Abstract
We demonstrate that in situ optical surface plasmon resonance spectroscopy can be used to monitor hybridization kinetics for unlabeled DNA in tethered monolayer nucleic acid films on gold in the presence of an applied electrostatic field. The dc field can enhance or retard hybridization and can also denature surface-immobilized DNA duplexes. Discrimination between matched and mismatched hybrids is achieved by simple adjustment of the electrode potential. Although the electric field at the interface is extremely large, the tethered single-stranded DNA thiol probes remain bound and can be reused for subsequent hybridization reactions without loss of efficiency. Only capacitive charging currents are drawn; redox reactions are avoided by maintaining the gold electrode potential within the ideally polarizable region. Because of potential-induced changes in the shape of the surface plasmon resonance curve, we account for the full curve rather than simply the shift in the resonance minimum.
Keywords: electrochemistry, DNA hybridization, DNA mismatch discrimination
Increasing research efforts are directed toward the detection of nucleic acid interactions with immobilized oligonucleotide probes for DNA microarray applications (1, 2). Limitations of most current technologies include complex DNA immobilization procedures, the need for fluorescence or other labeling, and slow hybridization kinetics that require long incubation times. In addition, those experimental conditions that optimize DNA duplex formation often also reduce the stringency of hybridization. That is, for oligonucleotides that are sufficiently long to distinguish a particular sequence in the presence of unrelated DNA, a mismatched base pair has only a marginal effect on the stability of the duplex (3). In DNA microarray applications, mismatched hybrids lead to “false positives.”
The use of an electric field to easily control the electrostatic forces on surface-immobilized polyelectrolytes such as DNA has not yet been fully exploited as a means for improving the speed and stringency of biomolecular interactions at interfaces.
In this paper, we investigate the effect of simple electrostatic charging on the interactions of surface-bound monolayer nucleic acid films with unlabeled DNA target oligonucleotides by using a combination of electrochemical control and in situ real-time surface plasmon resonance (SPR) spectroscopy detection.
The monolayer DNA thiol films used in this work are tethered directly to the SPR metal sensor surface through a gold–thiol covalent attachment. Therefore, the immobilized DNA hybrids are exposed to a field gradient at the metal/electrolyte interface on the order of 109 V/m. We show that this field can be used, in a reversible manner, to increase or decrease the rate of oligonucleotide hybridization. In addition, we show that a repulsive potential preferentially denatures mismatched DNA hybrids within a few minutes, while leaving the fully complementary hybrids largely intact. This sequence selectivity in hybrid denaturation imparts an extremely high stringency for assaying DNA interactions and represents an extremely simple method for mutation detection based purely on electrostatic charging effects.
This work is in contrast to the “electronic denaturation” of DNA hybrids reported by Heller and coworkers (4), in which an electric current is reported to denature fluorescent-labeled DNA oligonucleotides immobilized in a gel. In that case, denaturation is likely to arise through a complex mechanism, potentially involving Joule heating within the gel, formation of hydroxyl ion or radical intermediates, or other current-induced phenomena. Direct electric field denaturation cannot be the mechanism, as the potential gradient in the gel, on the order of a few hundred volts per meter, is insufficient to destabilize the hybrid.
Despite the extremely large field strengths at the electrochemical interfaces used here, the current is limited to capacitive, non-Faradaic charging current. Redox reactions are avoided by maintaining the gold electrode potential within the ideally polarizable region. The tethered single-stranded (ss)DNA thiol probes remain bound and can be reused for subsequent hybridization reactions without loss of efficiency. The robustness of these monolayer films is not surprising, given the well-known extensive use of alkanethiol self-assembled monolayer films in a wide range of electrochemical applications (5). The specific DNA thiol films used here have been extensively characterized in previous work by various techniques (6), including neutron scattering (7) and SPR surface melting experiments (8), and are known to be robust and reusable in nonelectrochemical experiments even after heating to 75°C. Previous in situ SPR experiments confirm that the thermal stability of double-stranded DNA on these surfaces is analogous to that observed in free solution (8, 9).
