Abstract
Eukaryotic genomes contain significant amounts of transposons and repetitive DNA elements, which, if transcribed, can be detrimental to the organism. Expression of these elements is suppressed by establishment of repressive chromatin modifications. In Arabidopsis thaliana, they are silenced by the siRNA–mediated transcriptional gene silencing pathway where long non-coding RNAs (lncRNAs) produced by RNA Polymerase V (Pol V) guide ARGONAUTE4 (AGO4) to chromatin and attract enzymes that establish repressive chromatin modifications. It is unknown how chromatin modifying enzymes are recruited to chromatin. We show through chromatin immunoprecipitation (ChIP) that SPT5L/KTF1, a silencing factor and a homolog of SPT5 elongation factors, binds chromatin at loci subject to transcriptional silencing. Chromatin binding of SPT5L/KTF1 occurs downstream of RNA Polymerase V, but independently from the presence of 24-nt siRNA. We also show that SPT5L/KTF1 and AGO4 are recruited to chromatin in parallel and independently of each other. As shown using methylation-sensitive restriction enzymes, binding of both AGO4 and SPT5L/KTF1 is required for DNA methylation and repressive histone modifications of several loci. We propose that the coordinate binding of SPT5L and AGO4 creates a platform for direct or indirect recruitment of chromatin modifying enzymes.
Author Summary
Transposons and other repetitive elements occupy vast areas of the eukaryotic genomes. They pose a threat to genome integrity but at the same time regulate expression of many genes and have been proposed to be a major factor contributing to genome evolution. One of the processes responsible for controlling activity of transposons and other repetitive elements is transcriptional gene silencing. This process uses small interfering RNA and long non-coding RNA to recruit enzymes that establish repressive chromatin modifications. Several proteins have been identified to be needed for siRNA–mediated transcriptional silencing in Arabidopsis thaliana, however for many of them their position in the silencing pathway is unknown. One of those proteins is SPT5L/KTF1, a homolog of an elongation factor associated with RNA Polymerase II. Here we establish the position of SPT5L in the silencing pathway and propose the molecular mechanism of its function. This gives further knowledge of the mechanism of transcriptional gene silencing and is important to understand how transposons are controlled.
Introduction
Eukaryotic genomes contain significant amounts of transposons and other repetitive DNA elements, which usually remain transcriptionally inactive. Efficient silencing of transposon transcription is essential for preventing their mobility and for maintaining genome integrity [1]. Transposon silencing has also been hypothesized to regulate expression of genes that contain transposable elements in their promoters and to facilitate the evolution of genomes [2].
Transposons are silenced at both transcriptional and post-transcriptional levels by mechanisms that involve small interfering RNAs (siRNAs) [3]. These 20–25-nt RNA molecules are generated by the RNase III enzyme Dicer and provide sequence specificity for effector complexes mediating RNA cleavage and/or the establishment of chromatin modifications that silence transcriptional activity [3]. In Arabidopsis thaliana, single-stranded RNA precursors for siRNA biogenesis are produced by RNA Polymerase II (Pol II) or RNA Polymerase IV (Pol IV), while the second strand is synthesized by RDR2 (RNA-Dependent RNA Polymerase 2). DCL3 (Dicer-like 3) cleaves double-stranded RNA into siRNAs that are then incorporated into ARGONAUTE4 (AGO4) [4], [5]. This mechanism seems to be similar in maize where homologs of RDR2 and Pol IV have been shown to be involved in transcriptional gene silencing [6]–[8].
Recognition of target loci by AGO4-siRNA complexes requires sequence identity between siRNAs and the genomic loci. These loci, however, are often actively transcribed, and it is not clear if siRNAs base-pair interact with DNA or nascent RNA transcripts [3], [9]. The latter possibility is well supported in Schizosaccharomyces pombe where loci subject to siRNA-mediated transcriptional silencing are actively transcribed by RNA Polymerase II [10]–[12]. The central role of nascent transcripts in recognition of siRNA targets in S. pombe was observed by the ability of Argonaute proteins to cleave RNA. This ability is required for the establishment of repressive chromatin modifications [13]. Moreover, tethering Argonaute and siRNA-containing RITS (RNA-induced initiation of transcriptional gene silencing) complex to nascent transcripts is sufficient for the initiation of repressive chromatin modifications and transcriptional silencing [14].
This mechanism may be similar in Arabidopsis where transcriptional silencing requires a specialized RNA Polymerase complex known as RNA Polymerase V (Pol V) [15]–[17]. Pol V produces non-coding transcripts in otherwise silent chromatin, and its activity is required for the establishment and maintenance of repressive chromatin modifications [18]. Pol V-produced non-coding transcripts physically interact with AGO4 and recruit siRNA-AGO4 complexes to their targets [19]. Additionally, transcriptional silencing of several loci needs AGO4 slicer activity [20], suggesting that in plants siRNAs may recognize their targets by base-pair interactions with Pol V transcripts [19].
RNA Polymerases and AGO4 are assisted in their functions by several other known protein components of the plant silencing system, all of which are required for efficient establishment and maintenance of transcriptional silencing [5]. One of them is SPT5L (Suppressor of Ty insertion 5 - like; also known as SPT5-like or KTF1), a homolog of SPT5 Pol II-associated elongation factor. It was shown to contain a domain rich in WG/GW repeats that facilitate physical interaction with AGO4 [21]–[23]. Because SPT5L interacts with RNA but is not required for the accumulation of Pol V-dependent transcripts, it was hypothesized to work downstream of Pol V and recruit AGO4 to Pol V-transcribed loci [22], [24].
Despite the recent progress in understanding the mechanisms of transcriptional gene silencing, it is not known how siRNAs work with Pol V transcripts, AGO4 and other proteins to recruit chromatin modifying enzymes to their target loci in chromatin. It is unknown how chromatin-bound AGO4 recruits enzymes that establish repressive chromatin modifications. It is unknown if other protein components of the silencing system help AGO4 recruit chromatin modifying enzymes. It is also unknown in what order proteins involved in silencing are recruited to chromatin. Here we try to resolve the mechanism of siRNA-mediated recruitment of chromatin modifying enzymes to chromatin and the function of SPT5L in this process. We show that SPT5L physically interacts with chromatin and that SPT5L works downstream of Pol V but does not require 24-nt siRNA. SPT5L and AGO4 are recruited to chromatin in parallel and at least partially independently of each other and both are needed for DNA methylation and repressive histone modifications at several loci. We propose that the coordinate binding of SPT5L and AGO4 creates a platform for direct or indirect recruitment of chromatin modifying enzymes.
