodles, Norwegian Elkhound, Irish Setter, and several genetically distinct lines of Beagle. DNA from the reference pedigrees is available through Ralston Purina: http://www.purina.com/dogs/index.html and http://www.petgenome.com. Markers were genotyped and double-scored as described previously (Mellersh et al. 1997).
Meiotic Linkage Map Construction
Genotyping data from our panel of chromosome-specific microsatellites were merged with data from previous studies (Werner et al. 1999; Mellersh et al. 2000). Linkage maps were constructed with the computer program MultiMap (Matise et al. 1994) as described previously (Werner et al. 1999; Mellersh et al. 2000). Markers were assigned to linkage groups using the find-all-linkage-groups function of MultiMap; markers in each group were linked to at least one other marker with a recombination fraction <0.4 supported by a LOD score of at least 5.0 (equivalent to odds of 100,000:1 in favor of linkage). A sex-averaged, framework map and a comprehensive map were constructed for each linkage group using MultiMap. The comprehensive map is shown in Figure 1.
Human/Dog Comparative Mapping
Orthologous human genes have been defined by BLAST searches (Altschul et al. 1990) against public databases (GenBank “nr” and “HTGS”) in February 2001; default BLAST criteria were used. Chromosomal locations have been found in GeneAtlas (http://www.citi2.fr/GENATLAS) and LocusLink (http://www.ncbi.nlm.nih.gov/LocusLink).
Web Sites
Comprehensive data of the integrated map (tables and figures) and the characteristics of all markers will appear on the following linked Web sites: http://www-recomgen.univ-rennes1.fr/doggy.html, http://www.aht.org.uk/cytogenetic-map/dog.html, and http://www.fhcrc.org/science/dog_genome/dog.html.
Acknowledgments
We thank Sally Debenham and Philip Ricketts (Animal Health Trust) for technical help. Canine genome analysis at the Animal Health Trust and the University of Leicester is generously supported by the Guide Dogs for the Blind Association. F.G. is supported by funds from CNRS, Conseil Regional de Bretagne, and the American Kennel Club/Canine Health Foundation. We thank Thierry Giffon and Edouard Cadieu for their help in RH mapping as well as Stephane Dréano for sequencing and Bruno Coutard, Valérie Lelaure, and Hervé Cartron for their help in microsatellite isolation, and Tara Matise for her help in the use of the MultiMap package. E.A.O. is supported by grants from the American Kennel Club/Canine Health Foundation, the American Cancer Society, and a Burroughs Wellcome Fund Award for Excellence in Innovative Genomics. C.S.M. is generously supported by a fellowship from Ralston Purina. We are grateful to the American Kennel Club Canine Health Foundation and Ralston Purina for sponsoring the production of the integrated map as a poster. We also thank the Geraldine Rockefeller Dodge Foundation.
Footnotes
E-MAIL mtbreen@hgmp.mrc.ac.uk; FAX 44-1638-750794
E-MAIL Francis.Galibert@univ-rennes1.fr; FAX 33-2993-36200.
Article and publication are at http://www.genome.org/cgi/doi/10.1101/gr.189401.
