Abstract
Eukaryotic nuclei contain regions of differentially staining chromatin (heterochromatin), which remain condensed throughout the cell cycle and are largely transcriptionally silent. RNAi knockdown of the highly conserved heterochromatin protein HP1 in Drosophila was previously shown to preferentially reduce male viability. Here we report a similar phenotype for the telomeric partner of HP1, HOAP, and roles for both proteins in regulating the Drosophila sex determination pathway. Specifically, these proteins regulate the critical decision in this pathway, firing of the establishment promoter of the masterswitch gene, Sex-lethal (Sxl). Female-specific activation of this promoter, SxlPe, is essential to females, as it provides SXL protein to initiate the productive female-specific splicing of later Sxl transcripts, which are transcribed from the maintenance promoter (SxlPm) in both sexes. HOAP mutants show inappropriate SxlPe firing in males and the concomitant inappropriate splicing of SxlPm-derived transcripts, while females show premature firing of SxlPe. HP1 mutants, by contrast, display SxlPm splicing defects in both sexes. Chromatin immunoprecipitation assays show both proteins are associated with SxlPe sequences. In embryos from HP1 mutant mothers and Sxl mutant fathers, female viability and RNA polymerase II recruitment to SxlPe are severely compromised. Our genetic and biochemical assays indicate a repressing activity for HOAP and both activating and repressing roles for HP1 at SxlPe.
Author Summary
Eukaryotic genomes are organized into two distinct classes of chromatin, euchromatin and heterochromatin. The former is less condensed to enable transcription, whereas heterochromatin, which is marked by Heterochromatin Protein 1 (HP1), remains compact and mostly transcriptionally silent throughout the cell cycle. The viability of Drosophila males is known to be preferentially compromised in mutants for HP1 and some HP1-associated proteins, suggesting more generalized roles for these proteins in sex-specific gene expression. We now describe a male viability defect for the telomeric partner of HP1, HOAP, and misregulation of the sex determination pathway. Key to the sex determination process is the activation of the X chromosome dose sensing promoter of Sex-lethal, SxlPe. We provide genetic and biochemical evidence that HOAP, the telomere binding partner of HP1, has repressing activity; while HP1 has both activating and repressing roles at this critical promoter. Chromatin immunoprecipitation assays show both proteins are associated with SxlPe sequences. Additionally, RNA polymerase II association with SxlPe shows a requirement for HP1, suggesting a transcription initiation role for HP1. Combined, our data implicate HP1 and HOAP at a euchromatic gene, functioning in a developmental context, and provide the first evidence for a non-telomeric function for HOAP.
Introduction
Eukaryotic genomes are organized into two distinct classes of chromatin [1]. The major class (euchromatin) can undergo decondensation to enable transcription during interphase, whereas a minor fraction (heterochromatin) remains compact and mostly transcriptionally silent throughout the cell cycle. Pericentric and telomeric regions of chromosomes from fungi to humans are organized into a constitutive form of heterochromatin, marked by heterochromatin protein 1 (HP1a in Drosophila [2] and humans [3], Swi6 in S. pombe [4]) and lysine 9-methylated histone H3 (MeK9H3) [5], [6]. Lysine 9 methylation of histone H3 is catalyzed by the Drosophila SU(VAR)3-9 protein [7] (human SUV39H1 [8], S. pombe clr4 [9]) and provides a chromatin-binding site for HP1. Both heterochromatin marks have also been observed in euchromatic genes [10], where their roles in gene activation, as well as repression, have recently been uncovered [11]–[13].
The question of how Drosophila HP1a (designated HP1 throughout this text) is targeted to specific chromosomal regions prompted our biochemical characterizations of HP1 complexes in the maternally loaded cytoplasm of early embryos [14], [15]. HP1/ORC-Associated Protein (HOAP) was identified as a component of a complex that also contains Drosophila origin recognition complex (ORC) subunits [16]. Similarity of the HOAP N-terminus to the HMG-box of mammalian SRY (sex-determining region of the Y chromosome) proteins suggested a role for its DNA-binding activity and that of the ORC, in targeting HP1 to constitutive heterochromatin. Recent data in Drosophila and S. pombe point to a role for small interfering RNAs (siRNA) from heterochromatin-enriched transposable elements in targeting SU(VAR)3-9 (clr4) and HP1 (Swi6) to these regions [17]–[21]. RNAi-independent mechanisms also operate in recruiting HP1 to heterochromatin and to euchromatic genes [5], [8], [22]–[25]. Indeed, recent data point to a role for the DNA binding activity of KAP-1 (TIF1-β) in targeting HP1 to SRY-regulated genes in repressing transcription of testis-specific genes in the ovary [26].
HOAP is best known for its cooperative role with HP1 in forming a capping complex over Drosophila telomeres [27]–[29]. Immunostaining for HOAP also shows the protein at multiple non-telomeric sites in both heterochromatin and euchromatin of larval salivary gland polytene chromosomes [16], [30]. This study was undertaken to examine the non-telomeric functions of HOAP through microarray expression profiling of a mutant for it in order to identify candidate HOAP-regulated genes in these regions [27]. Contrary to our expectation, the majority of genes with altered expression in the mutant had reduced, rather than elevated, transcript levels. The majority of those with reduced transcript levels were found to normally be expressed only in the testis. This led us to uncover an underlying effect of the mutation on male viability and a role for both HOAP and HP1 in regulating the establishment promoter for the master sex determination gene in Drosophila, Sex lethal (Sxl) [31]–[33].
This establishment promoter of Sxl, SxlPe, is critical to the sex determination decision which is made early in embryogenesis (see Figure 4A for an overview of the Sxl locus). SxlPe is only transcribed in females, which have two X chromosomes. In counting the X chromosome number, also known as the X∶A ratio, SxlPe responds to five X-linked activating genes (sisterless-a, sisterless-b, runt, myc and unpaired) working in conjunction with positive maternal factors such as Daughterless. These activating components have their dose measured against the negative effect of maternal factors, such as Groucho and Extramacrochetae, and genes on the autosomes (deadpan is the only known member). Firing of SxlPe generates functional SXL protein which initiates the female-mode of splicing of transcripts from the maintenance promoter, SxlPm. SxlPm is transcribed in both sexes, soon after SxlPe shuts down and its mRNAs are being turned over. In female embryos, the SXL protein from SxlPe transcripts inhibits inclusion of the male-specific exon, which would otherwise prematurely terminate translation, of SxlPm transcripts. This autoregulatory splicing loop maintains SXL expression for the rest of the female life cycle. As males do not activate SxlPe they make no SXL protein and the splicing of SxlPm transcripts includes the male exon by default. Through this autoregulation, the binary sex determination decision is maintained.
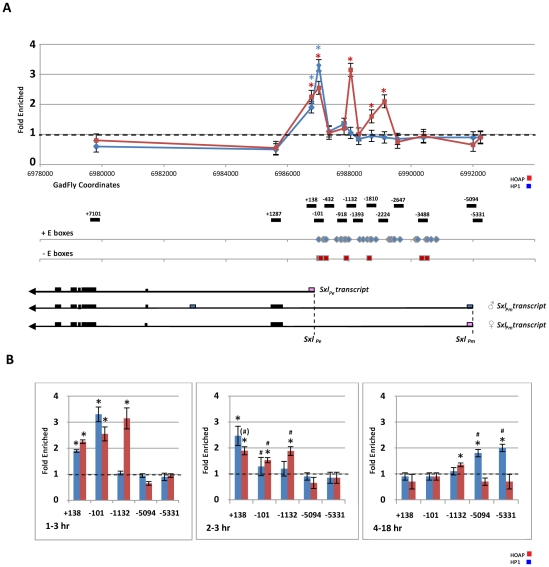
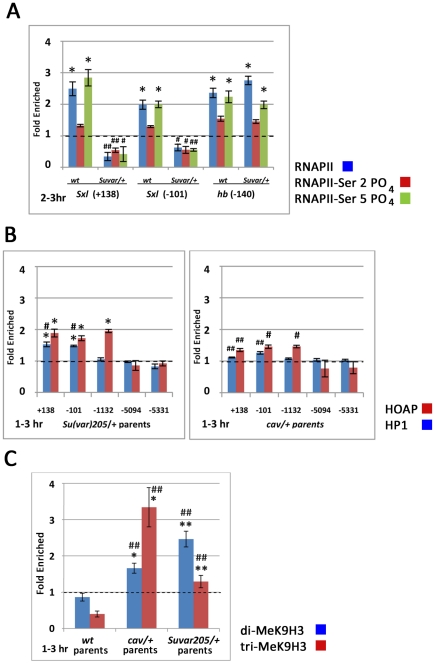
Figure 4. Chromatin immunoprecipitation (ChIP) assays show association of both HOAP and HP1 with the Sxl locus.
(A) Relative enrichment (ChIP/input-Sxl sequence/ChIP/input-RpA-70) of specific Sxl locus sequences in HOAP- and HP1-ChIP fractions from 1–3 hr embryos plotted against the molecular map of the Sxl locus (HOAP in red; HP1 in blue). The positions of Sxl fragments monitored for enrichment are shown relative to the SxlPe initiation site, below the map. The positions of E-box binding sites for positive (blue diamonds) and negative (red squares) bHLH factors [63] and Sxl transcripts are also indicated below the map. (p(no enrichment)<0.05*) (B) Bar graphs show enrichment of Sxl sequences in HOAP- (red bars) and HP1- (blue bars) ChIP fractions during development (1–3 hr, 2–3 hr, and 4–18 hr staged embryos). (p(no enrichment)<0.05*; change between 1–3 hr and 2–3 hr embryos, p(no effect) p(no change)<0.10(#), <0.05#).
SXL in females, through splicing and translational regulation, controls the downstream sex determination genes. A vital effect is turning off dosage compensation (DC), which equalizes X chromosome gene dose between the sexes by upregulating transcription of the male X by about two-fold [32], [34]. Failure to activate SxlPe thus leads to the improper, male mode of splicing of SxlPm transcripts and female lethality. Conversely, inappropriate activation of it in males results in the female mode of splicing of SxlPm transcripts and male lethality.