Results
SPT5L interacts with chromatin
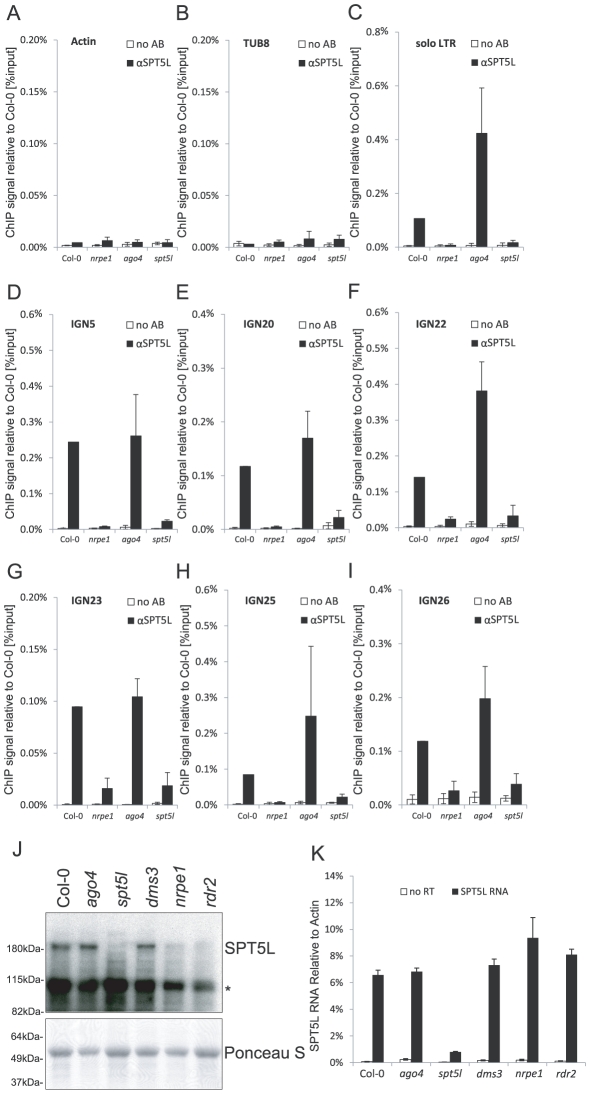
The interaction of SPT5L with AGO4 [21], [22] suggested that like AGO4 [19], SPT5L may bind loci targeted by siRNA-mediated transcriptional gene silencing. We first used chromatin immunoprecipitation (ChIP) with anti-SPT5L antibody to test if SPT5L binds chromatin. Subsequent real-time PCR demonstrated recovery of IGN5 and solo LTR DNA from Col-0 wild type at much higher levels than from spt5l mutant which represents the background level (Figure 1C, 1D). This shows that SPT5L physically interacts with IGN5 and solo LTR loci which are known to be transcribed by Pol V and silenced by the siRNA-mediated transcriptional gene silencing pathway [18], [19], [25], [26]. There was, however, no enrichment on the control Actin 2 and Tubulin 8 (TUB 8) loci (Figure 1A, 1B), which are transcribed by Pol II and not occupied by components of the silencing pathway [18], [27]. This suggests that SPT5L is present at the loci undergoing transcriptional silencing and that its function in silencing is most likely direct.
Figure 1. SPT5L interacts with chromatin in a Pol V-dependent and AGO4-independent manner.
(A–I) ChIP data showing SPT5L binding to chromatin in Col-0 wild type, nrpe1, ago4 and spt5l mutants at loci transcribed by Pol V and silenced by siRNA-mediated transcriptional silencing: solo LTR(C), IGN5 (D), IGN20 (E), IGN22 (F), IGN23 (G), IGN25 (H) and IGN26 (I). Two loci transcribed by Pol II are shown as controls: Actin 2 (A) and Tubulin 8 (B). No antibody controls (white bars) provide background level for ChIP samples (black bars). Bars represent mean value of ChIP signals normalized to Col-0 wild type. Error bars are standard deviations of three independent biological replicates. (J) Immunoblot detection of SPT5L in whole-cell protein extracts from Col-0 wild type, ago4, spt5l, dms3, nrpe1 and rdr2 mutants. Ponceau S staining of the membrane is a loading control. Asterisk denotes nonspecific bands. (K) Real time RT-PCR detection of SPT5L RNA in Col-0 wild type, ago4, spt5l, dms3, nrpe1 and rdr2 mutants. Bars represent average SPT5L mRNA accumulation relative to Actin 2 from three biological replicates. Error bars represent standard deviation.
Interaction of SPT5L with chromatin was also demonstrated at IGN20, IGN22, IGN23, IGN25 and IGN26 (Figure 1E–1I), which have been identified in a genome-wide screen of Pol V occupancy (A. Wierzbicki, R. Lister, B. Gregory, J. Ecker and C. Pikaard, unpublished data), suggesting that SPT5L binding may be a general feature of Pol V-transcribed loci.
SPT5L works downstream of Pol V
SPT5L interacts with chromatin (Figure 1C–1I) as well as with AGO4, Pol V complex and Pol V transcripts [21]–[23]. SPT5L is also not required for the accumulation of Pol V-dependent transcripts at IGN5, IGN6 or AtSN1 [22]. This suggests that SPT5L should work downstream of Pol V. To test this prediction we assayed Pol V binding to chromatin by ChIP with antibody against NRPE1, the largest subunit of Pol V. Subsequent real-time PCR demonstrated recovery of DNA from Col-0 wild type at much higher level than from the nrpe1 mutant at IGN5, solo LTR and AtSN1 loci but not at Actin 2 or Tubulin 8 loci (Figure 2A–2E) demonstrating that Pol V binds chromatin at IGN5, solo LTR and AtSN1 loci. DNA recovery from spt5l mutant was comparable to Col-0 wild type (Figure 2A–2E) showing that SPT5L is not needed for Pol V binding to chromatin. Interestingly, Pol V binding to chromatin was reproducibly increased at solo LTR locus in ago4 mutant (Figure 2C), indicating that AGO4 may inhibit Pol V binding to chromatin possibly by affecting initiation and/or elongation of Pol V transcription. We conclude that SPT5L does not work upstream of Pol V in siRNA-mediated transcriptional gene silencing pathway.
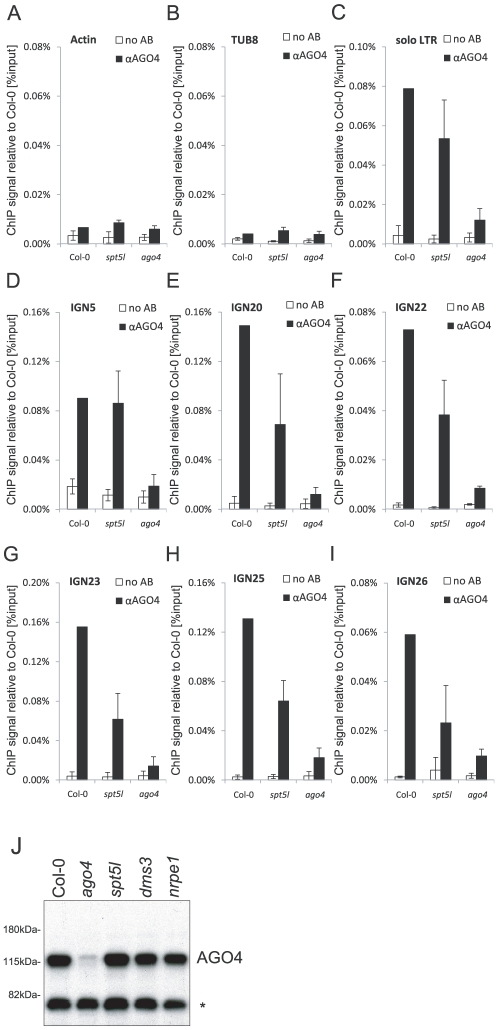
Figure 2. SPT5L and AGO4 are not required for Pol V binding to chromatin.