This work is distinguished by the use of label-free SPR detection of hybridization and denaturation achieved by monitoring gain or loss of DNA at the interface in the presence of applied electrochemical field. Although a wide body of literature describes the application of SPR to study the interactions of biologically active species with surfaces, films, and biomaterials, including DNA hybridization (6–8, 10), relatively few applications of electrochemical SPR have appeared in the literature. None of these previous electrochemical studies have used capacitive non-Faradaic charging of the sensor interface to control biomaterial interactions. The combination of SPR with electrochemistry has been limited to cases where electrochemical currents associated with oxidation/reduction reactions at the metal SPR substrate are monitored simultaneously. Previous work includes investigations of immobilized protein redox reactions (11, 12), selective desorption of monolayer films (13), reversible doping of organic conducting polymer films (14), and electrochemical release of surface bound ligands (15).
It is well known that the application of an interfacial electric field alters the ionic double layer at the metal/electrolyte interface, which in turn alters the surface electron distribution and therefore the optical properties of the metal. For bare metal surfaces, this effect has been measured by a number of different surface optical techniques including SPR (16), electroreflectance (17, 18), second harmonic generation (19), and ellipsometry (20–22). In SPR, potential-induced optical changes lead to substantial alteration in the shape of the reflectance spectrum; therefore, we account for the full curve rather than simply the shift in the resonance minimum.
Materials and Methods
The SPR optical instrumentation used for these experiments has been described previously (8, 23, 24). The electrochemical experiments use a potentiostat (EG & G, Salem, MA, 263A) to control the potential of a standard three-electrode cell, as described previously (14). The cell uses the gold SPR substrate as the working electrode, a platinum counter electrode in the bulk solution, and an Ag/AgCl reference electrode placed in a remote reservoir that is connected to the SPR cell through a salt bridge (Vycor plug, Corning). Preparation of the SPR gold substrate and subsequent self assembly of the ssDNA monolayer film was carried out as described previously (8). Gold substrates were cleaned with piranha solution before use, as described previously (23).
Oligonucleotides [thiol-derivatized ssDNA (HS-C6-ssDNA), underivatized ssDNA complement, and underivatized two-base mismatch] were synthesized and purified by reverse-phase HPLC by Synthegen (Houston, TX). The complementary target sequence used in this work was 3′-TCTAGTCACGCAGACATGATCGTGT-5′ and the two-base mismatched target sequence was 3′-TCTAGTCACACAGACATCATCGTGT-5′. The sequence for the fully noncomplementary control oligomer is 3′- AGATCAGTGCGTCTGTACTAGCACA-5′. Monobasic anhydrous sodium phosphate, potassium chloride, sodium perchlorate, sodium sulfate, 1-dodecanethiol, mercaptohexanol, and ethanol were purchased from the Aldrich and used without further purification. Sodium chloride was purchased from Fisher Scientific. Potassium phosphate (monobasic) was purchased from EM Science. All water was purified by using a Bamstead E-pure 18 MΩ cm−1 water purifier. Piranha solution was prepared as a 7:3 mixture of sulfuric acid (EM Science) and 30% hydrogen peroxide solution (Mallinckrodt).
Hybridization was achieved by exposing the immobilized ssDNA probe surface to 1-μM target DNA solution in 1 M NaCl for a total of 14 h for passive hybridization and 5 h for electric field-assisted hybridization. For all hybridization experiments performed under electrochemical control, the surface was first exposed to target DNA solution at open circuit. After 60 s, the potential was applied and immediately swept from the potential of zero charge (PZC) to the desired potential (+300 mV or −300 mV) at a rate of 50 mV s−1. For all electric field-assisted denaturation experiments, the applied field was maintained for 5 h.
All data presented here are obtained on the same regenerated probe surface; however, consistent results (within the noise) are seen on different gold/DNA thiol interfaces. Hot water (90°C) rinses were used to regenerate the ssDNA probe surface for sequential experiments. All potentials are reported with respect to an Ag/AgCl reference electrode. For all electrochemically assisted experiments, the applied potential remains within the ideally polarizable region, so as to avoid any oxidation/reduction reactions at the electrode surface.