Our data show roles for both HOAP and HP1 in regulating the critical decision of whether activation of SxlPe will occur. These data support HOAP acting as a repressor, and HP1 as an activator at this promoter. They also suggest that HP1 first cooperates with HOAP in repressing SxlPe before switching to an activation mode. This is the first report of a non-telomeric function for HOAP and the most precisely defined role to date for HP1 in developmental control of a euchromatic gene.
Results
Decrease in Testis-Specific Transcripts Reflects Under-Representation of cav − Males
In an effort to identify candidate HOAP-regulated genes, the Affymetrix Drosophila Genome Array 1 was used to compare the expression profile of wild type larvae to those that were mutant for the HOAP-encoding gene (caravaggio or cav). The original recessive lethal cav 1 allele was used in the study. This allele contains a 5 bp insertion that causes truncation of the HOAP protein after two of three copies of a C-terminal HP1-binding repeat [27], [30]. The y 1 w 67c23 stock, which provides the genetic background for all genetic manipulations in the lab, was used as the wild type control. Larvae of each genotype were collected at the first and second instar stage, prior to the lethal phase of the cav 1 mutant. Out of 13,500 transcription units represented on the array, 183 genes were found to have significantly altered expression (log2R+2.0, p<0.01) in cav1 mutant larvae. Within this set, 142 genes had reduced transcript levels and 41 had increased levels.
Two strategies were then used to catalogue the normal expression profiles of genes in each data set. A gene's relative representation in publically available tissue-specific cDNA libraries [35]–[37] provided the first method for assessing its normal tissue distribution. This analysis was later complemented by data from two published microarray profiling studies of Drosophila sex- or tissue-specific gene expression [38], [39]. The results of these analyses are summarized in Tables S1 and S2. Genes categorized as “multiple” were represented in cDNA libraries of at least three different developmental specificities and enriched in at least three different tissue types in the Chintapalli et al. study [38]. Those categorized as “rare” were represented by a single or few cDNA clones and detected at low levels in multiple tissues by Chintapalli et al. [38]. Those categorized as tissue-specific (e.g., testis or midgut) were represented only, or predominantly, in cDNA libraries of that tissue-specificity and also specifically enriched in that tissue in Chintapalli et al. [38]. The most striking pattern to emerge from these analyses was the relative enrichment of testis-enriched genes (67%) in gene set with reduced transcript levels in the cav1 mutant. The Parisi et al. [39] study of sex-specific gonad expression provided corroboration for the testis-specificity of 67% of these genes, as they were both enriched in testes relative to whole animals and in testes relative to ovaries. This is in contrast to a complete absence of testis-specific genes in the gene set with elevated transcript levels, and also far exceeds the ∼12% of Drosophila genes reported to have testis-specific expression [40].
The enrichment of testis-specific genes in the reduced transcript level data set could indicate a requirement for HOAP in testis-specific gene expression. Alternatively, it could reflect an early lethal phase for cav 1 mutant males and, thus, under-representation of male-specific transcripts in the cav 1 RNA sample. RNA interference (RNAi) against the HP1-encoding gene (Su(var)205) in Drosophila has revealed enhanced vulnerability of males to partial HP1 knockdown [41]. To determine if males were under-represented in our cav1 larval sample, we used the X-linked yellow (y +) marker to sex individual cav 1 mutant larvae, allowing us to sex the animals through a phenotype other than gonad size. By this criterion, 2.83-fold fewer male cav 1 larvae were observed than female cav 1 larvae. Using PCR with Y-linked primers to sex individual cav 1 homozygous embryos, we also observed approximately two-fold more male embryos fail to progress to the larval stage.
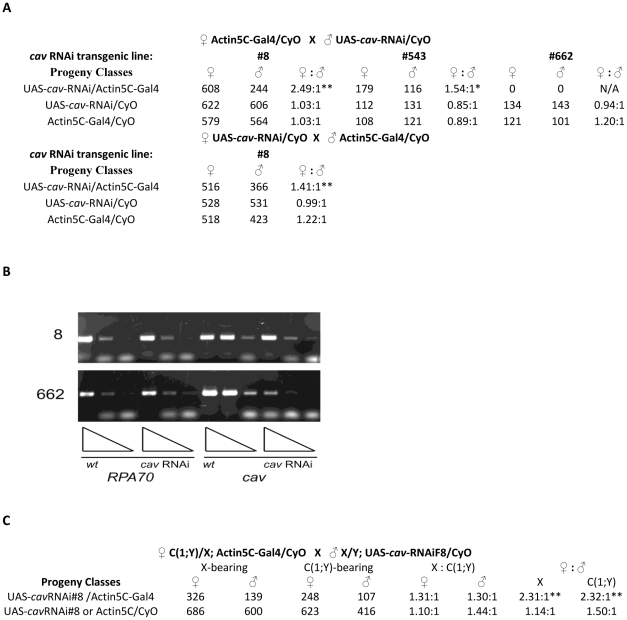
We then used RNAi to partially knockdown HOAP and determine its effect on adult male viability. The GAL4/UAS binary expression system was used to drive expression of a cav RNAi transgene (Figure 1). Ubiquitous expression of cav RNAi from two transgenic lines (F8 and 543) through a maternally introduced Actin5C GAL4 driver resulted in a 2.5- and 1.5-fold reduction in adult male viability relative to females, respectively (Figure 1A). A third cav RNAi transgenic line (662) resulted in lethality of both males and females. The severity in effect of cav RNAi expression in different transgenic lines correlated with the degree to which endogenous cav mRNA was reduced as monitored through semi-quantitative RT-PCR, with ∼80% cav knockdown resulting in a 2.5-fold reduction in male viability (Figure 1B). The dose-dependent effects of cav RNAi expression on male viability are similar to those observed with Su(var)205 RNAi expression (2.54-fold reduction with 60–90% Su(var)205 knockdown and lethality of both sexes with >90% knockdown) [41]. A less severe, but significant, reduction in viability of male progeny was also observed in the reciprocal cross in which the Actin5C GAL4 transgene was introduced through the fathers. Although the results in both sets of crosses indicate a zygotic requirement for HOAP in male viability, the more pronounced effect was with maternally contributed Actin5C GAL4. This suggests maternally expressed GAL4 is needed to drive earlier expression of the cav RNAi for maximum effect.
Figure 1. cav RNAi reduces adult male viability.
(A) The GAL4/UAS binary system using maternally contributed Actin-5C GAL4 to drive expression of cav RNAi shows reduced viability of males carrying the GAL4 driver and cav RNAi transgene #8; both sexes carrying the GAL4 driver and cav RNAi transgene #662 had reduced viability, in comparison to comparison to their control siblings containing either cav RNAi or Act5C-GAL4 transgene alone. (p<0.01**) Male viability was also significantly reduced (albeit less dramatically) in the reciprocal cross in which the Actin-5C GAL4 transgene was paternally derived. (p<0.01**) (B) RT-PCR assays of endogenous cav and RpA70 (normalizing standard) in larvae expressing cav RNAi transgenes from lines #8 and #662. The triangles represent a 5-fold dilution series of RNA from animals carrying either the cav RNAi transgene only (wt) or cav RNAi and Act5C-GAL4 (cav RNAi). (C) Effect of the compound X-Y chromosome, C(1;Y), in animals expressing cav RNAi. The presence of C(1;Y) did not significantly affect viability of adult females or males expressing cav RNAi, in comparison to their control siblings containing either cav RNAi or Act5C-GAL4 transgene alone (p>0.10). By contrast, the animal's sex significantly affected viability, only in the class of animals expressing cav RNAi, regardless of whether they did or did not carry C(1;Y). (p<0.01**).
A dominant effect on male viability was also observed for a newly recovered cav allele (cav 2248). Heterozygosity for this allele reduces male viability 1.84-fold (n≥312, p<0.01). The cav 2248 carries a nonsense mutation at nucleotide 111 of the cav-PB coding sequence within the region of similarity to the SRY HMG box. This allele appears to exert dominant negative activity on the wild type protein, as its effect on male viability is similar to that caused by RNAi-induced HOAP knockdown but is more pronounced than a deficiency for the locus (Df(3R)F89-4). The prematurely truncated protein in this mutant, or a reinitiation product from the next Met within the HMG box, is apparently responsible for this dominant negative effect. Smaller, but significant, reductions in male viability were also observed in progeny from wild type crossed to either parent carrying the cav 2248 allele (data not shown). The lethal phase for cav 2248 heterozygous males was determined to be late in embryogenesis after denticle belt formation; cav 2248 homozygous embryos had an earlier lethal phase which is apparently due to telomeric defects (Figure S1).
Presence of Y Chromosome Is Not Responsible for Reduced Male Viability
Although the Y chromosome is not required for viability or sexual differentiation in Drosophila males, its heterochromatic composition enables it to act as a sink for heterochromatin proteins in suppressing position effect variegation of euchromatic genes artificially juxtaposed to heterochromatin [42], [43]. A limited pool of heterochromatin proteins might then render males more vulnerable to reductions in HOAP (and other heterochromatin proteins). To test whether the Y chromosome has a role in reducing viability of males that are deficient for HOAP, a compound X-Y chromosome [C(1;Y)] was introduced into animals expressing a cav RNAi transgene (Figure 1C). While the C(1;Y) chromosome modestly reduced viability of both male and female progeny by 30% relative to siblings lacking it, the sex of the animal had a pronounced effect on viability. Males expressing cav RNAi showed a 2.3-fold decrease in viability, regardless of whether they also carried C(1;Y).
Defects in Sex Determination Pathway Are Responsible for Reduced Male Viability
Defects in the sex determination pathway, causing inappropriate dosage compensation in Drosophila, result in sex-specific lethality. Sex-lethal (SXL) protein acts as the master switch regulator of this pathway at the level of splicing and translation [44]. As described earlier, the critical decision is made early in embryogenesis by the activation of the Sxl establishment promoter (SxlPe) in only females, to generate functional SXL protein [32]. Failure to activate SxlPe, thus leads to improper male splicing of SxlPm transcripts and female lethality. Conversely, inappropriate activation of SxlPe in males results in the female mode of splicing of SxlPm transcripts and male lethality.