REFERENCES
- Acland GM, Ray K, Mellersh CS, Gu W, Langston AA, Rine J, Ostrander EA, Aguirre GD. Linkage analysis and comparative mapping of canine progressive rod-cone degeneration (prcd) establishes potential locus homology with retinitis pigmentosa (RP17) in humans. Proc Nat Acad Sci. 1998;95:3048–3053. doi: 10.1073/pnas.95.6.3048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Acland GM, Ray K, Mellersh CS, Langston AA, Rine J, Ostrander EA. A novel retinal degeneration locus identified by linkage and comparative mapping of canine early retinal degeration. Genomics. 1999;59:134–142. doi: 10.1006/geno.1999.5842. [DOI] [PubMed] [Google Scholar]
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Breen M, Reimann N, Bosma AA, Landon D, Zijlstra C, Bartnitzke S, Switonski M, Long SE, de Haan NA, Binns MM, et al. Proceedings of the 13th European Colloquium on Cytogenetics of Domestic Animals. Budapest. 1998. Standardisation of the chromosome nos. 22–38 of the dog (Canis familiaris) with the use of chromosome painting probes. [Google Scholar]
- Breen M, Bullerdiek J, Langford CF. The DAPI banded karyotype of the domestic dog (Canis familiaris) generated using chromosome-specific paint probes. Chrom Res. 1999a;7:401–406. doi: 10.1023/a:1009224232134. ; erratum 7: 575. [DOI] [PubMed] [Google Scholar]
- Breen M, Langford CF, Carter NP, Holmes NG, Dickens HF, Thomas R, Suter N, Ryder EJ, Pope M, Binns MM. FISH mapping and identification of canine chromosomes. J Hered. 1999b;90:27–30. doi: 10.1093/jhered/90.1.27. [DOI] [PubMed] [Google Scholar]
- Breen M, Thomas R, Binns MM, Carter NP, Langford CF. Reciprocal chromosome painting reveals detailed regions of synteny between the karyotypes of the domestic dog (Canis familiaris) and human. Genomics. 1999c;61:145–155. doi: 10.1006/geno.1999.5947. [DOI] [PubMed] [Google Scholar]
- Dodds WJ. Hemostasis. In: Kaneko JJ, Bruss ML, editors. Clinical biochemistry of domestic animals. San Diego: Academic Press; 1989. pp. 282–315. [Google Scholar]
- Galibert, F., André, C., Chéron, A., Chuat, J.C., Hitte, C., Jiang, Z., Jouquand, S., Priat, C., Rénier, C., and Vignaux, F. 1998. The importance of the canine model in medical genetics. Bull. Acad. Natl. Med. 182: 818–821. [PubMed]
- Henthorn PS, Somberg RL, Fimiani VM, Puck JM, Patterson DF, Felsburg PJ. IL-2Rγ gene microdeletion demonstrates that canine X-linked severe combined immunodeficiency is a homologue of the human disease. Genomics. 1994;23:69–74. doi: 10.1006/geno.1994.1460. [DOI] [PubMed] [Google Scholar]
- Holmes NG, Mellersh CS, Humphreys SJ, Binns MM, Holliman A, Curtis R, Sampson J. Isolation and characterization of microsatellites from the canine genome. Anim Genet. 1993;24:289–292. doi: 10.1111/j.1365-2052.1993.tb00313.x. [DOI] [PubMed] [Google Scholar]
- Jauch A, Wienberg J, Stanyon R, Arnold N, Tofanelli S, Ishida T, Cremer T. Reconstruction of genomic rearrangements in great apes and gibbons by chromosome painting. Proc Natl Acad Sci. 1992;89:8611–8615. doi: 10.1073/pnas.89.18.8611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang Z, Priat C, Galibert F. Traced orthologous amplified sequence tags (TOASTs) and mammalian comparative maps. Mamm Genome. 1998;9:577–587. doi: 10.1007/s003359900821. [DOI] [PubMed] [Google Scholar]
- Jónasdóttir TJ, Mellersh CS, Moe L, Heggebø R, Gamlem H, Ostrander EA, Lingaas F. Genetic mapping of a naturally occurring hereditary renal cancer syndrome in dogs. Proc Natl Acad Sci. 2000;97:4132–4137. doi: 10.1073/pnas.070053397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jouquand, S., Chéron, A., and Galibert, F. 1999. Microsatellite analysis using a two-step procedure for fluorescence labeling of PCR products. Biotechniques 526: 902–905. [DOI] [PubMed]