Results
SPR Detection in the Presence of an Applied Electrochemical Field.
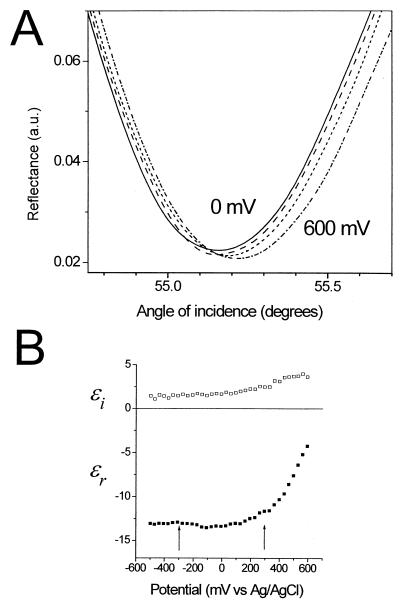
In this section, we establish the potential dependence of the optical properties for bare gold SPR substrates and compare the results with substrates coated with monolayer films. Potential-induced changes in the shape of the SPR curve (reflectance vs. angle of incidence) for bare gold are shown in Fig. 1A for a series of applied potentials (0, 200, 400, and 600 mV). The alterations in shape can be accounted for by changes in the optical constants of the gold substrate (Fig. 1B), in excellent agreement with results from previous work (18). Here, the complex dielectric constant of the gold as a function of applied potential was calculated from fitting the SPR reflectance data, assuming that only the outermost 2 Å of the gold are affected by the electric field. In agreement with previous optical studies of electrochemical interfaces, we find a strong potential dependence at potentials positive of the PZC and a negligible effect at potentials negative of the PZC (approximately +100 mV). As expected, the charging behavior depends on the nature of the electrolyte. Detailed studies in various electrolytes (data not shown) show that the strongest potential dependence at positive charging is observed for systems with the most negative value of the PZC. In the series of electrolytes studied, our results are consistent with the trend of PZC(chloride)<PZC(phosphate)≈PZC(sulfate)<PZC(perchlorate), in agreement with previous electrochemical determinations of the PZC. In chloride solutions that exhibit the lowest PZC, the potential dependence at the positive charging is about 20% larger than the magnitude shown in Fig. 1B.
Figure 1.
(A) The effect of charging a gold surface in 10 mM sodium perchlorate electrolyte on the SPR data for same gold substrate at a series of applied potentials (0, 200, 400, and 600 mV). For clarity, only a portion of the full reflectance curves are shown. (B) Potential dependence of the complex dielectric constants (real, ɛr, and imaginary, ɛi, components) of the gold, calculated by fitting the full SPR reflectance curves (see text for details). The arrows indicate the potentials at which hybridization experiments were undertaken.
The presence of a self-assembled monolayer film suppresses the potential dependence of the SPR response substantially (data not shown). For example, for a complete, well-packed monolayer of dodecanethiol, the potential dependence is essentially eliminated. For the two-component ssDNA thiol/mercaptohexanol monolayer film used in this work, the SPR response potential dependence is suppressed to ≈10% that of bare gold in the same electrolyte solution.
Hybridization Studies.
In this section, we examine the effect of electrostatic charging on target/probe hybridization. The DNA monolayer films used in this work have been extensively characterized by SPR, neutron scattering, and surface melting experiments and are found to be robust and reusable (8, 9, 25). For these experiments, hybridization was allowed to proceed with no potential applied, then the potential was swept at a rate of 50 mV s−1 to the desired potential (+300 mV or −300 mV), and hybridization allowed to continue.