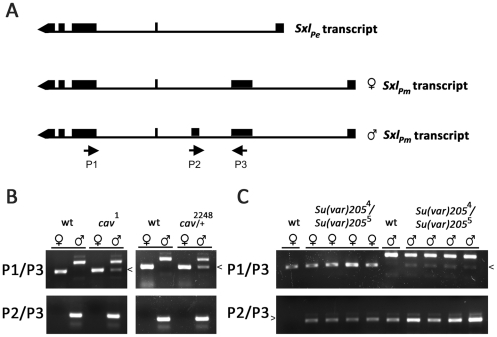
To determine if the enhanced male lethality of cav mutants is associated with inappropriate SXL activity in males, RT (reverse transcriptase)-PCR assays were used to monitor sex-specific splicing of SxlPm transcripts in heterozygous cav 2248 and homozygous cav 1 male embryos that failed to progress to the larval stage. Individual embryos of the appropriate genotype were identified through a GFP-marked wild type balancer chromosome (described more fully in Materials and Methods) and sexed by PCR with Y-linked primers (Figure S2). RNA was then purified from pools of male or female embryos and used in RT-PCR assays with Sxl primer pairs (P1/P3 and P2/P3) designed to discriminate between SxlPm transcripts spliced in the male vs. female mode (Figure 2A). Assays of RNA from wild type (y 1 w 67c23) embryos showed correct sex-specific splicing of SxlPm transcripts in both males and females (Figure 2B). By contrast, a minor fraction of transcripts had undergone the female-mode of splicing in heterozygous cav 2248 and homozygous cav 1 male embryos (arrowhead in Figure 2B).
Figure 2. Aberrant sex-specific SxlPm-derived transcripts are observed in mutants for HOAP and in mutants for HP1.
(A) RT-PCR was used to specifically amplify male- or female-specific SxlPm transcripts. The P1/P3 primer pair amplifies a 482 bp product from the female-specific transcript and a 672 bp product from the male-specific transcript in wild type animals (labeled wt in all three panels). The P2/P3 pair amplifies a 284 bp product from a male-specific transcript in males only. (B) Pools of RNA from individually sexed male or female cav 1 homozygous or cav 2248 heterozygous embryos failing to progress to the larval stage were used in RT-PCR assays with both P1/P3 (top panel) and P2/P3 (bottom panel) primer pairs, revealing the presence of aberrant female-specific SxlPm transcripts in male embryos (arrowhead). RT-PCR assays of RNA from four different individually sexed Su(var)205 4/Su(var)205 5 female (♀) or male (♂) larvae, each with both P1/P3 (top panel) and P2/P3 (bottom panel) primer pairs revealed the presence of aberrant sex-specific SxlPm transcripts in both sexes. The methods used to genotype and sex individual embryos and larvae are described in Materials and Methods.
If inappropriate SXL expression is the cause of the enhanced male lethality in the cav 2248 heterozygous males, we would expect a loss of function Sxl allele to rescue them. To this end, the Sxl f1 point null allele was introduced into heterozygous male and female cav 2248 progeny through their mothers. Closely linked recessive markers, cut and carmine, which flank Sxl were used to follow the Sxl f1 allele in males, while in females white+, which should segregate with Sxl f1 82.3% of the time, was used as a close approximation. As shown in Table 1, the viability of cav 2248 males carrying a defective Sxl allele (Sxl f1) was not significantly different from their female siblings, unlike their Sxl+ brothers (yw). It should be noted that in this cross, the effect of the cav 2248 mutation on male viability was somewhat reduced; we ascribe this difference to the temperature at which the cross was done. For reasons that are not clear at present, all progeny from this cross were nonviable at 25°C, so the cross was performed at room temperature. An overall rescue in male viability was also observed by the introduction of the Sxl fP7BO allele, although for this cross the markers on the deletion allele chromosome did not allow identification of the different progeny classes.
Table 1. Mutations in HOAP reduce male viability, and loss of Sxl partially rescues the lethality.
| ♀ yw; cav 2248/TM3 X ♂ yw; cav 2248/TM3 | ||
| ♀ | ♂ | ♀ ∶ ♂ |
| 202 | 110 | 1.84 ∶ 1** |
The ratio of male to female cav 2248 heterozygous progeny differs significantly from the expected ratio; introduction of the Sxl f1 allele to these progeny restored the expected ratio. (p<0.05**, <0.10*).
As HP1 and HOAP interact, and HP1 reduction also preferentially affects male viability, we wondered whether male larvae mutant for the HP1-coding gene [Su(var)205 5/Su(var)205 4] would also show altered splicing of Sxl Pm transcripts. GFP-marked balancer chromosomes were used to identify individual larvae of the correct genotype, and both an X-linked genetic marker (y+) and PCR assays of a Y-linked gene were used to sex them. Surprisingly, we found aberrant Sxl Pm transcripts in multiple individual Su(var)205 5/Su(var)205 4 larvae of both sexes (arrowheads in Figure 2C), suggesting HP1 has a dual role, of opposite consequence in each sex.
We therefore explored whether a maternal mutation in either cav or Su(var)205 would affect the viability of female progeny that also have a reduction in Sxl dose (Table 2). Female viability is not compromised in a cross between either wild type females and Sxl− males (fP7B0 or f1), or between cav mutant females and Sxl −/Y fathers (Note: the modest reduction in male viability in the progeny from cav 2248 females and Sxl− males is essentially the same as that observed in progeny from cav 2248 females and wild type males, as described earlier). However, in progeny from Su(var)205 heterozygous females crossed to Sxl−/Y males, female viability was dramatically reduced, particularly with the strong loss of function Su(var)205 5 allele. The effect of Su(var)205 mutations is strictly maternal; no significant reduction in female viability is observed in the reciprocal cross (Table 2). Interestingly, this effect is allele-specific. Whereas mothers carrying the Su(var)205 5 null allele or the Su(var)205 4 carboxyl-terminally deleted allele had greatly reduced viability in their female progeny, mothers carrying a point mutation in the MeK9-H3-binding chromodomain allele [Su(var)205 2] had a modest, but significant, effect. Although neither cav allele affected sex-specific viability of progeny from Sxl mutant fathers, the introduction of either allele into Su(var)205 mothers rescued the Su(var)205 maternal effect on female viability (Table 3). Both the dominant negative cav 2248 allele and the C-terminally truncated cav 1 allele, with compromised HP1-binding activity [27], [30], were capable of rescuing the Su(var)205 maternal effect, and this rescue was also strictly maternal.
Table 2. Mutations in HP1 interact with loss of function Sxl alleles to reduce female viability.
| ♀ Su(var)205−/CyO X ♂ Sxl fP7B0/Y | ||||||
| Progeny: | ♀ | ♂ | ♀ ∶ ♂ | |||
| maternal allele | Su(var)205 | CyO | Su(var)205 | CyO | Su(var)205 | CyO |
| Su(var)2055 | 14 | 31 | 231 | 230 | 1 ∶ 16.5** | 1 ∶ 7.42** |
| Su(var)205 4 | 82 | 88 | 325 | 281 | 1 ∶ 3.96** | 1 ∶ 3.19** |
| Su(var)205 2 | 154 | 152 | 224 | 192 | 1 ∶ 1.45* | 1 ∶ 1.26 |
Female viability is markedly reduced in progeny from Su(var)205 mothers and Sxl fathers (Maternal) but not in progeny from Su(var)205 mothers and Sxl− fathers (Paternal). (p<0.05**, <0.10*). For the third cross, numbers of FM7 classes not presented as the balancer also affects viability (males in particular) and these progeny were not factored in the female to male comparison.
Table 3. Maternal mutations in HOAP and HP1 have antagonistic effects on female viability.
| ♀ Su(var)2055/CyO; cav − X ♂ Sxl fP7B0/Y | |||
| maternal allele | ♀ | ♂ | ♀ ∶ ♂ |
| cav 2248 | 231 | 200 | 1.15 ∶ 1 |
| cav 1 | 42 | 172 | 1 ∶ 4.09 |
Maternal (Maternal), but not paternal (Paternal), introduction of a cav mutant allele to Su(var)205 mothers rescues viability of female progeny from Sxl− fathers. (p<0.01**).
Transcription of SxlPe Is Affected by Mutations in Either cav or Su(var)205
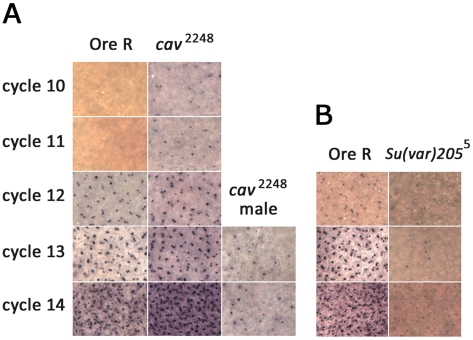
The rescue of cav 2248 male viability by the loss of Sxl and the antagonistic maternal effects of cav and Su(var)205 on the viability of their Sxl −-bearing female progeny, suggested a role for maternal HOAP and HP1 in regulating SxlPe. To directly assess the effect of reduced HOAP or HP1 on SxlPe, in situ hybridizations were performed with probes that distinguish the early from maintenance Sxl mRNAs on 0–4 hr embryos from cav 2248 and Su(var)205 5 heterozygous parents. For SxlPe, wild type embryos show two dots on the chromosomes, one for each female X, beginning in cycle 12 through to early cycle 14. Cycle 14 is also when the maintenance promoter, SxlPm, is activated and the autoregulatory splicing loop set in motion in females. In embryos from cav 2248 heterozygous parents, two key changes were observed (Figure 3A). First, expression of SxlPe in females began two cycles earlier than normal – cycle 10, and overall expression appeared more robust than in wild type embryos. Second, male embryos also showed SxlPe activation (single in situ dot), although the expression was not as strong or as early as in females. The level of expression in male embryos was more variable both between embryos and at the level of individual nuclei. The majority had sporadic or scattered positive nuclei, while others had much greater numbers, as shown in Figure 3A. A count of early cycle 14 male embryos suggests >95% of the male embryos (n = 33) had at least some positive nuclei. In embryos from Su(var)205 5 heterozygous parents, SxlPe expression was not observed in male embryos (single in situ signal) and was weaker and more variable than normal in females (Figure 3B). Analysis of the female embryos indicates that ∼85% express the promoter weakly during cycles 12 and 13 (n = 14–20 for each cycle) and less than half the embryos have normal levels of expression at cycle 14 (n>25). These results are consistent with the early expression of SxlPe being heavily reliant on the maternal deposit of HP1 protein and/or mRNA, with the zygotic contribution becoming more apparent at cycle 14. Expression of the maintenance SxlPm transcripts did not show significant changes in embryos from either genotype (Figure S3). Although HP1 binds to SxlPm in 4–18 hr embryos, at cellular blastoderm the promoter does not appear to be sensitive to a reduction in maternal HP1.