- Jouquand, S., André, C., Chéron, A., Hitte, C., Chuat, J.C., and Galibert, F. 2000a. Using the fluorogenic 5′ nuclease assay for high-throughput detection of (CA)n repeats in radiation hybrid mapping. Biotechniques 28: 754–758, 760–762, 764–765. [DOI] [PubMed]
- Jouquand S, Priat C, Hitte C, Lachaume P, André C, Galibert F. Identification and characterization of a set of 100 tri- and dinucleotide microsatellites in the canine genome. Animal Genet. 2000b;31:266–272. doi: 10.1046/j.1365-2052.2000.00642.x. [DOI] [PubMed] [Google Scholar]
- Langford CF, Fischer PE, Binns MM, Holmes NG, Carter NP. Chromosome-specific paints from a high resolution flow karyotype of the dog. Chrom Res. 1996;4:115–123. doi: 10.1007/BF02259704. [DOI] [PubMed] [Google Scholar]
- Lantinga-van Leeuwen IS, Kooistra HC, Moll JA, Renier C, Breen M, van Oost BA. The canine Prop1 gene: Genomic structure, mapping, and evaluation as a candidate gene for combined pituitary hormone deficiency in German Shepherd dogs. Cytogenet Cell Genet. 2000a;88:140–144. doi: 10.1159/000015507. [DOI] [PubMed] [Google Scholar]
- Lantinga-van Leeuwen IS, Kooistra HS, Moll JA, Rijnberk A, Breen M, Renier C, van Oost B. Cloning of the canine gene encoding transcription factor Pit-1 and mutation analysis in a canine model of pituitary dwarfism. Mamm Genome. 2000b;11:31–36. doi: 10.1007/s003350010006. [DOI] [PubMed] [Google Scholar]
- Leeb T, Kopp T, Deppe A, Breen M, Matis U, Brunnberg L, Brenig B. No vidence for protein C gene defects in canine Legg-Calve-Perthes disease. Mamm Genome. 1999;10:134–139. doi: 10.1007/s003359900958. [DOI] [PubMed] [Google Scholar]
- Lin L, Faraco J, Li R, Kadotani H, Rogers W, Lin X, Qiu X, de Jong PJ, Nishino S, Mignot E. The sleep disorder canine narcolepsy is caused by a mutation in the hypocretin (orexin) receptor 2 gene. Cell. 1999;98:365–376. doi: 10.1016/s0092-8674(00)81965-0. [DOI] [PubMed] [Google Scholar]
- Lingaas F, Sorensen A, Juneja RK, Johansson S, Fredholm M, Wintero AK, Sampson J, Mellersh CS, Curzon A, Holmes NG, et al. Towards construction of a canine linkage map: Establishment of 16 linkage groups. Mamm Genome. 1997;8:218–221. doi: 10.1007/s003359900393. [DOI] [PubMed] [Google Scholar]
- Lingaas F, Aarskaug T, Sletten M, Bjerkas I, Grimholt U, Moe L, Juneja RK, Wilton AN, Galibert F, Holmes NG, et al. Genetic markers linked to neuronal ceroid lipofuscinosis in English Setter dogs. Animal Genet. 1998;29:371–376. doi: 10.1046/j.1365-2052.1998.295358.x. [DOI] [PubMed] [Google Scholar]
- Matise TC, Perlin M, Chakravarti A. Automated construction of genetic linkage maps using an expert system (MultiMap): A human genome linkage map. Nat Genet. 1994;6:384–390. doi: 10.1038/ng0494-384. [DOI] [PubMed] [Google Scholar]
- McCarthy LC, Terrett J, Davis ME, Knights CJ, Smith AL. A first-generation whole genome-radiation hybrid map spanning the mouse genome. PCR Methods Appl. 1997;7:1153–1161. doi: 10.1101/gr.7.12.1153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mellersh CS, Langston AA, Acland GM, Fleming MA, Ray K, Wiegand NA, Fransisco LV, Gibbs M, Aguirre GD, Ostrander EA. A linkage map of the canine genome. Genomics. 1997;46:326–336. doi: 10.1006/geno.1997.5098. [DOI] [PubMed] [Google Scholar]
- Mellersh CS, Hitte C, Richman M, Vignaux F, Priat C, Jouquand S, Werner P, André C, DeRose S, Patterson DF, et al. An integrated linkage-radiation hybrid map of the canine genome. Mamm Genome. 2000;11:120–130. doi: 10.1007/s003350010024. [DOI] [PubMed] [Google Scholar]
- Menon KP, Tieu PT, Neufeld EF. Architecture of the canine IDUA gene and mutation underlying canine mucopolysaccharidosis. Genomics. 1992;14:763–768. doi: 10.1016/s0888-7543(05)80182-x. [DOI] [PubMed] [Google Scholar]