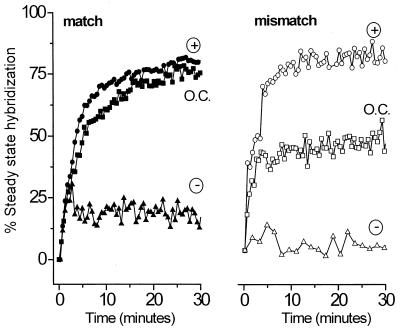
Fig. 2 shows the data for electrostatic controlled experiments compared with hybridization under passive open-circuit conditions for a fully complementary target (Fig. 2 Left) and a partially mismatched target (Right). The ordinate is given as percentage of steady-state hybridization. That is, the DNA coverage is calculated from analysis of the SPR reflectance curves as described previously (8, 9, 24, 25). The progress of hybridization is shown as the percent of the duplex DNA surface coverage relative to the steady state coverage achieved after ≈14 h of hybridization at open circuit.
Figure 2.
Comparison of the kinetics of hybridization at open circuit (squares) and under electrochemical control at an applied potential of +300 mV (circles) and −300 mV (triangles). Data are shown for the interaction of the fully complementary target (solid symbols) for the 2-bp mismatched target (open symbols). For each electrochemical experiment, hybridization proceeds for 3 minutes before the selected potential is applied (see text for details). Here only the first 30 min are shown, although each hybridization proceeds for at least 5 h. The magnitude of hybridization is compared with the steady-state value reached after 14 h of unassisted (open circuit) hybridization in an unstirred cell. All experiments were carried out in 1 M NaCl solution containing 1 μM oligonucleotide probe on the same immobilized ssDNA thiol probe film. The probe coverage remains constant (within 10%) when regenerated by rinsing with hot water between runs.
Application of an attractive potential (+300 mV) increases the rate of duplex formation for both the matched and mismatched target DNA compared with passive hybridization. Interestingly, the hybridization enhancement is much more pronounced for the mismatched target. The extent of hybridization with field assistance is never larger than when hybridization is carried out with overnight incubation at open circuit. Application of a repulsive potential (−300 mV) immediately halts hybridization.
We have reported previously that, at open circuit, the kinetics of hybridization provide a means of discriminating against mismatched DNA duplex formation (9). That is, the kinetics of hybridization for matched duplexes (filled squares in Fig. 2) are clearly distinct from those of mismatched DNA hybridization (unfilled squares in Fig. 2). Here, we note an alternative means of mismatch discrimination on the basis of the more pronounced response of the mismatched target to an attractive field relative to passive conditions. This different response, seen when comparing matched and mismatched kinetics at open circuit and +300 mV, could have application in real-time detection of mismatch without the need to monitor the full kinetic isotherm.
Electrostatic Denaturation.
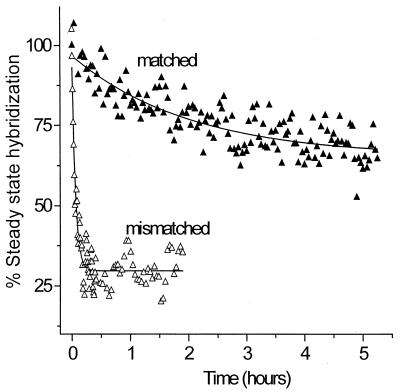
DNA films containing fully complementary target/probe duplexes or two-base mismatch target/probe duplexes were formed under passive hybridization conditions and then subjected to a repulsive applied potential (−300 mV). The effect of the applied electrostatic potential on the stability of the duplex films was determined by monitoring loss of ssDNA strands from the surface (Fig. 3) by in situ SPR detection in the presence of hybridization buffer (1 M NaCl solution containing 1 μM oligonucleotide target). The data (Fig. 3) are reported relative to the level of steady-state hybridization present at the onset of the applied field. For the mismatched hybrid, application of a repulsive potential results in rapid denaturation of most (≈75%) of the immobilized duplex within a few minutes, whereas for the fully complementary hybrid, relatively little loss of ssDNA is observed even after many hours of exposure to the electrostatic field. Exponential fits to the data (solid lines) indicate that the time constant for denaturation of the mismatch is ≈35 times smaller than for the full complement (Fig. 3).