Figure 3. In situs for SxlPe transcripts in embryos from wild type, cav2248/TM3, Sb, or Su(var)205 5/CyO parents.
Comparisons of the same-sized area of images taken at 40×. (A) SxlPe transcripts are present only in Ore R wild type females (containing 2 dots per nucleus) during cycle 12 to 14. Sxl Pe transcripts are present in female embryos from cav2248/TM3 parents as early as cycle 10. SxlPe transcripts are also frequently present in male embryos from cav2248/TM3 parents (single dot per nucleus, usually near nuclear periphery where the dosage compensated X chromosome resides [62]. Not much signal is detected before cycle 13 in males. (B) Poor SxlPe expression is observed in embryos from Su(var)205 5/CyO parents in comparison to simultaneously stained embryos from Ore R wild type parents.
Both HOAP and HP1 Are Associated with the Sxl Pe Regulatory Region
Chromatin immunoprecipitation assays were then used to determine if the effects of cav 2248 and Su(var)205 5 on SxlPe activity are mediated through direct physical association of the proteins with the Sxl locus (Figure 4). Cross-linked chromatin was prepared from developmentally staged collections of wild type (y 1 w 67c23) embryos and immunoprecipitated with antibodies against HP1, HOAP, and non-immune IgG. Embryos were staged to monitor both Sxl promoters: before and during SxlPe activation (1–3 hr), during SxlPe activation (2–3 hr), and during SxlPm expression (4–18 hr). Quantitative real time PCR was then used to measure enrichment of Sxl sequences in each ChIP fraction. As summarized in Figure 4A, significant enrichment of sequences in the immediate vicinity of SxlPe (+138 and −101 fragments) was observed in the HP1 and HOAP ChIP fractions but not in the IgG ChIP fraction from 1–3 hr embryos. No enrichment of SxlPm sequences was observed at this stage. Significant enrichment of sequences upstream of SxlPe (−1132,−1800, and −2224) was also observed in the HOAP ChIP fraction. Using an anti-Myc antibody to probe chromatin from a stock expressing Myc-tagged HOAP from the endogenous cav locus [29], similar results were obtained (data not shown).
A comparison of the ChIP data from 1–3 hr versus 2–3 hr embryos shows a significant decrease (p<0.5) in the associations of both HOAP and HP1 with the −101 region of SxlPe . Both proteins retained significant associations with the +138 region of SxlPe in 2–3 hr embryos at a time that is coincident with SxlPe expression. HOAP enrichment in both the +138 region and the −1132 region appeared reduced at this time, although only at the 90% confidence level. No enrichment of either protein was found more 3′ in Sxl gene coding sequences or in the SxlPm region in either the 1–3 hr or 2–3 hr embryos. Also, SxlPe sequences were not enriched in either the HP1 or HOAP ChIP fractions from 4–18 hr embryos. However, HP1 enrichment in the vicinity of SxlPm (−5094 and −5331) and a slight, but significant, enrichment of HOAP in the −1132 region was observed in later staged embryos.
HOAP and HP1 Affect SxlPe Chromatin
The ChIP assays described above confirmed physical association of HOAP and HP1 with the SxlPe promoter, but do not distinguish between the sexes. We, therefore, took advantage of the feminizing and masculinizing effects of maternal mutations in cav and Su(var)205 to gain insight into the requirement for each protein in regulating SxlPe activity.
The in situs, as well as the lethal effect of a maternal mutation in Su(var)205 on female progeny lacking one functional copy of Sxl, indicated an activation function for HP1 at SxlPe. To further test this hypothesis and examine its nature, we determined the effect of reducing maternal HP1 on RNA polymerase II (RNAP II) association with SxlPe (Figure 5A). Chromatin was prepared from 2–3 hr. embryos from wild type or heterozygous Su(var)205 5 mothers mated to Sxl f1 fathers, as SxlPe is active at this stage in wild type embryos. Antibodies that recognize three different phosphoisoforms of RNAP II, which mark distinct stages of transcription initiation and elongation, were used to gain insight into the level of transcriptional activation at SxlPe at which HP1 is functioning. In embryos from wild type mothers, both unphosphorylated RNAP II (blue bar) and Ser 5-phorphorylated carboxyterminal domain RNAP II (green bar), which marks early steps of transcription elongation, were enriched in the proximal regions of SxlPe and the hunchback (hb) promoter. Both genes are active at this stage. Ser 2-phosphorylated RNAP II (red bar) which identifies polymerase in its elongation phase was mostly absent from both promoters. All RNAP II isoforms, particularly those associated with transcription initiation (unphosphorylated RNAP II, blue bars) and early events in transcription elongation (Ser 5-phosphorylated RNAP II, green bars), were drastically reduced from SxlPe proximal sequences in embryos from Su(var)205 mutant mothers and Sxl f1 /Y fathers (Figure 5A). By contrast, the Su(var)205 mutation did not affect RNA polymerase II association with the hb promoter (Figure 5A).
Figure 5. Chromatin immunoprecipitation (ChIP) assays show perturbations in Sxl Pe chromatin in embryos from HOAP and HP1 mutant mothers.
(A) Bar graphs show SxlPe (Sxl +138 and −101) and hunchback (hb −140) promoter sequences in RNA polymerase II ChIP fractions from embryos produced by either wild type (wt) or Su(var)205 5/+ mothers crossed to Sxl f1/Y fathers. ChIP assays for unphosphorylated (blue bar), Ser2-phosphorylated (red bar), or Ser5-phosphorylated (green bar) RNA polymerase II are shown. (p(no enrichment)<0.05*; change between wild type and mutant embryos, p(no change)<0.05#, <0.01##) (B) Bar graphs show changes in the enrichment of Sxl sequences in HOAP- (red bars) and HP1- (blue bars) ChIP fractions in embryos produced by Su(var)205 5/CyO parents (left) and by cav 2248/TM3, Sb parents (right). (p(no enrichment)<0.05*; change between wild type and mutant embryos, p(no change)<0.05#, <0.01##) (C) Bar graphs show changes in the enrichment of Sxl sequences in di-MeK9H3- (blue bars) and tri-MeK9H3- (red bars) ChIP fractions in embryos produced by wt, Su(var)205 5/CyO and cav 2248/TM3, Sb parents. (p(no enrichment)<0.05*; change between wild type and mutant embryos, p(no change)<0.05#, <0.01##).
The feminizing effects of reducing maternal HOAP, most notably the effect of the cav 2248 mutation on transcription from SxlPe, support a repressive role for HOAP at this promoter. The partial rescue of the maternal Su(var)205 masculinizing effect by the HP1-binding-defective cav 1 allele implicates HP1 in this repression as well. To examine the interdependency of HOAP and HP1 in their association with SxlPe sequences, we performed ChIP assays of each protein in 1–3 hr embryos with a reduced maternal dose of the other protein. As shown in Figure 5B, a maternal mutation in one copy of the gene for either protein significantly reduced association of its encoded protein with SxlPe sequences. These data also indicate a strong reliance of HP1 on HOAP for its association with Sxl Pe proximal sequences in embryos of this stage. HP1 association with these sequences was significantly reduced in embryos from cav 2248 heterozygous parents. HOAP association with SxlPe sequences, also appeared educed in embryos from Su(var)205 5 heterozygous parents, but this reduction was not statistically significant. These data support a role for HOAP in recruiting HP1 to SxlPe prior to the time of SxlPe activation; failure to form this repressive chromatin in embryos from heterozygous cav 2248 mothers apparently negates the need for HP1 in Sxl Pe activation slightly later in embryonic development.
K9-methylated histone H3 has a well known conserved role in HP1 association with constitutive pericentric heterochromatin [7]–[9] and some euchromatic genes [5]. The activating function of HP1 at Sxl Pe does not appear to rely on this histone modification, as a maternal mutation for the Su(var)205 2 allele, which encodes a protein that is defective for MeK9H3-binding, does not strongly affect the viability of female progeny from Sxl mutant fathers. It is still possible, however, that HP1-binding to MeK9H3 has a role in HP1 repression prior to the time of Sxl Pe activation. We, therefore, used ChIP assays to monitor the presence of both di- and tri-methylated histone H3 at the major site of HP1enrichment in the −101 region of SxlPe. The hb promoter, which is active at the same time as SxlPe but does not appear to require HP1 for activation, was used as the normalizing standard in these experiments.
Di-MeK9H3 was detected at above background levels (p<0.05) at SxlPe, but not at the simultaneously expressed hb promoter, in embryos from wild type mothers (Figure 5C). Although no significant enrichment was observed at SxlPe relative to hb in wild type embryos, a maternal cav 2248 or Su(var)205 5 mutation resulted in significant enrichment of both di- and tri-MeK9H3 at SxlPe (Figure 5C). Assuming, as in pericentric heterochromatin, a repressive activity for these histone modifications at SxlPe, their increased enrichment in this region in mutants for HP1 and HOAP may reflect a feedback mechanism to compensate the loss of HP1.