- Miller AB, Breen M, Murphy KE. Chromosomal localization of acidic and basic keratin gene clusters of Canis familiaris. Mamm Genome. 1999;10:371–375. doi: 10.1007/s003359901004. [DOI] [PubMed] [Google Scholar]
- Nash WG, Wienberg J, Ferguson-Smith MA, Menninger JC, O’Brien SJ. Comparative genomics: Tracking chromosome evolution in the family Ursidae using reciprocal chromosome painting. Cytogenet Cell Genet. 1998;83:182–192. doi: 10.1159/000015176. [DOI] [PubMed] [Google Scholar]
- Neff MW, Bromann KW, Mellersh CS, Ray K, Acland GM, Aguirre GD, Ziegle JS, Ostrander EA, Rine J. A second-generation genetic linkage map of the domestic dog, Canis familiaris. Genetics. 1999;151:803–820. doi: 10.1093/genetics/151.2.803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olivier M, Breen M, Binns MM, Lust G. Localization and characterisation of nucleotide sequences from the canine Y chromosome. Chrom Res. 1999;7:223–233. doi: 10.1023/a:1009203500926. [DOI] [PubMed] [Google Scholar]
- Ostrander EA, Giniger E. Semper fidelis: What man's best friend can teach us about human biology and disease. Am J Hum Genet. 1997;61:47–80. doi: 10.1086/515522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ostrander EA, Galibert F, Patterson DF. Canine genetics comes of age. Trends Genet. 2000;16:117–124. doi: 10.1016/s0168-9525(99)01958-7. [DOI] [PubMed] [Google Scholar]
- Patterson DF, Haskins ME, Jezyk PF, Giger U, Meyers-Wallen VN, Aguirre G, Fyfe JC, Wolfe JH. Research on genetic diseases: Reciprocal benefits to animals and man. J Am Vet Med Assoe. 1988;193:1131–1144. [PubMed] [Google Scholar]
- Patterson DF. Companion animal medicine in the age of medical genetics. J Vet Internal Med. 2000;14:1–9. [PubMed] [Google Scholar]
- Patterson DF. Canine genetic disease information system: A computerized knowledge base of genetic diseases in the dog. St. Louis: Mosby-Harcourt; 2001. (in press). [Google Scholar]
- Priat, C., Hitte, C., Vignaux, F., Renier, C., Jiang, Z., Jouquand, S., Chéron, A., André, C., and Galibert, F. 1998. A whole-genome radiation hybrid map of the dog genome. Genomics 54: 361–378. [DOI] [PubMed]
- Priat C, Jiang ZH, Renier C, André C, Galibert F. Characterization of 463 type I markers suitable for dog genome mapping. Mamm Genome. 1999;10:803–813. doi: 10.1007/s003359901095. [DOI] [PubMed] [Google Scholar]
- Reimann N, Bartnitzke S, Bullerdiek J, Schmitz U, Rogalla P, Nolte I, Ronne M. An extended nomenclature of the canine karyotype. Cytogenet Cell Genet. 1996;73:140–144. doi: 10.1159/000134326. [DOI] [PubMed] [Google Scholar]
- Ryder EJ. “Genetic mapping in the dog towards localizing the genes responsible for progressive retinal atrophy in Miniature Long-Haired Dachshunds and Labrador Retrievers.” Ph.D. thesis. Kentford, UK: Animal Health Trust, Open University; 2000. [Google Scholar]
- Sargan DR, Yang F, Squire M, Milne BS, O'Brien PC, Ferguson-Smith MA. Use of flow-sorted canine chromosomes in the assignment of canine linkage, radiation hybrid, and syntenic groups to chromosomes: Refinement and verification of the comparative chromosome map for dog and human. Genomics. 2000;69:182–195. doi: 10.1006/geno.2000.6334. [DOI] [PubMed] [Google Scholar]
- Schatzberg SJ, Olby NJ, Breen M, Anderson LVB, Langford CF, Dickens HF, Wilton SD, Zeiss CJ, Binns MM, Kornegay JN, et al. Molecular analysis of a spontaneous dystrophin “knockout” dog. Neuro Disorders. 1999;9:289–295. doi: 10.1016/s0960-8966(99)00011-5. [DOI] [PubMed] [Google Scholar]
- Sharp NJH, Kornegay JN, van Camp SD, Herbstreith HH, Secore SL, Kettle S, Hung WY, Constantinou CD, Dykstra MJ, Roses AD, et al. An error in dystrophin mRNA processing in Golden Retriever muscular dystrophy, an animal homologue of Duchenne muscular dystophy. Genomics. 1992;13:115–121. doi: 10.1016/0888-7543(92)90210-j. [DOI] [PubMed] [Google Scholar]