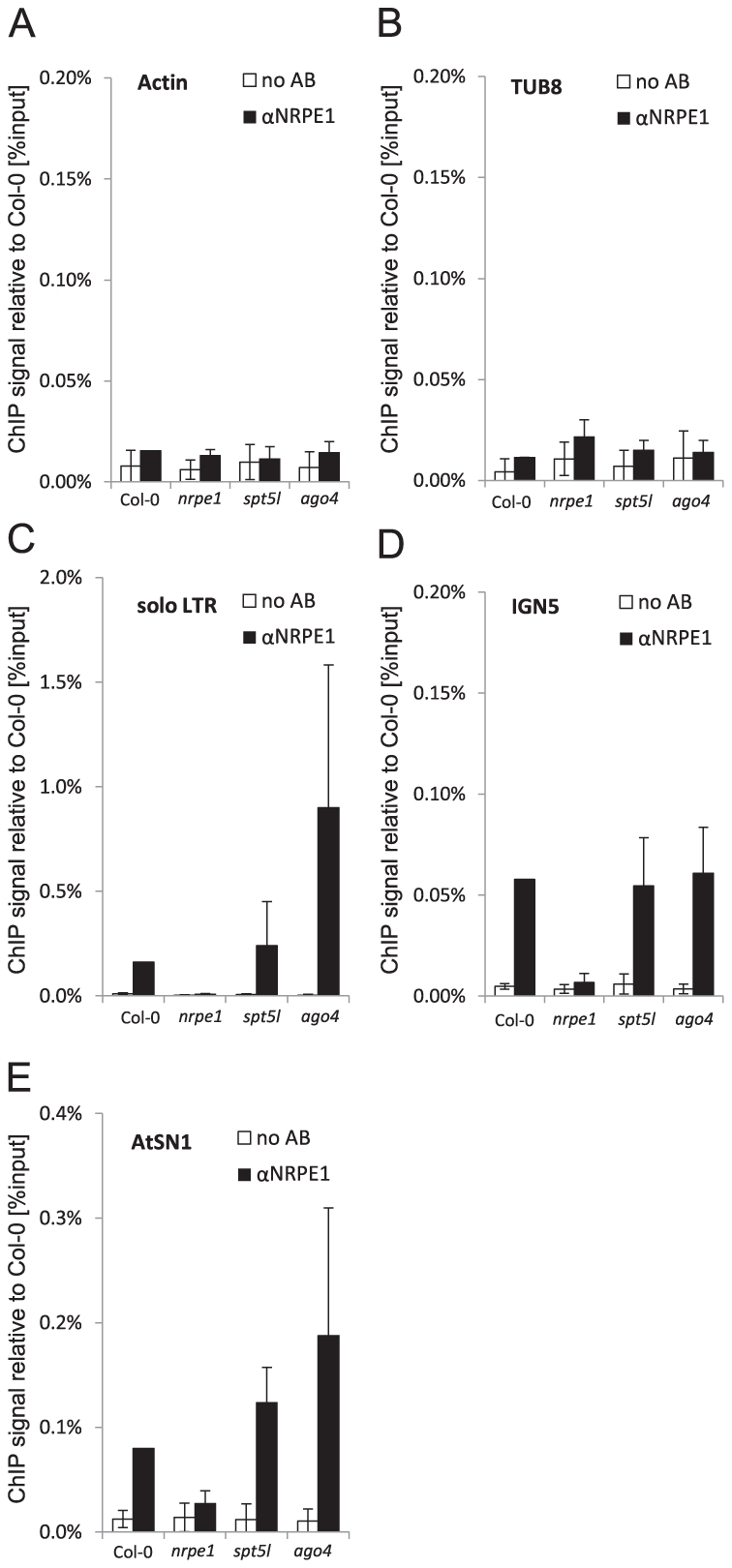
(A–E) Pol V occupancy of Actin 2 (A) and Tubulin 8 (B) control loci, solo LTR (C), IGN5 (D) and AtSN1 (E) assayed by ChIP in Col-0 wild type, nrpe1, spt5l and ago4. No antibody controls (white bars) provide background level for ChIP samples (black bars). Bars represent mean value of ChIP signals normalized to Col-0 wild type. Error bars are standard deviations of three independent biological replicates.
Because both Pol V and SPT5L are required for DNA methylation at several silenced loci [21]–[23], SPT5L may be functionally dependent on Pol V and/or Pol V transcription. To test this possibility we performed western blot with anti-SPT5L antibody in nrpe1 mutant background. Accumulation of SPT5L was strongly reduced in the nrpe1 mutant (Figure 1J). To test if nrpe1 mutation affects accumulation of SPT5L mRNA or SPT5L protein stability, we assayed SPT5L RNA using real time RT-PCR. Accumulation of SPT5L RNA was not reduced in the nrpe1 mutant (Figure 1K) indicating that Pol V is needed for SPT5L protein stability. This behavior of SPT5L in nrpe1 mutant is reminiscent of reduced AGO4 protein stability in mutants that reduce siRNA production [28]. Interestingly, we observed a slight increase in SPT5L RNA level in the nrpe1 mutant which may be explained by the presence of an AtMU10 transposon in SPT5L coding region. Overall, these results suggest that SPT5L is functionally dependent on Pol V.
We further tested the functional relationship between Pol V and SPT5L by performing ChIP with anti-SPT5L antibody in nrpe1 mutant background. Consistent with the reduced stability of SPT5L in nrpe1 (Figure 1J), DNA recovery from Pol V-transcribed loci was reduced to the level observed in the spt5l mutant (Figure 1C–1I). This result may be explained by the overall reduction in the amount of SPT5L. However, a similar reduction in the SPT5L protein accumulation in rdr2 mutant (Figure 1J) did not affect SPT5L binding to chromatin (see below). This suggests that nrpe1 may affect the ChIP signal not only by destabilizing SPT5L, but also by affecting its ability to bind chromatin. Because SPT5L does not work upstream of Pol V and is functionally dependent on Pol V, we conclude that SPT5L works downstream of Pol V and/or Pol V transcription and may be recruited to chromatin by Pol V.
SPT5L binds chromatin independently of AGO4
The recruitment of SPT5L to chromatin by Pol V (Figure 1, Figure 2) is consistent with the interaction of SPT5L with Pol V transcripts and AGO4 [21], [22]. There are at least two explanations of SPT5L function in the establishment of siRNA-mediated transcriptional gene silencing. SPT5L may be recruited by Pol V and then help recruit AGO4-siRNA complexes. Alternatively, AGO4-siRNA may recognize target loci and then recruit SPT5L which further recruits chromatin modifying enzymes. To test the latter possibility we performed ChIP with αSPT5L antibody in the ago4 mutant. DNA recovery of all tested Pol V-transcribed loci was comparable from Col-0 wild type and the ago4 mutant (Figure 1C–1I). This shows that binding of SPT5L to chromatin was not affected in the ago4 mutant, and suggests that SPT5L is not recruited to its target loci by AGO4-siRNA complexes. We conclude that SPT5L does not work downstream of AGO4 in the siRNA-mediated transcriptional gene silencing pathway.