Discussion
The canonical heterochromatin protein HP1 is most commonly associated with constitutive heterochromatin and gene repression. Here we report a critical role for it in regulating one of the earliest decisions in metazoan development, whether to embark on a female or male path of sexual differentiation and dosage compensation. The role of heterochromatin in mammalian dosage compensation has been recognized since the early work of Lyon [45]. Although Drosophila utilizes a different mechanism to equalize X-linked gene dose, through hyper-activation of the single male X chromosome via chromatin modification [46], this study provides the first evidence of a role for heterochromatin proteins in the early events of Drosophila sex determination. HP1, together with its telomere partner HOAP, influence the critical decision in sex determination - activation of SxlPe, the Sxl establishment promoter.
Heterochromatin Proteins in Sex-Specific Viability
We find that reductions in HOAP preferentially compromise male viability. This was observed for two different cav mutant alleles and by reducing HOAP through RNAi. The presence of SxlPm-derived transcripts that have been spliced in the female mode in cav mutant males suggested inappropriate Sxl activation to be responsible for this reduced viability. In situ data indicating inappropriate firing of SxlPe in male embryos from cav 2248 heterozygous parents support this view, as does the rescue of the cav 2248 male viability defect by Sxl loss of function mutations. The more pronounced male lethality observed from reducing HOAP by RNAi expression driven by maternal, versus paternal, contribution of Actin5C GAL4 is consistent with such an early requirement for HOAP for male viability (Figure 1).
Previous reports have shown that reducing HP1 by RNAi similarly reduces male viability preferentially [41]. RT-PCR assays of SxlPm transcripts in HP1 mutants, however, suggested a more complex scenario as incorrect sex specific transcripts were observed in both sexes (Figure 2C). This pointed to an activation, as well as repressor, role for HP1. Consistent with an activation role, reduction of maternal HP1 severely compromised female viability when the dose of Sxl was also reduced in the progeny (Table 2), and ChIP assays of embryos from this cross showed recruitment of RNAP II to SxlPe to be impaired (Figure 5A). This effect of reducing HP1 on female viability was strictly maternal, as was the antagonizing effect of simultaneously reducing maternal HOAP (Table 3). Moreover, the partial rescue of the Su(var)205 maternal effect by the C-terminally truncated cav 1 allele, which produces a protein that is compromised for HP1-binding [27], [30], points to an involvement of HP1 in the antagonizing activity of HOAP. Finally, ChIP assays show a dependence of HP1 on HOAP for its association with SxlPe. Combined, these data indicate both antagonistic and cooperative roles for these heterochromatin proteins in regulating SxlPe, whereby HOAP acts as a repressor and HP1 acts as both an activator and repressor. The reliance of HP1 on HOAP for recruitment to the promoter would suggest HOAP may also have a role in the activation function of HP1 at the promoter, although this was not readily apparent in our assays.
Although our data clearly show maternal roles for HOAP and HP1 in regulating the activity of SxlPe, for both HOAP and HP1 [41] the RNAi knockdown data indicate a substantial zygotic component in their effects on male viability. These zygotic effects, observed only in progeny carrying both an interference RNA transgene and a GAL4 driver transgene (Figure 1A), suggest additional later sex-specific roles for both proteins. Such roles could be related to those observed for HP1 and SU(VAR)3-7 in male dosage compensation [47]. Because the effect of reducing these proteins on the chromosomal distribution of DCC proteins [47] is the opposite of those observed for males that are deficient for DCC proteins [48], as predicted to occur with inappropriate SxlPe expression, the activities of heterochromatin proteins in dosage compensation appear to be distinct from the early roles of HP1 and HOAP at SxlPe. In addition, there may be zygotic roles for heterochromatin proteins in sex-specific gene expression, as proposed for HP1 by Liu et al. [41].
Dynamic Roles of HOAP and HP1 at SxlPe
Previous analysis of SxlPe indicated that 400 bp immediately upstream of the promoter are sufficient for sex-specific regulation, but distal sequences, extending to −1700 bp, are required for wild type levels of expression [33]. As shown in Figure 4A, E-box binding sites for antagonistically acting bHLH proteins, which are encoded by zygotically expressed X-linked and autosomal signal elements (XSE and ASE) and direct an X counting mechanism, are distributed throughout both regions [49].
Both HP1 and HOAP are enriched in the region proximal to SxlPe which contains binding sites for both positive and negative E-box proteins. Within the SxlPe promoter distal region, HOAP alone is enriched in two peaks where there is a striking relationship with E-box binding sites for positive factors, but those for negative factors appear essentially devoid of HOAP. HOAP may antagonize positive factors but permit negative factors to bind in the SxlPe distal region, in an HP1-independent repressing role. Whereas loss of HOAP de-represses SxlPe in males, the strength and uniformity of expression does not approach that in wild type females. This indicates continued influence from the X counting mechanism in cav mutant males. SxlPe is also expressed prematurely in female embryos. This de-repression by reduced levels of maternal HOAP in both sexes indicates that HOAP is present at SxlPe in both sexes of wild type embryos. However, whether the proximal and distal SxlPe regions have the same or different compositions of HOAP and HP1 in the two sexes cannot be determined from our ChIP assays, as the embryos are of mixed sexual identity.
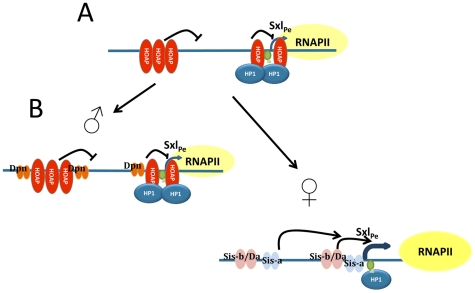
The interdependency of HOAP and HP1 for their binding to the SxlP e proximal region, most notably the dependence of HP1 on HOAP, also indicates both proteins are in this region in, at least, wild type female embryos. In spite of this interdependency, the genetic data show HOAP repression antagonizes HP1 activation. HOAP repression appears to also be partly HP1-dependent; the mutant HOAP protein from the cav 1 allele which lacks HP1-binding also antagonizes HP1 activation. This combination of antagonistic and cooperative interactions suggests a model in which maternal HOAP and HP1 first cooperate to repress SxlPe prior to its activation (Figure 6A). The repressive structure formed by maternal HOAP and HP1 likely serves to reduce the sensitivity of SxlPe to spurious fluctuations in zygotic XSE levels, ensuring it is only activated in females where an effective ratio of activating to repressing transcription factors exists (Figure 6B). HP1 is retained at SxlPe during its activation in females, where it presumably switches into an activation role. In early embryos constitutive heterochromatin proteins may be more appropriate for such regulation than the Polycomb Group of facultative heterochromatin proteins, as they would not be subject to cross regulatory signals from body plan specification pathways.
Figure 6. Model of the interactions between HOAP and HP1 at SxlPe.
(A) Maternal HOAP and HP1 cooperate to form a repressive complex which serves to reduce the sensitivity of SxlPe to spurious fluctuations in zygotic expression of positive (e.g. Sis-a, Sis-b) and negative (e.g. Dpn) regulatory proteins. Pre-loading of RNA pol II requires HP1. (B) Binding of bHLH proteins to SxlPe sequences is dependent on their zygotic dose and binding sites relative to those for HOAP. At 2–3 hr of development, the two X chromosome dose of positive factors in females, is able to result in activation of SxlPe, and allow the pre-loaded RNA pol II to extend. In males, the single X chromosome presumably does not produce enough positive factors to displace (all of) the negative components. For simplicity not all known X∶A factors are shown. The triangle depicts low levels of H3K9 methylation at the promoter.
How HP1 switches over to transcriptional activation mode in the Sxl Pe proximal region is unclear. Changes in HP1 phosphorylation and/or association with other factors could alter its activity [50], [51]. Several XSE binding sites are nearby, making them strong candidates. Presumably, this would only occur in females where the XSE dose surpasses a threshold and SxlPe is activated.
Mechanism of HP1 in Transcriptional Activation at SxlPe
This report provides the most clearly defined role for HP1 in developmental control of a euchromatic gene in a metazoan species, and the first evidence of a bifunctional regulatory role for it in such a context. Prior reports describing HP1 in transcriptional activation have focused on it in the context of transcription elongation [11]–[13], [52], [53]. Our ChIP data at SxlPe, however, show a requirement of it for association of RNAP II with the promoter, more consistent with a role in transcription initiation. A role in initiation is also in keeping with the position of HP1 on the gene; we find very little HP1 elsewhere on the Sxl gene, even during the time of SxlPe activity. This dependence of RNAP II association on HP1 is similar to what is observed in the accumulation of noncoding RNAs at S. pombe centromeric repeats and mating type locus [54]. Nonetheless, it is possible that the loss of RNAP II at SxlPe reflects reduced stability of all RNAP II isoforms as a consequence of an early defect in transcription elongation, rather than a defect in RNAP II recruitment to the promoter.
Pausing of RNAP II in promoter proximal regions prior to activation has been observed in a high proportion of genes under developmental control in Drosophila embryos [55], and such pauses have also been implicated in regulation of alternative splicing [56]. While SxlPm appears to have the features of a promoter with paused RNAP II in a genome wide RNAP II ChIP study of 0–4 hr embryos (Flybase MODENCODE), RNAP II was absent from SxlPe. It is likely that the collection window for this study did not precisely coincide with the time of SxlPe activity. Our more narrowly timed collection indicates paused RNAP II at SxlPe, suggesting that, like SxlPm, it is a pre-loaded promoter. A preloaded SxlPe also readily explains how generalized up-regulation of phosphorylation of the RNAP II CTD by the loss of Nanos, causes SxlPe activation in males with an unchanged X∶A ratio [57].
Evolutionary Implications of HOAP, Similarity to SRY?
Finally, the dominant negative activity of the cav 2248 allele suggests a role for the partially deleted SRY-like HMG box in HOAP association with SxlPe. Our ChIP data show HOAP association with the SxlPe proximal region is required for HP1 association. This proposed role for the HMG box of HOAP in SxlPe regulation is of particular interest with regards to a recent report linking HP1 and KAP-1 (TIF1β) to SRY-dependent repression of testis-specific genes in the ovary [26]. Because mammalian sex determination is inextricably linked to gonad sex determination, SRY and HOAP each appear to constitute early decision points in their respective sex determination pathways. There are, perhaps, unexpected parallels between these divergent pathways.
