Abstract
Background
The effectiveness of knowledge-based word sense disambiguation (WSD) approaches depends in part on the information available in the reference knowledge resource. Off the shelf, these resources are not optimized for WSD and might lack terms to model the context properly. In addition, they might include noisy terms which contribute to false positives in the disambiguation results.
Methods
We analyzed some collocation types which could improve the performance of knowledge-based disambiguation methods. Collocations are obtained by extracting candidate collocations from MEDLINE and then assigning them to one of the senses of an ambiguous word. We performed this assignment either using semantic group profiles or a knowledge-based disambiguation method. In addition to collocations, we used second-order features from a previously implemented approach.
Specifically, we measured the effect of these collocations in two knowledge-based WSD methods. The first method, AEC, uses the knowledge from the UMLS to collect examples from MEDLINE which are used to train a Naïve Bayes approach. The second method, MRD, builds a profile for each candidate sense based on the UMLS and compares the profile to the context of the ambiguous word.
We have used two WSD test sets which contain disambiguation cases which are mapped to UMLS concepts. The first one, the NLM WSD set, was developed manually by several domain experts and contains words with high frequency occurrence in MEDLINE. The second one, the MSH WSD set, was developed automatically using the MeSH indexing in MEDLINE. It contains a larger set of words and covers a larger number of UMLS semantic types.
Results
The results indicate an improvement after the use of collocations, although the approaches have different performance depending on the data set. In the NLM WSD set, the improvement is larger for the MRD disambiguation method using second-order features. Assignment of collocations to a candidate sense based on UMLS semantic group profiles is more effective in the AEC method.
In the MSH WSD set, the increment in performance is modest for all the methods. Collocations combined with the MRD disambiguation method have the best performance. The MRD disambiguation method and second-order features provide an insignificant change in performance. The AEC disambiguation method gives a modest improvement in performance. Assignment of collocations to a candidate sense based on knowledge-based methods has better performance.
Conclusions
Collocations improve the performance of knowledge-based disambiguation methods, although results vary depending on the test set and method used. Generally, the AEC method is sensitive to query drift. Using AEC, just a few selected terms provide a large improvement in disambiguation performance. The MRD method handles noisy terms better but requires a larger set of terms to improve performance.
Introduction
Word sense disambiguation (WSD) is an intermediate task within information retrieval and information extraction, attempting to select the proper sense of ambiguous words. For instance, the word cold could either refer to low temperature or the viral infection.
Existing knowledge sources, such as the Unified Medical Language System (UMLS)® [1,2], are used to annotate terms in text. The UMLS 2009AB version has at least 24,000 ambiguous terms, i.e. where a given term is part of more than one concept unique identifier (CUI) in the Metathesaurus. These ambiguous cases increase if we consider term variability introduced by matching algorithms. An example of an automatic text annotation tool is MetaMap [3], which annotates spans of text with UMLS Concept Unique Identifiers (CUIs). Ambiguity of terms in knowledge repositories pose a challenge to these tools which rely primarily on string matching techniques to map the candidate concepts to the terms in the text. Failure to properly disambiguate ambiguous annotations has a negative impact in tasks such as information retrieval, information extraction or text summarization.
Among the available approaches to perform WSD, statistical learning approaches achieve better performance [4-6]. On the other hand, statistical learning approaches require manually annotated training data for each ambiguous word to be disambiguated. The preparation of this data is very labor intensive and therefore scarce. Manual annotation to cover all of the ambiguous cases of a large resource like the UMLS is infeasible.
Knowledge-based methods do not require manual annotation and are an alternative to statistical learning methods but typically result in a lower performance [5]. These methods compare the overlap of the context of the ambiguous word to candidate senses in the reference knowledge base.
In some cases, the reference resource used in knowledge-based methods might lack content to properly differentiate the senses of an ambiguous word. We are interested in identifying this missing content automatically and transferring contextual information of ambiguous words to existing resources. Specifically, we are interested in improving the content of the UMLS Metathesaurus® to enhance WSD based on knowledge-based methods. In this work, we focus on the first task which collects collocations using several heuristics.
Several approaches have been proposed in the literature to collect collocation information for the purpose of aiding disambiguation methods. In the biomedical domain, Stevenson et al. [7] use a relevance feedback method to extract terms which could be used to further identify relevant examples for disambiguation. They found that there was a small decrease in performance compared to the baseline approach.
In addition, preliminary work that we have done using similar approaches to extract from an automatically generated corpus for each one of the senses of the ambiguous word decreased the quality of the final corpus. One of the problems is that the original query retrieves non-relevant documents which added noisy terms to the expanded query. To alleviate this problem, we propose a method to reduce the noise returned by the query in order to increase the accuracy of the disambiguation model. First, we identify terms which form a collocation with the ambiguous word; and second, we assign one of the senses to the collocation using several disambiguation approaches. We compare these collocations with second-order features using a method implemented for WSD [8]. The presented methods rely on the extraction of terms from MEDLINE® [9] related to the ambiguous word and then on its categorization into candidate senses. This article is organized as follows. In the next section, we introduce: the UMLS, used as a knowledge source for WSD methods; MEDLINE, used as a resource to identify collocations; and finally the word sense disambiguation methods used to evaluate the extraction of collocations. Then, we describe the methods used in this work, which includes collocation extraction methods, the changes done to the disambiguation methods to accommodate the collocations and the evaluation test sets. Finally, we show the results and conclusions and propose direction for future work.