AGO4 binds chromatin partially independently of SPT5L
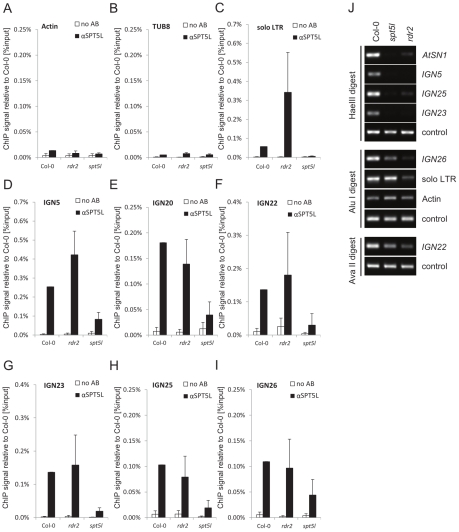
Having concluded that SPT5L does not work downstream of AGO4, we tested the alternative hypothesis that SPT5L may work upstream of AGO4 by binding Pol V and/or Pol V transcripts and recruiting AGO4 to chromatin. To test this possibility we performed ChIP with anti-AGO4 antibody. As demonstrated by real-time PCR we recovered DNA from wild type plants above the background level observed in the ago4 mutant at IGN5 and solo LTR (Figure 3C, 3D) as well as at IGN20, IGN22, IGN23, IGN25 and IGN26 loci (Figure 3E–3I). This indicates that AGO4 binds chromatin at all tested Pol V-transcribed loci. In the spt5l mutant total accumulation of AGO4 protein was not affected (Figure 3J). At all assayed Pol V-transcribed loci AGO4 binding to chromatin in the spt5l mutant was reproducibly above the background level observed in the ago4 mutant indicating that AGO4 is able to bind chromatin in the absence of SPT5L (Figure 3C–3I). Interestingly, we observed that the intensity of AGO4 binding to chromatin is slightly reduced in the spt5l mutant at solo LTR, IGN20, IGN22, IGN23, IGN25 and IGN26 (Figure 3C–3I). This indicates that although SPT5L is not required for AGO4 recruitment to chromatin, it enhances AGO4 chromatin binding. Alternatively, most loci may be occupied by two pools of AGO4. One being SPT5L-dependent and other recruited to chromatin independently of SPT5L.
Figure 3. AGO4 can bind chromatin independently of SPT5L.
(A–I) ChIP data showing AGO4 binding to chromatin in Col-0 wild type, spt5l and ago4 mutants at Actin 2 (A) and Tubulin 8 (B) control loci, solo LTR (C), IGN5 (D), IGN20 (E), IGN22 (F), IGN23 (G), IGN25 (H) and IGN26 (I). No antibody controls (white bars) provide background level for ChIP samples (black bars). Bars represent mean value of ChIP signals normalized to Col-0 wild type. Error bars are standard deviations of three independent biological replicates. (J) Immunoblot detection of AGO4 in whole-cell protein extracts from Col-0 wild type, ago4, spt5l, dms3 and nrpe1 mutants using anti-AGO4 antibody. Asterisk denotes a nonspecific band. Ponceau S staining of the membrane shown in Figure 1J is a loading control.
We conclude that SPT5L is not required for recruitment of a pool of AGO4 to specific loci in chromatin and therefore does not work upstream of AGO4 in the siRNA-mediated transcriptional gene silencing pathway. Since SPT5L also does not work downstream of AGO4, they are most likely recruited in parallel and at least partially independently of each other.
SPT5L binds chromatin independently of 24-nt siRNA
The parallel and independent recruitment of SPT5L and AGO4 to chromatin suggests that they are both guided by the interactions with Pol V complex and/or Pol V transcripts. To test if SPT5L is also guided by siRNA we used ChIP to assay SPT5L binding to chromatin in rdr2, a mutant in an RNA-dependent RNA polymerase responsible for production of the majority of 24-nt siRNA [29]. The rdr2 mutation reduced the stability of SPT5L protein (Figure 1J, 1K) but did not cause reduction in DNA recovery of the tested loci after ChIP (Figure 4C–4I). This suggests that although RDR2 increases the amount of SPT5L protein, the chromatin-bound fraction of SPT5L is not affected by the rdr2 mutation. This also suggests the presence of siRNA-dependent pool of SPT5L that does not physically interact with assayed Pol V-transcribed loci.
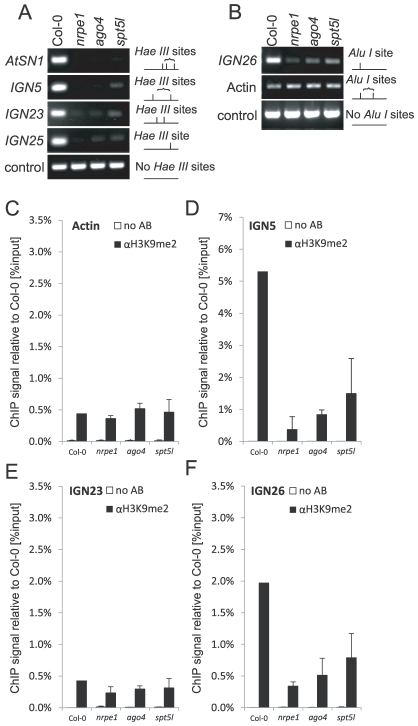
Figure 4. SPT5L interacts with chromatin in an siRNA–independent manner.
(A–I) ChIP data showing SPT5L binding to chromatin in Col-0 wild type, rdr2 and spt5l mutants at loci transcribed by Pol V and silenced by siRNA-mediated transcriptional silencing: solo LTR(C), IGN5 (D), IGN20 (E), IGN22 (F), IGN23 (G), IGN25 (H) and IGN26 (I). Two loci transcribed by Pol II are shown as controls: Actin 2 (A) and Tubulin 8 (B). No antibody controls (white bars) provide background level for ChIP samples (black bars). Bars represent mean value of ChIP signals normalized to Col-0 wild type. Error bars are standard deviations of three independent biological replicates. (J) DNA methylation analysis of AtSN1, IGN5, IGN23 and IGN25 performed by digestion with HaeIII restriction endonuclease, IGN26 and solo LTR performed by digestion with AluI restriction endonuclease and IGN22 performed by digestion with AvaII restriction endonuclease. Digested genomic DNA was amplified by PCR. Sequences lacking HaeIII (Actin 2), AluI (IGN5) or AvaII (Actin 2) were used as loading controls.
These results demonstrate that binding of SPT5L to chromatin is not affected in the rdr2 mutant and suggest that RDR2-dependent siRNA is not required for SPT5L binding to chromatin. In contrast, RDR2 is necessary for proper establishment of DNA methylation at AtSN1, IGN5, IGN25, IGN23, IGN26, solo LTR and IGN22 (Figure 4J); demonstrating that all assayed loci are in fact targets of the siRNA-mediated transcriptional gene silencing pathway. We conclude that SPT5L is recruited to chromatin in a manner independent of 24-nt siRNA.
Both AGO4 and SPT5L are needed for repressive chromatin modifications
Parallel and at least partially independent recruitment of SPT5L and AGO4 by Pol V suggests that at Pol V-transcribed loci none of them is sufficient for the establishment and maintenance of silent chromatin modifications. To further test this possibility we assayed several Pol V-transcribed loci for DNA methylation side-by-side in nrpe1, ago4 and spt5l mutants using DNA methylation-sensitive restriction endonucleases. Methylation of cytosines in HaeIII, AluI or AvaII restriction sites blocks the enzymes from cutting and allows amplification of the genomic region by PCR. However, unmethylated sites are cleaved and PCR amplification fails. All three enzymes recognize asymmetric (CHH) methylation at tested loci. Consistently with previous reports, DNA methylation was strongly reduced at AtSN1 locus in both ago4 [19], [24], [30] and spt5l mutants [21]–[23] and at IGN5 locus in ago4 mutant [19] (Figure 5A). DNA methylation was also reduced at IGN5 locus in spt5l mutant and at IGN23, IGN25 and IGN26 loci in both ago4 and spt5l mutants (Figure 5A, 5B). Importantly, in all these cases reduction of DNA methylation was comparable in ago4 and spt5l mutants (Figure 5A, 5B) suggesting that neither AGO4 nor SPT5L is sufficient for the establishment of asymmetric DNA methylation at Pol V-transcribed loci.