Materials and Methods
Genetic Analyses
The cav RNAi lines were obtained by germline transformation of the pUAST vector containing an inverted repeat of a near full length cDNA sequence for the cav-RB transcript (See Table S3 for primer sequences). The cav 2248 allele was recovered in a screen of progeny from ethane methyl sulfonate (EMS)-mutagenized males which failed to complement the original cav 1 allele, and the sequence of the allele was determined from PCR products obtained from homozygous cav 2248 embryos identified through their lack of the homologous twi-GAL4, UAS-GFP marked balancer chromosome.
Flies in all crosses were reared under uncrowded conditions on standard cornmeal medium enriched with active dry yeast. Unless otherwise noted, all crosses were done at 25°C; Ore R or y 1 w 67c23 were used as the wild type control. Progeny were counted for 8 days beginning on the first day of eclosion. The Z test was used in statistical analyses of distributions in two populations [58]. Description of genes can be found in Flybase (http://www.flybase.org/).
Microarray Expression Profiling Study
RNA was TRIzol-extracted from y 1 w 67c23; cav 1 homozygous (those lacking the Tm6B,Tb balancer chromosome used to maintain the cav 1 mutation in heterozygous condition) and wild type first and second instar larvae and purified through Qiagen RNeasy columns. The RNA was then used by the University of Kentucky Microarray Facility to prepare biotin-labeled cRNA that was hybridized to separate Affymetrix Drosophila Genome microarrays (version 1). The y 1 w 67c23 stock to which all mutant stocks are out-crossed during genetic manipulations was used as the wild type control in these experiments. The data were obtained from two biological replicate samples from each stock. The statistical analyses of the arrays were carried out by A.J. Stromberg, PhD (UK Dept. of Statistics) associated with this facility according to standard Affymetrix specifications. Of 13,982 affytags to Drosophila genes on the array, an average of 46% and 53% were present in the cav 1 and wild type y 1 w 67c23 samples, respectively. Data from publically available tissue-specific cDNA libraries [35]–[37] and from two published microarray profiling studies of Drosophila sex- or tissue-specific gene expression [38], [39] were used to catalogue normal gene expression profiles of genes with reduced or elevated transcript levels (log2R>2.0, p<0.01).
RT-PCR Analysis
RNA was similarly prepared from pools of male and female cav 2248 heterozygous and cav 1 homozygous embryos and from individual Su(var)205 4/Su(var)205 5 larvae. The cav 2248 heterozygous embryos were identified from a pool of tightly staged embryos that failed to progress to the larval stage and contained intermediate levels of GFP expression from the P{GAL4-twi.C}2.3, P{UAS-2xEGFP}AH2.3-marked TM3Sb balancer chromosome. The cav 1 homozygous embryos were similarly identified through their lack of GFP expression from this balancer chromosome. Su(var)205 4/Su(var)205 5 larvae were identified through their lack of GFP expression from the P{Act5C-GAL4}25F01-marked CyO balancer chromosome. The sex of individual cav 2248 heterozygous and cav 1 homozygous embryos was first determined through PCR of the Y-linked kl-2 gene from a total nucleic acid extraction from individual embryos (See Table S3 for primer sequences). RNA was then purified from pools of nucleic acids from individual male or female embryos. The sex of individual Su(var)205 4/Su(var)205 5 larvae was first determined through the presence or absence of the y +-marker from the paternally derived X chromosome and then substantiated through PCR assays of the Y-linked kl-2 gene. RT-PCR assays were carried out with RNA purified from these pools using the Qiagen One-Step RT-PCR kit (Qiagen 210212). The sequences of all primers used are shown in Table S3. Primers for the Drosophila RpA-70 were used in PCR reactions to determine that all RNA samples were DNA-free before they were used in RT-PCR as a positive control and in normalization of all RT-PCR data (Table S3).
In Situ Hybridization
These were done as described in [59]. Digoxygenin labeled RNA probes complementary to Sxl exon E1 or L1 region were prepared using T7 RNA polymerase in vitro transcription of plasmid- or PCR-derived templates. The establishment (407 nt) and maintenance (1039 nt) transcript specific probes were generated by the primers shown in Table S3. All in situs were repeated at least once. Each batch was done simultaneously with an Ore R control and had sufficient embryos so that several representatives of each cycle could be examined.
Chromatin Preparation
A modification of the protocol described in [60] was used to prepare cross-linked chromatin from embryonic progeny from parents of the following genotypes: wild type (y 1 w 67c23 or yw), yw; cav 2248/TM3Sb, yw; Su(var)205 5/CyO, yw females×Sxl f1/Y males, and yw; Su(var)205 5/CyO females×Sxl f1/Y males. Embryos from yw parents were collected at the following developmental stages: 0.75 to 2.75 hr (labeled 1–3 hr), 2–3 hr, and 4–18 hr. Embryos were collected from yw; Su(var)205 5/CyO females crossed to Sxl f1/Y males in parallel to those from yw females crossed to Sxl f1/Y males (at 2–3 hr stage). Embryos from yw; cav 2248/TM3Sb and from yw; Su(var)205 5/CyO parents were also collected in parallel (at 0.75 to 2.75 stage). All embryo collections and staging were done at 22°C. Chromatin was prepared from 6.0 g yw embryos in 6.0 ml homogenization buffer (50 mM Hepes at pH 7.6, 60 mM potassium chloride, 0.25 M sucrose, protease inhibitor cocktail [15]). The homogenate was first clarified by centrifugation at 500× g for 10 minutes before the addition of formaldehyde to 2%. Cross-linked chromatin was then washed 3 times in phosphate buffered saline (150 mM sodium chloride (NaCl), 10 mM sodium phosphate, pH 7.6) (with centrifugation at 3,000× g for 10 minutes after each wash) and re-suspended in 6.0 ml RIPA buffer (50 mM Tris-HCl, pH 7.6, 1 mM ethylene diamine tetraacetic acid (EDTA), 0.5 mM ethylene glycol tetraacetic acid (EGTA), 140 mM NaCl, 1% Triton X-100, 0.1% Na-deoxycholate, 0.1% SDS) and sonnicated to an average length of 500 bp. A scaled down version of the protocol was carried out with 0.5 g mutant embryos in 1.5 ml homogenization buffer.
Chromatin Immunoprecipitations
Chromatin immunoprecipitations were performed with 0.2 ml clarified chromatin, 20–40 µg antibody [anti-HOAP [61], anti-HP1 [15], anti-RNA polymerase II antibody (Covance MMS-126R, MMS-134, or MMS-129R), anti-di and tri MeK9 histone H3 (Millipore 05-1249 and 05-1242) or non-immune IgG (Santa Cruz Biotech. SC-2027 or SC-2025)] and 100 µl anti-rabbit (Sigma A914) or anti-mouse (Sigma A6531) IgG agarose in 1.5 ml RIPA buffer. Washes were performed as described in Alekseyenko et al., [60]. The immunoprecipitated material was eluted from the beads by incubation at 37°C for 1 hr in 500 µl TE (1 mM EDTA, 50 mM Tris, pH 8.0) containing 0.5% sodium dodecyl sulfate (SDS) and proteinase K (0.1 mg/ml), followed by 12 hr at 65°C after the addition of NaCl to 0.3 M and SDS to 1%. The samples were then extracted once with phenol/chloroform, once with chloroform before ethanol precipitation in the presence of glycogen.
Protocol for Quantitative Real-Time PCR
The iCycler iQ real-time PCR detection system (Bio Rad) was used to quantitate Sxl sequences in the precipitated DNA from each ChIP fraction. The primer pairs shown in Table S3 were used to amplify fragments spanning the Sxl locus as shown in Figure 4 and RpA-70 normalizing standard (average length 288 bp). Similar enrichment values were obtained for all data when calculated as % of total in ChIP vs. input fraction. PCR amplification was performed in duplicate in 50 µl SYBR Green qPCR SuperMix (Bio-Rad 170–8880) on two biological replicates of each ChIP fraction. Dissociation curve analysis was performed at the end of 40 cycles, and quantification was carried out by Bio-Rad comparative CT methodology with standard curves constructed for each primer pair with a serial dilution of input DNA having PCR efficiencies of 80–120%. A one sample t-test was performed to identify sequences that were enriched in ChIP fractions above background (i.e., >1); a student's t test was used to determine significance of differences between two samples of equal variance.
Supporting Information
Characterizations of cav 2248 mutant embryos. (A) Cuticle preparations of genotyped cav 2248 homozygous and heterozygous embryos. (B) DAPI-staining of presumed pre-cycle 14 cav 2248 homozygous embryo and genotyped late-staged cav 2248 heterozygous embryo (Defects in cav 2248 homozygous embryos appear before expression of GFP marker used in genotyping.)
(TIF)
Representative PCR assays used to sex mutant animals. PCR assays of nucleic acids extracted from individual cav 2248 heterozygous (cav 2248/twi-GAL4, UAS-2x-EGFP TM3) or cav 1 homozygous (cav 1/cav 1) embryos or Su(var)205 5/Su(var)205 4 larvae using primers for kl-2 Y-linked gene to sex animals and RpA-70 as a positive control. Male embryos were identified as those yielding products with both sets of primers; female embryos were identified as those yielding products with RpA-70 primers only. RNA was then isolated from pools of individually sexed cav mutant embryos or individual Su(var)205 mutant larvae, and RT-PCR assays with RpA-70 primers were used to assess the quality of each RNA template before using them in RT-PCR assays of Sxl transcripts.
(TIF)
SxlPm transcription is essentially unchanged in embryos from cav2248/TM3, Sb or Su(var)205 5/CyO parents. Comparisons of the same-sized area of images taken at 40×. Early to mid-cycle 14 female embryos from Ore R, cav2248/TM3 (cav 2248) or Su(var)205 5/CyO (Su(var)205 5) parents show essentially equivalent signal. Probe is specific to SxlPm transcripts, spanning exon 1 and small section of adjacent intron.