Background
In this section, we introduce the components required by the experiments described in the methods section: the knowledge source used (UMLS), the corpus used to extract collocations (MEDLINE) and the knowledge-based WSD methods used to evaluate the impact of the distilled collocations.
UMLS
The National Library of Medicine’s (NLM) UMLS provides a large resource of knowledge and tools to create, process, retrieve, integrate and/or aggregate biomedical and health data. The UMLS has three main components:
• Metathesaurus, a compendium of biomedical and health content terminological resources under a common representation which contains lexical items for each one of the concepts and relations among them. In the 2009AB version, it contains over a million concepts.
• Semantic network, which provides a categorization of Metathesaurus concepts into semantic types. In addition, it includes relations among semantic types.
• SPECIALIST lexicon, containing lexical information required for natural language processing which covers commonly occurring English words and biomedical vocabulary.
Concepts are assigned a unique identifier (CUI) which has linked to it a set of terms that denotes alternative ways to represent the concept in text. These terms, depending on the availability, are represented in several languages, although only English terms are used in this work. Concepts are assigned one or more semantic types. Concepts may have a definition linked to them and sometimes more than one from multiple sources. Relations between concepts are often available. All the information about a concept can be traced back to the resource from where it was collected.
For example, the concept with CUI C0009264 denotes the idea of cold temperature. According to the Metathesaurus, terms like cold, cold temperature and low temperature could be used to express this idea. In addition, two definitions are available for this concept (from MeSH and from the NCI Thesaurus), e.g. An absence of warmth or heat or a temperature notably below an accustomed norm. Several related concepts can be found for this concept. For instance, sibling concepts (heat), hypernyms (temperature) and non-taxonomically related concepts (cold storage, cryotherapy).
MEDLINE
MEDLINE is an abbreviation for Medical Literature Analysis and Retrieval System Online. It is a bibliographic database containing over 18 million citations to journal articles in the biomedical domain and is maintained by NLM. Currently the citations come from approximately 5,200 journals in 37 different languages starting from 1949. The majority of the publications are scholarly journals but a small number of newspapers, magazines, and newsletters have been included. MEDLINE is the primary component of PUBMED® [10] which is a free online repository allowing access to MEDLINE as well as other citations and abstracts in the fields of medicine, nursing, dentistry, veterinary medicine, health care systems, and pre-clinical sciences.
Word sense disambiguation methods
We have considered two knowledge-based disambiguation methods which have already been compared in previous work [5,6]. These methods are supported by different assumptions, so the collocations they produce will have differences, which we are interested to compare. The first method, AEC, uses UMLS knowledge to build queries to collect training data for a statistical learning method. The learned model is, then, used to disambiguate the context of the ambiguous word. The second method, MRD, builds a concept profile which is compared to the context of the ambiguous word.
The Automatic Extracted Corpus (AEC) Method
The Automatic Extracted Corpus (AEC) Method attempts to alleviate the problem of requiring manually annotated training data for supervised learning algorithms. In this method, training data is automatically created for a statistical learning algorithm; this automatically generated data is used to train the learning algorithm to disambiguate ambiguous terms.
The training data is automatically generated using documents from MEDLINE. To create the training data, we automatically generate queries using English monosemous relatives11 of the candidate concepts which, potentially, have an unambiguous use in MEDLINE. The list of candidate relatives includes synonyms and terms from related concepts. Documents retrieved using PUBMED are assigned to the concept which was used to generate the query. The retrieved documents are used to create training examples for each sense.
This training data is used to train a Naïve Bayes classifier using the words surrounding the ambiguous words as features. Disambiguation is performed using the trained model with new examples where the ambiguous word has to be disambiguated. The trained model is evaluated against a manually annotated set from which accuracy values are recorded.
In some cases, automatically generated queries retrieved no citations for a given sense of an ambiguous term. In the experiments reported in this study, we have randomly selected documents from MEDLINE for the senses in which no citation is retrieved. This has shown to improve the results for ambiguous terms like determination and growth. This also explains the differences with the results reported in [5,12].
The Machine Readable Dictionary (MRD) Method
In this method, context words surrounding the ambiguous word are compared to a profile built from each of the UMLS concepts linked to the ambiguous term being disambiguated. Vectors of concept profiles linked to an ambiguous word and word contexts are compared using cosine similarity. The concept with the highest cosine similarity is selected. This method has been previously used by McInnes [13] in the biomedical domain with the NLM WSD data set.