Figure 5. Both SPT5L and AGO4 are required for silencing at certain loci.
(A) DNA methylation analysis of AtSN1, IGN5, IGN23 and IGN25 performed by digestion with HaeIII restriction endonuclease. Digested genomic DNA was amplified by PCR. Sequence lacking HaeIII (Actin 2) sites was used as a loading control. (B) DNA methylation analysis of IGN26 performed by digestion with AluI restriction endonuclease. Digested genomic DNA was amplified by PCR. Sequence lacking AluI (IGN5) sites was used as a loading control. (C–F) Analysis of H3K9me2 at actin 2 (C), IGN5 (D), IGN23 (E) and IGN26 (F) loci performed by ChIP with anti-H3K9me2 antibody in Col-0 wild type, nrpe1, ago4 and spt5l. No antibody controls (white bars) provide background level for ChIP samples (black bars). Bars represent mean value of ChIP signals normalized to Col-0 wild type. Error bars are standard deviations of three independent biological replicates.
We also tested the effect of nrpe1, ago4 and spt5l mutations on dimethylation of lysine 9 of histone H3 (H3K9me2). At IGN5 and IGN26 loci, H3K9me2 was reduced in all three mutants (Figure 5D, 5F) showing that both AGO4 and SPT5L are required not only for the establishment and/or maintenance of DNA methylation but also H3K9me2. We conclude that at least at a subset of loci SPT5L and AGO4 work together to recruit repressive chromatin modifications. We propose that it is the coordinate action of SPT5L and AGO4 that directly or indirectly recruits de novo DNA methyltransferase DRM2 and H3K9 methyltransferases.
SPT5L contributes to repressive chromatin modifications in a locus-specific manner
While AtSN1, IGN5, IGN23, IGN25 and IGN26 loci require both AGO4 and SPT5L for repressive chromatin modifications (Figure 5), soloLTR has been shown to be methylated independently of SPT5L [21], [23]. We confirm this result and further show that solo LTR and IGN22 which, like other Pol V-transcribed loci, are methylated in a Pol V and AGO4-dependent manner (Figure 6A) did not show reduction of DNA methylation on AluI or AvaII sites in the spt5l mutant (Figure 6A). This suggests that there is some significant locus specificity in SPT5L contributions to DNA methylation. Furthermore, H3K9me2 was reduced at both soloLTR and IGN22 in nrpe1 and ago4 mutants but not in the spt5l mutant (Figure 6B, 6C). Also acetylation of histone H3 (H3Ac) at solo LTR was increased in nrpe1 and ago4 but not in spt5l (Figure 6D). This demonstrates that the locus-specific function of SPT5L affects not only DNA methylation but also H3K9me2 and H3Ac.
Figure 6. Locus-specific effects of SPT5L on silencing.
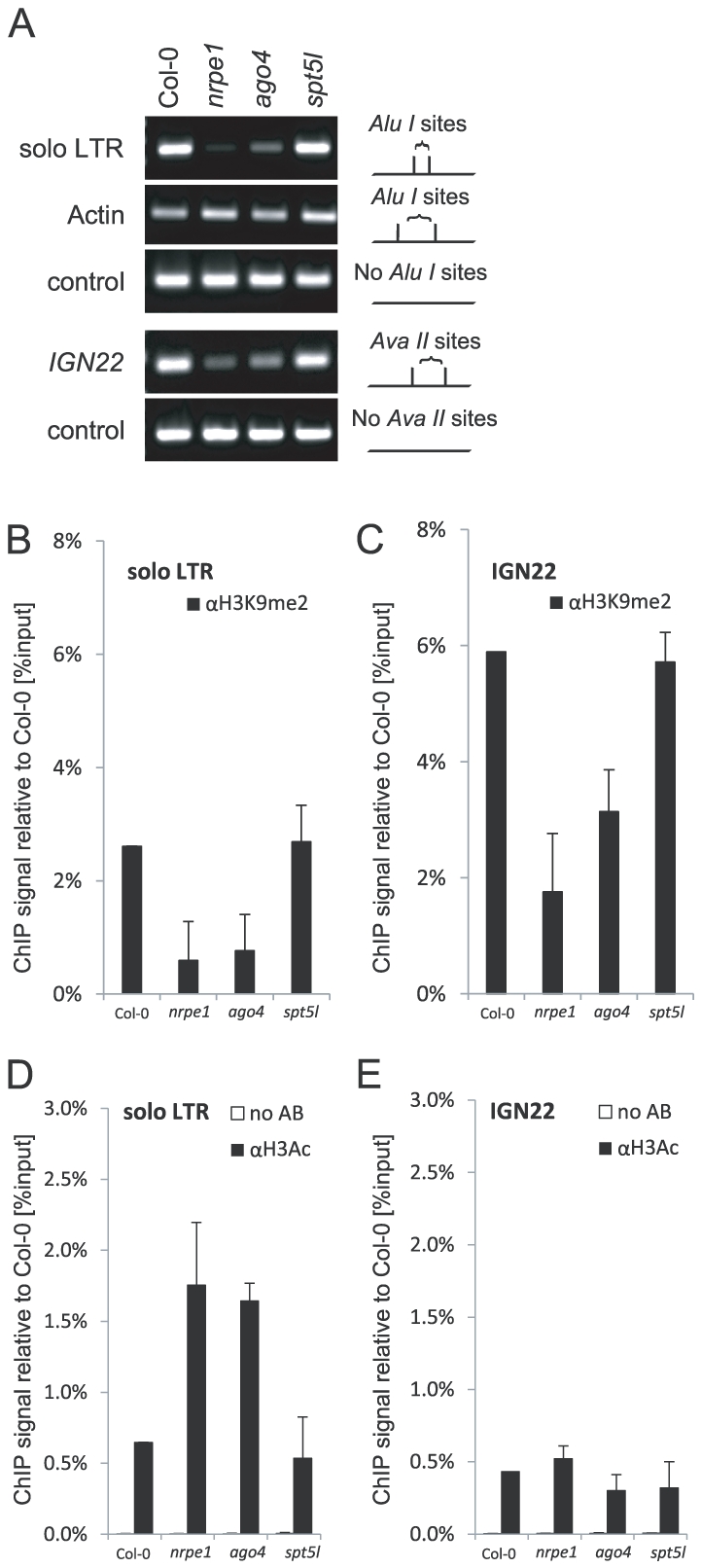
(A) DNA methylation analysis of solo LTR performed by digestion with AluI restriction endonuclease and IGN22 performed by digestion with AvaII restriction endonuclease. Digested genomic DNA was amplified by PCR. Sequences lacking AluI (IGN5) or AvaII (Actin 2) sites were used as loading controls. (B–C) ChIP analysis of H3K9me2 at solo LTR (B) and IGN22 (C) in Col-0 wild type, nrpe1, ago4 and spt5l. Corresponding no antibody controls are shown in panels D and E. Bars represent mean value of ChIP signals normalized to Col-0 wild type. Error bars are standard deviations of three independent biological replicates. (D–E) ChIP analysis of H3Ac at solo LTR (D) and IGN 22 (E) in Col-0 wild type, nrpe1, ago4 and spt5l. No antibody controls (white bars) provide background level for ChIP samples (black bars). Bars represent mean value of ChIP signals normalized to Col-0 wild type. Error bars are standard deviations of three independent biological replicates.