Figure 3.
Discrimination between the fully complementary hybrid (closed triangles) and the 2-bp mismatched hybrid (open triangles) by electric field-induced denaturation on the same surface. After passive hybridization for 14 h, a dc electrochemical potential of −300 mV vs. Ag/AgCl was applied and the percent hybridization monitored as loss of ssDNA from the interface by in situ SPR. The same discrimination is also observed for hybrids formed by electrostatically assisted hybridization. Experimental conditions were 1 M NaCl solution containing 1 μM oligonucleotide target. Data were obtained by using the same regenerated probe surface.
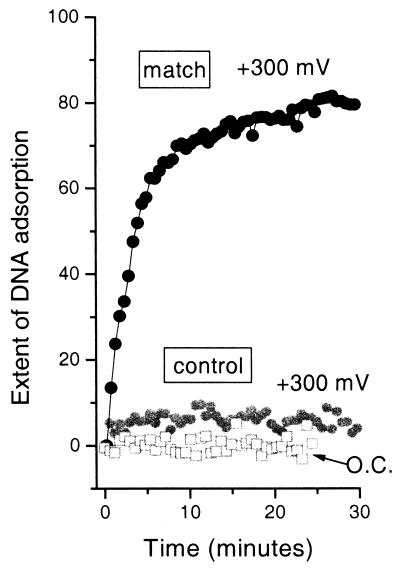
Control experiments with the noncomplementary DNA oligomer show that nonspecific binding is negligible even in the presence of an applied electrostatic field of +300 mV (Fig. 4). These results are in agreement with previous work under open circuit conditions (8, 9, 25), which showed that the mercaptohexanol component appears to help block nonspecific adsorption of DNA. As with previous work, the kinetics of hybridization for a specific sequence and probe density are reproducible within the noise for other gold substrates (8, 9, 25).
Figure 4.
Results of control experiments with fully noncomplementary DNA. The extent of DNA adsorption, measured by in situ SPR under the same experimental conditions as for Fig. 2 (1 μM oligomer in 1 M NaCl) for the fully noncomplementary control oligomer at open circuit and at +300 mV (gray open squares and gray solid circles, respectively). The data are shown on the same scale as the electrostatically assisted hybridizations of Fig. 2. For comparison, the data for the fully complementary match sequence at +300 mV (same data as in Fig. 2) are also shown (black solid circles).
Conclusions
We have used electrostatic charging to increase and decrease the rate of target DNA adsorption onto a probe surface. Under electrostatic assistance, the initial rate of target adsorption is greater than for passive hybridization. This effect is especially pronounced for the mismatch target. Interestingly, the steady-state coverage reached by electrostatic assistance never exceeds that for passive hybridization, in agreement with previous work (8), which showed that, for these two-component DNA films, the mercaptohexanol component is a diluent that serves to block nonspecific adsorption of DNA.
Application of a repulsive potential preferentially denatures mismatched hybrids within a few minutes, leaving the fully complementary 25-mer hybrids largely intact even after many hours. This sequence selectivity in hybrid denaturation imparts an extremely high stringency for assaying DNA interactions and represents an extremely simple method for mutation detection based purely on electrostatic charging effects. This method can be applied in a microarray format.
In contrast with previous “electronic denaturation” of DNA hybrids reported previously by Heller and coworkers (4), here the DNA is immobilized directly at the metal/electrolyte interface and therefore is exposed to a field gradient on the order of 109 V/m. This electrostatic field is sufficient to induce denaturation of the hybrids. Because no Faradaic current flows, the ssDNA probes remain covalently bound to the gold and are stable to prolonged exposures at these electrode potentials. Finally, the simple planar geometry of our SPR substrate and the use of DNA monolayer films of controlled coverage mean that electric field induced denaturing at these simple interfaces are likely to be theoretically tractable. Recent theoretical work by Vainrub and Pettitt (26) has predicted strong electrostatic destabilization effects for DNA immobilized in electric double layers. It is important to compare theoretical predictions directly with the results of electrostatic SPR experiments.