(TIF)
Summary of tissue distribution of genes with decreased or increased transcript levels in cav 1 mutants.
(DOC)
Categorization of genes with decreased or increased transcript levels in cav 1 mutant larvae. The normal tissue distribution of each gene set was assessed through a combination of data on its relative representation in tissue-specific cDNA libraries (cDNA Library Representation) [35], [36], [37]. Data of tissue-specific expression in adults by Chintapalli et al. [38] (tissue/adult enrichment) and sex-specific gonad expression by Parisi et al. [39] (testis/ovary enrichment).
(DOC)
Oligonucleotides used as primers in this study.
(PDF)
Acknowledgments
We thank Dr. D. Harrison, and we thank members of the Kellum and Horabin labs for helpful discussions.
Footnotes
The authors have declared that no competing interests exist.
This work was supported in part by funds from NIGMS Grant GM08516 to JIH and GM059765 to RK. It was also supported by funds from the University of Kentucky College or Arts and Sciences and Office of Research Affairs to RK. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Heitz E. Das Heterochromatin der Moose. Jahrb Wissensch Bot. 1928;69:762–818. [Google Scholar]
- 2.James TC, Elgin SC. Identification of a nonhistone chromosomal protein associated with heterochromatin in Drosophila melanogaster and its gene. Mol Cell Biol. 1986;6:3862–3872. doi: 10.1128/mcb.6.11.3862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wreggett KA, Hill F, James PS, Hutchings A, Butcher GW, et al. A mammalian homologue of Drosophila heterochromatin protein 1 (HP1) is a component of constitutive heterochromatin. Cytogenet Cell Genet. 1994;66:99–103. doi: 10.1159/000133676. [DOI] [PubMed] [Google Scholar]
- 4.Allshire RC, Nimmo ER, Ekwall K, Javerzat JP, Cranston G. Mutations derepressing silent centromeric domains in fission yeast disrupt chromosome segregation. Genes Dev. 1995;9:218–233. doi: 10.1101/gad.9.2.218. [DOI] [PubMed] [Google Scholar]
- 5.Nielsen SJ, Schneider R, Bauer UM, Bannister AJ, Morrison A, et al. Rb targets histone H3 methylation and HP1 to promoters. Nature. 2001;412:561–565. doi: 10.1038/35087620. [DOI] [PubMed] [Google Scholar]
- 6.Lachner M, O'Carroll D, Rea S, Mechtler K, Jenuwein T. Methylation of histone H3 lysine 9 creates a binding site for HP1 proteins. Nature. 2001;410:116–120. doi: 10.1038/35065132. [DOI] [PubMed] [Google Scholar]
- 7.Schotta G, Ebert A, Krauss V, Fischer A, Hoffmann J, et al. Central role of Drosophila SU(VAR)3-9 in histone H3-K9 methylation and heterochromatic gene silencing. Embo J. 2002;21:1121–1131. doi: 10.1093/emboj/21.5.1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nielsen AL, Oulad-Abdelghani M, Ortiz JA, Remboutsika E, Chambon P, et al. Heterochromatin formation in mammalian cells: interaction between histones and HP1 proteins. Mol Cell. 2001;7:729–739. doi: 10.1016/s1097-2765(01)00218-0. [DOI] [PubMed] [Google Scholar]
- 9.Ekwall K, Ruusala T. Mutations in rik1, clr2, clr3 and clr4 genes asymmetrically derepress the silent mating-type loci in fission yeast. Genetics. 1994;136:53–64. doi: 10.1093/genetics/136.1.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hwang KK, Eissenberg JC, Worman HJ. Transcriptional repression of euchromatic genes by Drosophila heterochromatin protein 1 and histone modifiers. Proc Natl Acad Sci U S A. 2001;98:11423–11427. doi: 10.1073/pnas.211303598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cryderman DE, Grade SK, Li Y, Fanti L, Pimpinelli S, et al. Role of Drosophila HP1 in euchromatic gene expression. Dev Dyn. 2005;232:767–774. doi: 10.1002/dvdy.20310. [DOI] [PubMed] [Google Scholar]
- 12.Fanti L, Berloco M, Piacentini L, Pimpinelli S. Chromosomal distribution of heterochromatin protein 1 (HP1) in Drosophila: a cytological map of euchromatic HP1 binding sites. Genetica. 2003;117:135–147. doi: 10.1023/a:1022971407290. [DOI] [PubMed] [Google Scholar]
- 13.Vakoc CR, Mandat SA, Olenchock BA, Blobel GA. Histone H3 lysine 9 methylation and HP1gamma are associated with transcription elongation through mammalian chromatin. Mol Cell. 2005;19:381–391. doi: 10.1016/j.molcel.2005.06.011. [DOI] [PubMed] [Google Scholar]
- 14.Pak DT, Pflumm M, Chesnokov I, Huang DW, Kellum R, et al. Association of the origin recognition complex with heterochromatin and HP1 in higher eukaryotes. Cell. 1997;91:311–323. doi: 10.1016/s0092-8674(00)80415-8. [DOI] [PubMed] [Google Scholar]
- 15.Huang DW, Fanti L, Pak DT, Botchan MR, Pimpinelli S, et al. Distinct cytoplasmic and nuclear fractions of Drosophila heterochromatin protein 1: their phosphorylation levels and associations with origin recognition complex proteins. J Cell Biol. 1998;142:307–318. doi: 10.1083/jcb.142.2.307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Reinhart BJ, Bartel DP. Small RNAs correspond to centromere heterochromatic repeats. Science. 2002;297:1831. doi: 10.1126/science.1077183. [DOI] [PubMed] [Google Scholar]
- 18.Volpe TA, Kidner C, Hall IM, Teng G, Grewal SI, et al. Regulation of heterochromatic silencing and histone H3 lysine-9 methylation by RNAi. Science. 2002;297:1833–1837. doi: 10.1126/science.1074973. [DOI] [PubMed] [Google Scholar]
- 19.Haynes KA, Caudy AA, Collins L, Elgin SC. Element 1360 and RNAi components contribute to HP1-dependent silencing of a pericentric reporter. Curr Biol. 2006;16:2222–2227. doi: 10.1016/j.cub.2006.09.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Riddle NC, Leung W, Haynes KA, Granok H, Wuller J, et al. An investigation of heterochromatin domains on the fourth chromosome of Drosophila melanogaster. Genetics. 2008;178:1177–1191. doi: 10.1534/genetics.107.081828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hall IM, Shankaranarayana GD, Noma K, Ayoub N, Cohen A, et al. Establishment and maintenance of a heterochromatin domain. Science. 2002;297:2232–2237. doi: 10.1126/science.1076466. [DOI] [PubMed] [Google Scholar]
- 22.Ryan RF, Schultz DC, Ayyanathan K, Singh PB, Friedman JR, et al. KAP-1 corepressor protein interacts and colocalizes with heterochromatic and euchromatic HP1 proteins: a potential role for Kruppel-associated box-zinc finger proteins in heterochromatin-mediated gene silencing. Mol Cell Biol. 1999;19:4366–4378. doi: 10.1128/mcb.19.6.4366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Aguilar-Arnal L, Marsellach FX, Azorin F. The fission yeast homologue of CENP-B, Abp1, regulates directionality of mating-type switching. Embo J. 2008;27:1029–1038. doi: 10.1038/emboj.2008.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nakagawa H, Lee JK, Hurwitz J, Allshire RC, Nakayama J, et al. Fission yeast CENP-B homologs nucleate centromeric heterochromatin by promoting heterochromatin-specific histone tail modifications. Genes Dev. 2002;16:1766–1778. doi: 10.1101/gad.997702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Petrie VJ, Wuitschick JD, Givens CD, Kosinski AM, Partridge JF. RNA interference (RNAi)-dependent and RNAi-independent association of the Chp1 chromodomain protein with distinct heterochromatic loci in fission yeast. Mol Cell Biol. 2005;25:2331–2346. doi: 10.1128/MCB.25.6.2331-2346.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Peng H, Ivanov AV, Oh HJ, Lau YF, Rauscher FJ., 3rd Epigenetic gene silencing by the SRY protein is mediated by a KRAB-O protein that recruits the KAP1 co-repressor machinery. J Biol Chem. 2009;284:35670–35680. doi: 10.1074/jbc.M109.032086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cenci G, Siriaco G, Raffa GD, Kellum R, Gatti M. The Drosophila HOAP protein is required for telomere capping. Nat Cell Biol. 2003;5:82–84. doi: 10.1038/ncb902. [DOI] [PubMed] [Google Scholar]
- 28.Bi X, Srikanta D, Fanti L, Pimpinelli S, Badugu R, et al. Drosophila ATM and ATR checkpoint kinases control partially redundant pathways for telomere maintenance. Proc Natl Acad Sci U S A. 2005;102:15167–15172. doi: 10.1073/pnas.0504981102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gao G, Walser JC, Beaucher ML, Morciano P, Wesolowska N, et al. HipHop interacts with HOAP and HP1 to protect Drosophila telomeres in a sequence-independent manner. Embo J. 2010;29:819–829. doi: 10.1038/emboj.2009.394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Badugu R, Shareef MM, Kellum R. Novel Drosophila heterochromatin protein 1 (HP1)/origin recognition complex-associated protein (HOAP) repeat motif in HP1/HOAP interactions and chromocenter associations. J Biol Chem. 2003;278:34491–34498. doi: 10.1074/jbc.M305262200. [DOI] [PubMed] [Google Scholar]
- 31.Cline TW. A female-specific lethal lesion in an X-linked positive regulator of the Drosophila sex determination gene, Sex-lethal. Genetics. 1986;113:641–663. doi: 10.1093/genetics/113.3.641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Keyes LN, Cline TW, Schedl P. The primary sex determination signal of Drosophila acts at the level of transcription. Cell. 1992;68:933–943. doi: 10.1016/0092-8674(92)90036-c. [DOI] [PubMed] [Google Scholar]