The requirement of SPT5L for repressive chromatin modifications (Figure 5, Figure 6) does not correlate with the extent of partial SPT5L-dependency of AGO4 binding to chromatin (Figure 3). It suggests that the pool of AGO4 that is bound to chromatin in an SPT5L-dependent manner is not required for silencing. This is consistent with our interpretation that AGO4 and SPT5L are recruited to chromatin in parallel and independently of each other.
Discussion
Order of events in siRNA–mediated silencing
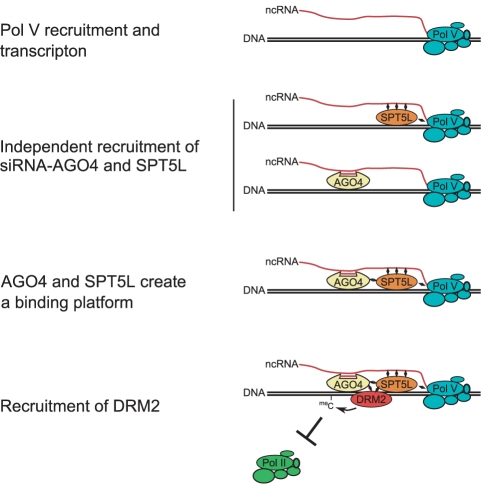
Our findings establish the order of events leading to siRNA-mediated establishment of transcriptional silencing. This process is initiated by recognition of silencing targets and production of two classes of non-coding RNA. The first class is siRNA which is produced from double-stranded RDR2 products by DCL3 and becomes incorporated into AGO4 and possibly also AGO6 and AGO9 [4], [5], [24]. The second class is long non-coding RNA produced by Pol V and/or Pol II [18], [26]. Pol V transcription is initiated independently of siRNA and Pol V transcripts most likely are not precursors for siRNA biogenesis [18], [31]. Pol V recruitment to chromatin and transcription requires the presence of DMS3, DRD1 and RDM1, which either help initiate Pol V transcription or assist elongation of Pol V transcripts [18], [19], [32].
Pol V transcription is followed by association of two RNA-binding proteins with chromatin (Figure 7). First is AGO4 which is recruited to chromatin by Pol V transcripts and uses the incorporated siRNA to provide sequence-specificity of silencing [19]. The second is SPT5L (Figure 1, Figure 2), which is recruited to chromatin by an unknown mechanism, possibly involving interactions between SPT5L and Pol V complex and/or with Pol V transcripts [22], [23]. SPT5L binds chromatin independently of 24-nt siRNA (Figure 4) and is likely a general factor associated with transcribing Pol V and its transcripts [21]–[23]. Since SPT5L binds chromatin in the absence of AGO4 (Figure 1), and the functional pool of AGO4 is able to bind chromatin in the absence of SPT5L (Figure 3), we concluded that they are recruited to chromatin in parallel and independently of each other. Both AGO4 and SPT5L are required for the establishment and/or maintenance of DNA methylation and repressive histone modifications at the majority of tested loci (Figure 5). This suggests that both are needed for the recruitment of enzymes establishing repressive chromatin modifications.
Figure 7. Model of SPT5L involvement in the recruitment of chromatin modifying enzymes.
Pol V produces intergenic non-coding transcripts which are the binding points for SPT5L and AGO4-siRNA complex. Both AGO4 and SPT5L may interact with both Pol V transcripts and Pol V complex itself. SPT5L and AGO4 are recruited to Pol V transcripts in parallel and independently of each other. When both AGO4 and SPT5L are present they create a binding platform for direct or indirect recruitment of DRM2 de novo DNA methyltransferase and other chromatin modifying enzymes. Establishment of chromatin modifications represses Pol II transcription.
Mechanism recruiting chromatin modifying enzymes
Because AGO4 and SPT5L bind chromatin independently of each other, and, at the majority of tested loci both are required for establishment and maintenance of silencing, we propose that AGO4 and SPT5L create a binding platform for the recruitment of chromatin modifying proteins. One possibility is that both weakly interact with a downstream protein but the interaction becomes strong enough to recruit chromatin modifying enzymes only when both are present. Alternatively, AGO4 may be a sole interacting partner of downstream proteins but SPT5L, which has a C-terminal domain rich in WG/GW motifs, interacts with AGO4 and alters its conformation to facilitate the recruitment of chromatin modifying enzymes.
Our results show that there are loci where DNA methylation is established in a Pol V, AGO4 and SPT5L-dependent manner (Figure 5A), but these loci have an overall low level of H3K9me2 and no change in the histone modifications in tested mutants (IGN23 in Figure 5E). It suggests that the de novo DNA methyltransferase DRM2 is likely the chromatin modifying enzyme directly recruited by the AGO4-SPT5L platform. It is also possible that DRM2 may be recruited indirectly by another protein that binds the AGO4-SPT5L platform.
Assembly of the silencing complexes
Binding of AGO4 and SPT5L to chromatin is mediated by multiple protein-protein and protein-RNA interactions. These interactions may mediate recruitment of proteins to specific genomic regions and/or stabilize binding after recruitment by an independent mechanism.
SPT5L binding to chromatin occurs downstream of Pol V and is most likely mediated by protein-RNA interaction between SPT5L and Pol V transcripts [22]. Like canonical SPT5, SPT5L may also form a heterodimer with SPT4 [33]. Alternatively, SPT5L may be recruited to chromatin by protein-protein interaction with Pol V complex as suggested by identification of SPT5L in Pol V holoenzyme [23] and interactions between yeast SPT5 as well as bacterial homolog of SPT5, nusG, with RNA polymerases [34], [35]. It is also possible that SPT5L is recruited to chromatin by interacting with both Pol V transcripts and Pol V complex. All these mechanisms explain the AGO4-independent binding of SPT5L to Pol V-transcribed loci.
Interaction with Pol V transcripts seems to be the major factor recruiting AGO4-siRNA to chromatin [19]. AGO4 also interacts with WG/GW-rich C-terminal domains of Pol V and SPT5L [21], [22], [36]. Because Argonautes contain only one WG/GW binding pocket [37] these interactions may be employed sequentially. First, they help recruit AGO4 to chromatin by interaction with Pol V and then they stabilize the binding of AGO4 to chromatin on its target loci by interaction with SPT5L. It is consistent with our observation that AGO4 binding to chromatin is slightly reduced in the spt5l mutant (Figure 3).
Locus-specific regulation of silencing
We show that SPT5L contributes to regulation of siRNA-mediated transcriptional silencing in a highly locus-specific manner. This is demonstrated by the observation that two of the tested loci require Pol V and AGO4 but not SPT5L for establishment and/or maintenance of repressive chromatin modifications (Figure 6). It could be explained by presence of the canonical SPT5 at a subset of silenced loci. However, both loci are occupied by SPT5L in wild type plants (Figure 1) suggesting that SPT5L is in fact involved in their silencing. Only when SPT5L is mutated, the canonical SPT5 is able to compensate the deficiency at these particular loci. Alternatively, it is possible that the observed locus-specificity of SPT5L is caused by the presence of both Pol V and Pol II at a subset of loci [26]. Pol II-bound canonical SPT5 may be able to compensate the lack of Pol V-bound SPT5L. The mechanism deciding locus specificity of the SPT5L function remains unknown.