Acknowledgments
We gratefully acknowledge the support of the National Science Foundation, CHE 9709347.
Abbreviations
- SPR
surface plasmon resonance
- ssDNA
single-stranded DNA
- PZC
potential of zero charge
References
- 1.Lockhart D J, Winzeler E A. Nature (London) 2000;405:827–836. doi: 10.1038/35015701. [DOI] [PubMed] [Google Scholar]
- 2.Fodor S P A. Science. 1997;277:393. [Google Scholar]
- 3.Cantor C R, Smith C L. Genomics: The Science and Technology Behind the Human Genome. New York: Wiley; 1999. [Google Scholar]
- 4.Sosnowski R G, Tu E, Butler W F, O'Connell J P, Heller M J. Proc Natl Acad Sci USA. 1997;94:1119–1123. doi: 10.1073/pnas.94.4.1119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Finklea H O. Electroanal Chem. 1996;19:109–335. [Google Scholar]
- 6.Herne T M, Tarlov M J. J Am Chem Soc. 1997;119:8916–8920. [Google Scholar]
- 7.Levicky R, Herne T M, Tarlov M J, Satija S K. J Am Chem Soc. 1998;120:9787–9792. [Google Scholar]
- 8.Peterlinz K A, Georgiadis R M. J Am Chem Soc. 1997;119:3401–3402. [Google Scholar]
- 9.Peterson A W, Heaton R J, Georgiadis R. J Am Chem Soc. 2000;122:7837–7838. [Google Scholar]
- 10.Steel A B, Herne T M, Tarlov M J. Bioconj Chem. 1999;10:419–423. doi: 10.1021/bc980115g. [DOI] [PubMed] [Google Scholar]
- 11.Schlereth D D. J Electroanal Chem. 1999;464:198–207. [Google Scholar]
- 12.Boussaad S, Pean J, Tao N J. Anal Chem. 2000;72:222–226. doi: 10.1021/ac990947n. [DOI] [PubMed] [Google Scholar]
- 13.Badia A, Arnold S, Scheumann V, Zizlsperger M, Mack J, Jung G, Knoll W. Sens Actuators B. 1999;54:145–165. [Google Scholar]
- 14.Georgiadis R, Peterlinz K A, Rahn J R, Peterson A W, Grassi J H. Langmuir. 2000;16:6759–6762. [Google Scholar]
- 15.Hodneland C D, Mrksich M. J Am Chem Soc. 2000;122:4235–4236. [Google Scholar]
- 16.Abeles F, Lopez-Rios R, Tadjeddine A. Solid State Commun. 1975;16:843–847. [Google Scholar]
- 17.McIntyre J D E. Surface Sci. 1973;37:658–682. [Google Scholar]
- 18.McIntyre J D E, Kolb D M. Symp Faraday Soc. 1970;4:99–113. [Google Scholar]
- 19.Corn R M, Romagnoli M, Levenson M D, Philpott M R. J Chem Phys. 1984;81:4127–4132. [Google Scholar]
- 20.Chao F, Costa M, Tadjeddine A. Surface Sci. 1974;46:265–281. [Google Scholar]
- 21.Chao F, Costa M, Tadjeddine A. J. Phys. (French) 1977. 97–107. [Google Scholar]
- 22.Chao F, Costa M, Tadjeddine A, Abeles F, Lopez-Rios T, Theye M L. J Electroanal Chem. 1977;83:65–86. [Google Scholar]
- 23.Peterlinz K A, Georgiadis R. Langmuir. 1996;12:4731–4740. [Google Scholar]
- 24.Peterlinz K A, Georgiadis R. Opt Commun. 1996;130:260–266. [Google Scholar]
- 25.Georgiadis R, Peterlinz K P, Peterson A W. J Am Chem Soc. 2000;122:3166–3173. [Google Scholar]
- 26.Vainrub A, Pettitt B M. Chem Phys Lett. 2000;323:160–166. [Google Scholar]