- 33.Estes PA, Keyes LN, Schedl P. Multiple response elements in the Sex-lethal early promoter ensure its female-specific expression pattern. Mol Cell Biol. 1995;15:904–917. doi: 10.1128/mcb.15.2.904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Salz HK, Erickson JW. Sex determination in Drosophila: The view from the top. Fly (Austin) 2010:4. doi: 10.4161/fly.4.1.11277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Andrews J, Bouffard GG, Cheadle C, Lu J, Becker KG, et al. Gene discovery using computational and microarray analysis of transcription in the Drosophila melanogaster testis. Genome Res. 2000;10:2030–2043. doi: 10.1101/gr.10.12.2030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Stapleton M, Carlson J, Brokstein P, Yu C, Champe M, et al. A Drosophila full-length cDNA resource. Genome Biol. 2002;3:RESEARCH0080. doi: 10.1186/gb-2002-3-12-research0080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Stapleton M, Liao G, Brokstein P, Hong L, Carninci P, et al. The Drosophila gene collection: identification of putative full-length cDNAs for 70% of D. melanogaster genes. Genome Res. 2002;12:1294–1300. doi: 10.1101/gr.269102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chintapalli VR, Wang J, Dow JA. Using FlyAtlas to identify better Drosophila melanogaster models of human disease. Nat Genet. 2007;39:715–720. doi: 10.1038/ng2049. [DOI] [PubMed] [Google Scholar]
- 39.Parisi M, Nuttall R, Edwards P, Minor J, Naiman D, et al. A survey of ovary-, testis-, and soma-biased gene expression in Drosophila melanogaster adults. Genome Biol. 2004;5:R40. doi: 10.1186/gb-2004-5-6-r40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Boutanaev AM, Kalmykova AI, Shevelyov YY, Nurminsky DI. Large clusters of co-expressed genes in the Drosophila genome. Nature. 2002;420:666–669. doi: 10.1038/nature01216. [DOI] [PubMed] [Google Scholar]
- 41.Liu LP, Ni JQ, Shi YD, Oakeley EJ, Sun FL. Sex-specific role of Drosophila melanogaster HP1 in regulating chromatin structure and gene transcription. Nat Genet. 2005;37:1361–1366. doi: 10.1038/ng1662. [DOI] [PubMed] [Google Scholar]
- 42.Dimitri P, Pisano C. Position effect variegation in Drosophila melanogaster: relationship between suppression effect and the amount of Y chromosome. Genetics. 1989;122:793–800. doi: 10.1093/genetics/122.4.793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Spofford JB, editor. Position-effect variegation in Drosophila. London: Academic Press; 1976. pp. 955–1018. [Google Scholar]
- 44.Lucchesi JC, Kelly WG, Panning B. Chromatin remodeling in dosage compensation. Annu Rev Genet. 2005;39:615–651. doi: 10.1146/annurev.genet.39.073003.094210. [DOI] [PubMed] [Google Scholar]
- 45.Lyon MF. Gene action in the X-chromosome of the mouse (Mus musculus L.). Nature. 1961;190:372–373. doi: 10.1038/190372a0. [DOI] [PubMed] [Google Scholar]
- 46.Lucchesi JC. The structure-function link of compensated chromatin in Drosophila. Curr Opin Genet Dev. 2009;19:550–556. doi: 10.1016/j.gde.2009.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Spierer A, Begeot F, Spierer P, Delattre M. SU(VAR)3-7 links heterochromatin and dosage compensation in Drosophila. PLoS Genet. 2008;4:e1000066. doi: 10.1371/journal.pgen.1000066. doi: 10.1371/journal.pgen.1000066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Demakova OV, Kotlikova IV, Gordadze PR, Alekseyenko AA, Kuroda MI, et al. The MSL complex levels are critical for its correct targeting to the chromosomes in Drosophila melanogaster. Chromosoma. 2003;112:103–115. doi: 10.1007/s00412-003-0249-1. [DOI] [PubMed] [Google Scholar]
- 49.Lu H, Kozhina E, Mahadevaraju S, Yang D, Avila FW, et al. Maternal Groucho and bHLH repressors amplify the dose-sensitive X chromosome signal in Drosophila sex determination. Dev Biol. 2008;323:248–260. doi: 10.1016/j.ydbio.2008.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Badugu R, Yoo Y, Singh PB, Kellum R. Mutations in the heterochromatin protein 1 (HP1) hinge domain affect HP1 protein interactions and chromosomal distribution. Chromosoma. 2005;113:370–384. doi: 10.1007/s00412-004-0324-2. [DOI] [PubMed] [Google Scholar]
- 51.Shimada A, Dohke K, Sadaie M, Shinmyozu K, Nakayama J, et al. Phosphorylation of Swi6/HP1 regulates transcriptional gene silencing at heterochromatin. Genes Dev. 2009;23:18–23. doi: 10.1101/gad.1708009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lin CH, Li B, Swanson S, Zhang Y, Florens L, et al. Heterochromatin protein 1a stimulates histone H3 lysine 36 demethylation by the Drosophila KDM4A demethylase. Mol Cell. 2008;32:696–706. doi: 10.1016/j.molcel.2008.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Piacentini L, Fanti L, Negri R, Del Vescovo V, Fatica A, et al. Heterochromatin protein 1 (HP1a) positively regulates euchromatic gene expression through RNA transcript association and interaction with hnRNPs in Drosophila. PLoS Genet. 2009;5:e1000670. doi: 10.1371/journal.pgen.1000670. doi: 10.1371/journal.pgen.1000670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zofall M, Grewal SI. Swi6/HP1 recruits a JmjC domain protein to facilitate transcription of heterochromatic repeats. Mol Cell. 2006;22:681–692. doi: 10.1016/j.molcel.2006.05.010. [DOI] [PubMed] [Google Scholar]
- 55.Zeitlinger J, Stark A, Kellis M, Hong JW, Nechaev S, et al. RNA polymerase stalling at developmental control genes in the Drosophila melanogaster embryo. Nat Genet. 2007;39:1512–1516. doi: 10.1038/ng.2007.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kornblihtt AR. Chromatin, transcript elongation and alternative splicing. Nat Struct Mol Biol. 2006;13:5–7. doi: 10.1038/nsmb0106-5. [DOI] [PubMed] [Google Scholar]
- 57.Deshpande G, Calhoun G, Jinks TM, Polydorides AD, Schedl P. Nanos downregulates transcription and modulates CTD phosphorylation in the soma of early Drosophila embryos. Mech Dev. 2005;122:645–657. doi: 10.1016/j.mod.2004.12.009. [DOI] [PubMed] [Google Scholar]
- 58.Sprinthall RC. 2003. Basic Statistical Analysis, Seventh Edition: Pearson Education Group.
- 59.Erickson JW, Cline TW. A bZIP protein, sisterless-a, collaborates with bHLH transcription factors early in Drosophila development to determine sex. Genes Dev. 1993;7:1688–1702. doi: 10.1101/gad.7.9.1688. [DOI] [PubMed] [Google Scholar]
- 60.Alekseyenko AA, Larschan E, Lai WR, Park PJ, Kuroda MI. High-resolution ChIP-chip analysis reveals that the Drosophila MSL complex selectively identifies active genes on the male X chromosome. Genes Dev. 2006;20:848–857. doi: 10.1101/gad.1400206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Gladstein N, McKeon MN, Horabin JI. Requirement of male-specific dosage compensation in Drosophila females—implications of early X chromosome gene expression. PLoS Genet. 2010;6:e1001041. doi: 10.1371/journal.pgen.1001041. doi: 10.1371/journal.pgen.1001041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Jinks TM, Calhoun G, Schedl P. Functional conservation of the sex-lethal sex determining promoter, Sxl-Pe, in Drosophila virilis. Dev Genes Evol. 2003;213:155–165. doi: 10.1007/s00427-003-0304-1. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Characterizations of cav 2248 mutant embryos. (A) Cuticle preparations of genotyped cav 2248 homozygous and heterozygous embryos. (B) DAPI-staining of presumed pre-cycle 14 cav 2248 homozygous embryo and genotyped late-staged cav 2248 heterozygous embryo (Defects in cav 2248 homozygous embryos appear before expression of GFP marker used in genotyping.)
(TIF)
Representative PCR assays used to sex mutant animals. PCR assays of nucleic acids extracted from individual cav 2248 heterozygous (cav 2248/twi-GAL4, UAS-2x-EGFP TM3) or cav 1 homozygous (cav 1/cav 1) embryos or Su(var)205 5/Su(var)205 4 larvae using primers for kl-2 Y-linked gene to sex animals and RpA-70 as a positive control. Male embryos were identified as those yielding products with both sets of primers; female embryos were identified as those yielding products with RpA-70 primers only. RNA was then isolated from pools of individually sexed cav mutant embryos or individual Su(var)205 mutant larvae, and RT-PCR assays with RpA-70 primers were used to assess the quality of each RNA template before using them in RT-PCR assays of Sxl transcripts.
(TIF)
SxlPm transcription is essentially unchanged in embryos from cav2248/TM3, Sb or Su(var)205 5/CyO parents. Comparisons of the same-sized area of images taken at 40×. Early to mid-cycle 14 female embryos from Ore R, cav2248/TM3 (cav 2248) or Su(var)205 5/CyO (Su(var)205 5) parents show essentially equivalent signal. Probe is specific to SxlPm transcripts, spanning exon 1 and small section of adjacent intron.
(TIF)
Summary of tissue distribution of genes with decreased or increased transcript levels in cav 1 mutants.
(DOC)
Categorization of genes with decreased or increased transcript levels in cav 1 mutant larvae. The normal tissue distribution of each gene set was assessed through a combination of data on its relative representation in tissue-specific cDNA libraries (cDNA Library Representation) [35], [36], [37]. Data of tissue-specific expression in adults by Chintapalli et al. [38] (tissue/adult enrichment) and sex-specific gonad expression by Parisi et al. [39] (testis/ovary enrichment).
(DOC)
Oligonucleotides used as primers in this study.
(PDF)