Our results also suggest the presence of two pools of AGO4: SPT5L-dependent and SPT5L-independent. Because both pools are detectable at loci that are silenced in a SPT5L-independent manner, the SPT5L-dependent pool of AGO4 is likely not required for silencing. It may be recruited independently of siRNA by direct interaction with SPT5L and may have some other, yet unknown and locus-specific functions.
Materials and Methods
Plant lines and antibodies
Arabiodopsis thaliana nrpe1 (nrpd1b-11), dms3-4, and ago4-1 introgressed into Col-0 background were described previously [19], [38]. rdr2-1 mutant was obtained from J. Carrington. spt5l-1 (rdm3-3; SALK_001254) mutant line, affinity-purified anti-SPT5L (anti-KTF1), affinity-purified anti-Pol V (anti-NRPE1) and affinity-purified anti-AGO4 antibodies were described previously [19], [22], [39]. Mouse monoclonal anti-H3K9me2 antibody (cat. #ab1220) was obtained from Abcam, rabbit polyclonal anti-H3Ac antibody (cat. #06-599) was obtained from Millipore.
Chromatin immunoprecipitation
ChIP was performed essentially as described [18], [19]. Detailed ChIP protocol is included in the Text S1. ChIP samples were amplified in triplicate in Applied Biosystems 7500 real time PCR machine and obtained data were analyzed using comparative CT method relative to inputs [40]. All ChIP experiments were performed in three independent biological replicates. Results from every biological replicate were normalized to Col-0 wild type and normalized data were used to obtain averages and standard deviations that show fold difference between analyzed strains. Normalized data were subsequently multiplied by average ChIP signal level of Col-0 wild type. This way data are corrected for variability in overall signal strength between independent experiments, the unit is %input and presented data reflect the relative signal strength observed at particular loci. Standard deviations for Col-0 wild type are not available because Col-0 wild type was used to normalize data.
DNA and RNA analysis
For DNA methylation analysis genomic DNA was extracted from above-ground tissue of 2-week old plants using DNeasy Plant Mini Kit (Qiagen). 100 ng of genomic DNA was digested with 10u of HaeIII, AluI or AvaII restriction enzymes (NEB) for 20 min. After heat-inactivation of the enzyme DNA was amplified using 0.75u Platinum Taq (Invitrogen).
Total RNA was extracted from 2-week old plants using RNeasy Plant Mini Kit (Qiagen) and amplified using SuperScript III Platinum SYBR Green One-Step qRT-PCR Kit (Invitrogen) in Applied Biosystems 7500 real time PCR machine.
Oligonucleotide primers used in this study are shown in Table S1.
Supporting Information
Loci assayed in this study, their accession numbers and oligonucleotide primers.
(PDF)
Detailed Chromatin Immunoprecipitation (ChIP) protocol.
(PDF)
Acknowledgments
We thank Eran Pichersky and David Engelke for critical reading of the manuscript.
Footnotes
The authors have declared that no competing interests exist.
This work was supported by the University of Michigan and the NIH National Research Service Award #5-T32-GM07544 from the National Institute of General Medicine Studies. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Girard A, Hannon GJ. Conserved themes in small-RNA-mediated transposon control. Trends Cell Biol. 2008;18:136–48. doi: 10.1016/j.tcb.2008.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Faulkner GJ, Kimura Y, Daub CO, Wani S, Plessy C, et al. The regulated retrotransposon transcriptome of mammalian cells. Nat Genet. 2009;41:563–571. doi: 10.1038/ng.368. [DOI] [PubMed] [Google Scholar]
- 3.Moazed D. Small RNAs in transcriptional gene silencing and genome defence. Nature. 2009;457:413–420. doi: 10.1038/nature07756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Matzke M, Kanno T, Daxinger L, Huettel B, Matzke AJM. RNA-mediated chromatin-based silencing in plants. Curr Opin Cell Biol. 2009;21:367–376. doi: 10.1016/j.ceb.2009.01.025. [DOI] [PubMed] [Google Scholar]
- 5.Law JA, Jacobsen SE. Establishing, maintaining and modifying DNA methylation patterns in plants and animals. Nat Rev Genet. 2010;11:204–220. doi: 10.1038/nrg2719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Alleman M, Sidorenko L, McGinnis K, Seshadri V, Dorweiler JE, et al. An RNA-dependent RNA polymerase is required for paramutation in maize. Nature. 2006;442:295–298. doi: 10.1038/nature04884. [DOI] [PubMed] [Google Scholar]
- 7.Erhard KF, Stonaker JL, Parkinson SE, Lim JP, Hale CJ, et al. RNA polymerase IV functions in paramutation in Zea mays. Science. 2009;323:1201–1205. doi: 10.1126/science.1164508. [DOI] [PubMed] [Google Scholar]
- 8.Sidorenko L, Dorweiler JE, Cigan AM, Arteaga-Vazquez M, Vyas M, et al. A dominant mutation in mediator of paramutation2, one of three second-largest subunits of a plant-specific RNA polymerase, disrupts multiple siRNA silencing processes. PLoS Genet. 2009;5:e1000725. doi: 10.1371/journal.pgen.1000725. doi: 10.1371/journal.pgen.1000725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cam HP, Chen ES, Grewal SIS. Transcriptional scaffolds for heterochromatin assembly. Cell. 2009;136:610–614. doi: 10.1016/j.cell.2009.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Volpe TA, Kidner C, Hall IM, Teng G, Grewal SIS, et al. Regulation of heterochromatic silencing and histone H3 lysine-9 methylation by RNAi. Science. 2002;297:1833–1837. doi: 10.1126/science.1074973. [DOI] [PubMed] [Google Scholar]
- 11.Cam HP, Sugiyama T, Chen ES, Chen X, FitzGerald PC, et al. Comprehensive analysis of heterochromatin- and RNAi-mediated epigenetic control of the fission yeast genome. Nat Genet. 2005;37:809–819. doi: 10.1038/ng1602. [DOI] [PubMed] [Google Scholar]
- 12.Djupedal I, Portoso M, Spåhr H, Bonilla C, Gustafsson CM, et al. RNA Pol II subunit Rpb7 promotes centromeric transcription and RNAi-directed chromatin silencing. Genes Dev. 2005;19:2301–6. doi: 10.1101/gad.344205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Irvine DV, Zaratiegui M, Tolia NH, Goto DB, Chitwood DH, et al. Argonaute Slicing Is Required for Heterochromatic Silencing and Spreading. Science. 2006;313:1134–1137. doi: 10.1126/science.1128813. [DOI] [PubMed] [Google Scholar]
- 14.Bühler M, Verdel A, Moazed D. Tethering RITS to a nascent transcript initiates RNAi- and heterochromatin-dependent gene silencing. Cell. 2006;125:873–86. doi: 10.1016/j.cell.2006.04.025. [DOI] [PubMed] [Google Scholar]