- Stewart EA, McKusick KB, Aggarwal A, Bajorek E, Brady S, Chu A, Fang N, Hadley D, Harris M, Hussain S, et al. An STS-based radiation hybrid map of the human genome. Genome Res. 1997;7:42–33. doi: 10.1101/gr.7.5.422. [DOI] [PubMed] [Google Scholar]
- Stolzfus LJ, Sosa-Pineda B, Moskowitz SM, Menon KP, Dlott B, Hooper L, Teplow DB, Shull RM, Neufeld EF. Cloning and characterisation of cDNA encoding canine α-L-iduronidase. J Biol Chem. 1992;267:6570–6575. [PubMed] [Google Scholar]
- Switonski M, Reimann N, Bosma AA, Long S, Bartnitzke S, Pienkowska A, Moreno-Milan MM, Fischer P. Report on the progress of standardisation of the G-banded canine (Canis familiaris) karyotype. Chrom Res. 1996;4:306–309. doi: 10.1007/BF02263682. [DOI] [PubMed] [Google Scholar]
- Thomas R, Breen M, Binns MM. Chromosome assignment of six dog genes by FISH, and correlation with dog–human Zoo-FISH data. Animal Genetics. 2001a;32:148–151. doi: 10.1046/j.1365-2052.2001.00731.x. [DOI] [PubMed] [Google Scholar]
- Thomas R, Breen M, Deloukas P, Holmes NG, Binns MM. An integrated cytogenetic, radiation-hybrid and comparative map of dog chromosome 5. Mamm Genome. 2001b;12:371–375. doi: 10.1007/s003350010287. [DOI] [PubMed] [Google Scholar]
- van der Sluis BJA, Breen M, Nanji M, van Wolferen M, de Jong P, Binns MM, Pearson PL, Kuipers J, Rothuizen J, Cox DW, et al. Genetic mapping of the copper toxicosis locus in Bedlington Terriers to dog chromosome 10, in a region syntenic to human chromosome region 2p13–16. Hum Mol Genet. 1999;8:501–507. doi: 10.1093/hmg/8.3.501. [DOI] [PubMed] [Google Scholar]
- Veske A, Nilsson SEG, Narfstrom K, Gal A. Retinal dystrophy of Swedish Briards/Briard-Beagle dogs is due to a 4bp deletion in RPE65. Genomics. 1999;57:57–61. doi: 10.1006/geno.1999.5754. [DOI] [PubMed] [Google Scholar]
- Vignaux F, Hitte C, Priat C, Chuat JC, André C, Galibert F. Construction and optimization of a dog whole-genome radiation hybrid panel. Mamm Genome. 1999;10:888–894. doi: 10.1007/s003359901109. [DOI] [PubMed] [Google Scholar]
- Werner P, Raducha MG, Prociuk U, Henthorn PS, Patterson DF. Physical and linkage mapping of human chromosome 17 loci to dog chromosomes 9 and 5. Genomics. 1997;42:74–82. doi: 10.1006/geno.1997.4723. [DOI] [PubMed] [Google Scholar]
- Werner P, Mellersh CS, Raducha MG, DeRose S, Acland GM, Prociuk U, Wiegand N, Aguirre GD, Henthorn PS, Patterson DF, et al. Anchoring of canine linkage groups with chromosome-specific markers. Mamm Genome. 1999;10:814–823. doi: 10.1007/s003359901096. [DOI] [PubMed] [Google Scholar]
- Wienberg J, Stanyon R, Jauch A, Cremer T. Homologies in human and Macaca fuscata chromosomes revealed by in situ suppression hybridisation with human chromosome specific DNA libraries. Chromosoma. 1992;101:265–270. doi: 10.1007/BF00346004. [DOI] [PubMed] [Google Scholar]
- Yang F, O'Brien PCM, Milne BS, Graphodatsky AS, Solanky N, Trifonov V, Rens W, Sargan D, Ferguson-Smith MA. A complete comparative chromosome map for the dog, red fox, and human and its integration with canine genetic maps. Genomics. 1999;62:189–202. doi: 10.1006/geno.1999.5989. [DOI] [PubMed] [Google Scholar]
- Yuzbasiyan-Gurkan V, Blanton SH, Cao Y, Ferguson P, Li J, Venta PJ, Brewer GJ. Linkage of a microsatellite marker to the canine copper toxicosis locus in Bedlington Terriers. Am J Vet Res. 1997;58:23–27. [PubMed] [Google Scholar]
- Zheng K, Thorner PS, Marrano P, Baumal R, McInnes RR. Canine X chromosome-linked hereditary nephritis: A genetic model for human X-linked hereditary nephritis resulting from a single base mutation in the gene encoding the α5 chain of collagen type IV. Proc Natl Acad Sci. 1994;91:3989–3993. doi: 10.1073/pnas.91.9.3989. [DOI] [PMC free article] [PubMed] [Google Scholar]