- 15.Pontier D, Yahubyan G, Vega D, Bulski A, Saez-Vasquez J, et al. Reinforcement of silencing at transposons and highly repeated sequences requires the concerted action of two distinct RNA polymerases IV in Arabidopsis. Genes Dev. 2005;19:2030–40. doi: 10.1101/gad.348405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kanno T, Huettel B, Mette MF, Aufsatz W, Jaligot E, et al. Atypical RNA polymerase subunits required for RNA-directed DNA methylation. Nat Genet. 2005;37:761–5. doi: 10.1038/ng1580. [DOI] [PubMed] [Google Scholar]
- 17.Ream TS, Haag JR, Wierzbicki AT, Nicora CD, Norbeck AD, et al. Subunit compositions of the RNA-silencing enzymes Pol IV and Pol V reveal their origins as specialized forms of RNA polymerase II. Mol Cell. 2009;33:192–203. doi: 10.1016/j.molcel.2008.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wierzbicki AT, Haag JR, Pikaard CS. Noncoding transcription by RNA Polymerase Pol IVb/ Pol V mediates transcriptional silencing of overlapping and adjacent genes. Cell. 2008;135:635–648. doi: 10.1016/j.cell.2008.09.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wierzbicki AT, Ream TS, Haag JR, Pikaard CS. RNA polymerase V transcription guides ARGONAUTE4 to chromatin. Nat Genet. 2009;41:630–634. doi: 10.1038/ng.365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Qi Y, He X, Wang X-J, Kohany O, Jurka J, et al. Distinct catalytic and non-catalytic roles of ARGONAUTE4 in RNA-directed DNA methylation. Nature. 2006;443:1008–12. doi: 10.1038/nature05198. [DOI] [PubMed] [Google Scholar]
- 21.Bies-Etheve N, Pontier D, Lahmy S, Picart C, Vega D, et al. RNA-directed DNA methylation requires an AGO4-interacting member of the SPT5 elongation factor family. EMBO Rep. 2009;10:649–654. doi: 10.1038/embor.2009.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.He X-J, Hsu Y-F, Zhu S, Wierzbicki AT, Pontes O, et al. An effector of RNA-directed DNA methylation in arabidopsis is an ARGONAUTE 4- and RNA-binding protein. Cell. 2009;137:498–508. doi: 10.1016/j.cell.2009.04.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Huang L, Jones AME, Searle I, Patel K, Vogler H, et al. An atypical RNA polymerase involved in RNA silencing shares small subunits with RNA polymerase II. Nat Struct Mol Biol. 2009;16:91–93. doi: 10.1038/nsmb.1539. [DOI] [PubMed] [Google Scholar]
- 24.Havecker ER, Wallbridge LM, Hardcastle TJ, Bush MS, Kelly KA, et al. The Arabidopsis RNA-Directed DNA Methylation Argonautes Functionally Diverge Based on Their Expression and Interaction with Target Loci. Plant Cell. 2010;22:231–334. doi: 10.1105/tpc.109.072199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Huettel B, Kanno T, Daxinger L, Aufsatz W, Matzke AJM, et al. Endogenous targets of RNA-directed DNA methylation and Pol IV in Arabidopsis. EMBO J. 2006;25:2828–36. doi: 10.1038/sj.emboj.7601150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zheng B, Wang Z, Li S, Yu B, Liu J-Y, et al. Intergenic transcription by RNA polymerase II coordinates Pol IV and Pol V in siRNA-directed transcriptional gene silencing in Arabidopsis. Genes Dev. 2009;23:2850–2860. doi: 10.1101/gad.1868009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Numa H, Kim J-M, Matsui A, Kurihara Y, Morosawa T, et al. Transduction of RNA-directed DNA methylation signals to repressive histone marks in Arabidopsis thaliana. EMBO J. 2010;29:352–362. doi: 10.1038/emboj.2009.374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li CF, Pontes O, El-Shami M, Henderson IR, Bernatavichute YV, et al. An ARGONAUTE4-containing nuclear processing center colocalized with Cajal bodies in Arabidopsis thaliana. Cell. 2006;126:93–106. doi: 10.1016/j.cell.2006.05.032. [DOI] [PubMed] [Google Scholar]
- 29.Kasschau KD, Fahlgren N, Chapman EJ, Sullivan CM, Cumbie JS, et al. Genome-wide profiling and analysis of Arabidopsis siRNAs. PLoS Biol. 2007;5:e57. doi: 10.1371/journal.pbio.0050057. doi: 10.1371/journal.pbio.0050057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zilberman D, Cao X, Jacobsen SE. ARGONAUTE4 control of locus-specific siRNA accumulation and DNA and histone methylation. Science. 2003;299:716–9. doi: 10.1126/science.1079695. [DOI] [PubMed] [Google Scholar]
- 31.Mosher RA, Schwach F, Studholme D, Baulcombe DC. PolIVb influences RNA-directed DNA methylation independently of its role in siRNA biogenesis. Proc Natl Acad Sci U S A. 2008;105:3145–50. doi: 10.1073/pnas.0709632105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Law JA, Ausin I, Johnson LM, Vashisht AA, Zhu J-K, et al. A protein complex required for polymerase V transcripts and RNA- directed DNA methylation in Arabidopsis. Curr Biol. 2010;20:951–956. doi: 10.1016/j.cub.2010.03.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schwer B, Schneider S, Pei Y, Aronova A, Shuman S. Characterization of the Schizosaccharomyces pombe Spt5-Spt4 complex. RNA. 2009;15:1241–1250. doi: 10.1261/rna.1572709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hartzog GA, Wada T, Handa H, Winston F. Evidence that Spt4, Spt5, and Spt6 control transcription elongation by RNA polymerase II in Saccharomyces cerevisiae. Genes Dev. 1998;12:357–369. doi: 10.1101/gad.12.3.357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Li J, Horwitz R, McCracken S, Greenblatt J. NusG, a new Escherichia coli elongation factor involved in transcriptional antitermination by the N protein of phage lambda. J Biol Chem. 1992;267:6012–6019. [PubMed] [Google Scholar]
- 36.El-Shami M, Pontier D, Lahmy S, Braun L, Picart C, et al. Reiterated WG/GW motifs form functionally and evolutionarily conserved ARGONAUTE-binding platforms in RNAi-related components. Genes Dev. 2007;21:2539–44. doi: 10.1101/gad.451207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Eulalio A, Huntzinger E, Izaurralde E. GW182 interaction with Argonaute is essential for miRNA-mediated translational repression and mRNA decay. Nat Struct Mol Biol. 2008;15:346–353. doi: 10.1038/nsmb.1405. [DOI] [PubMed] [Google Scholar]
- 38.Pontes O, Li CF, Nunes PC, Haag J, Ream T, et al. The Arabidopsis chromatin-modifying nuclear siRNA pathway involves a nucleolar RNA processing center. Cell. 2006;126:79–92. doi: 10.1016/j.cell.2006.05.031. [DOI] [PubMed] [Google Scholar]
- 39.Onodera Y, Haag JR, Ream T, Nunes PC, Pontes O, et al. Plant nuclear RNA polymerase IV mediates siRNA and DNA methylation-dependent heterochromatin formation. Cell. 2005;120:613–22. doi: 10.1016/j.cell.2005.02.007. [DOI] [PubMed] [Google Scholar]
- 40.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−Delta Delta C(T)) Method. Methods. 2001;25:402–8. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Loci assayed in this study, their accession numbers and oligonucleotide primers.
(PDF)
Detailed Chromatin Immunoprecipitation (ChIP) protocol.
(PDF)