A concept profile vector has as dimensions the tokens obtained from the concept definition (or definitions) if available, synonyms and related concepts excluding siblings. Stop words are discarded, and Porter stemming is used to normalize the tokens. In addition, the token frequency is normalized based on the inverted concept frequency so that tokens which are repeated many times within the UMLS will have less relevance.
Methods
As introduced above, we would like to improve the matching of the contextual features of ambiguous terms to the information available in the UMLS Metathesaurus.
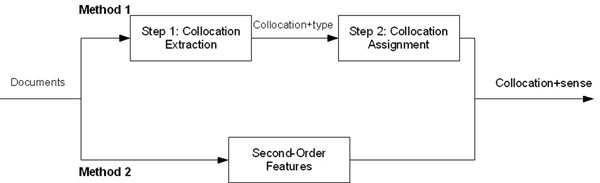
Figure 1 shows a diagram with different methods which are described in this section. In the first method, we describe the process used to extract collocation features from text and how these collocations are assigned to the senses of the ambiguous word. In the second method, we describe a process which extracts second-order features which is combined with the disambiguation algorithms presented above.
Figure 1.
Process diagram
The disambiguation methods presented above add the terms extracted by these methods to the set of terms obtained from the related concepts for each one of the candidate concepts. The changes required to accommodate the collocations into each one of the disambiguation methods are presented in this section.
Method 1: Collocation features
For our processing, we assume one-sense-per-collocation and one-sense-per-document as suggested by Yarowsky [14]. In our study, collocations present one more difficulty since the collocations have to be assigned to one sense, or none if it can co-occur with more than one candidate sense.
The process used to obtain collocations associated to one of the senses is split into two main steps. First, collocations are obtained from MEDLINE from a set of retrieved citations per ambiguous words. These citations are processed to extract different types of collocations. Then, collocations are assigned to one of the candidate senses of the ambiguous word.
Step 1: Collocation extraction
Extraction of collocations from MEDLINE is performed in several steps. First, 1,000 citations are retrieved containing one of the ambiguous terms using PUBMED. Then, several collocation types are used to perform term extraction. These collocation types are:
• Left side collocations
Left side collocations are terms which act as modifiers of the ambiguous term and which occur to the left of it. This combination with the ambiguous word will produce a hyponym which will have a lower chance of being ambiguous. Left side collocations have been explored by Rosario et al. [15], even though her approach had problems when dealing with ambiguous terms.
• Co-occurrence collocations
In Yarowsky’s work [14], the term collocation does not mean words which appear one adjacent to the other but words co-occurring in the same document. We use this definition in this type of collocation. This will produce a larger set of terms which might be noisier compared to the other groups.
• Syntactic dependent collocations
We have considered words occurring within a MEDLINE citation text and we have selected terms, on which a dependency is identified using a syntactic parser. To extract the dependent terms the citations are parsed using the Stanford Parser [16]. This method might extract terms which are less noisy that the ones obtained using co-occurrence collocations.
Once we have extracted these candidate terms, we assess if two words occur together more often than chance using hypothesis testing. In this case, the null hypothesis H0 is formulated as no association between the words other than chance occurrences. We determine if the collocation is statistically significant using the t-test as the statistical hypothesis test [17] with a confidence level of α = 0.005. Some of these collocations are general terms (e.g., age, study, results) which might be related to any of the senses of an ambiguous term. These non-discriminant terms might cause problems, like query drift, to methods like AEC. In addition, some of the terms are very frequent with high probability of occurrence in MEDLINE. We filter out terms with more than 400k occurrences in MEDLINE. This threshold has been established using as reference a standard information retrieval stop word list.
Tables 1, 2 and 3 show examples of collocations, where the headers of the table are ambiguous terms.
Table 1.
Left side collocation examples
| Adjustment | Determination | Repair |
|---|---|---|
| psychosocial | quantitative | dna |
| psychological | spectrophotometric | excision |
| social | photometric | mismatch |
| marital | potentiometric | surgical |
| occlusal | accurate | hernia |
Table 2.
Collocation examples based on co-occurrences
| Adjustment | Determination | Repair |
|---|---|---|
| age | chromatography | damage |
| study | liquid | injury |
| results | standard | defect |
| women | chromatographic | strand |
| data | quantitative | excision |
Table 3.
Collocation examples filtered using the Stanford parser
| Adjustment | Determination | Repair |
|---|---|---|
| measures | assay | damage |
| illness | procedure | injury |
| parents | paper | dna damage |
| social support | techniques | |
| recurrence |
Step 2: Collocation assignment to ambiguous term sense
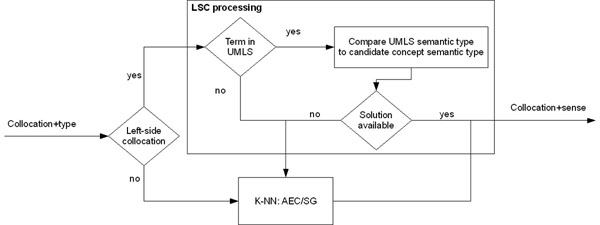
Extracted collocations are assigned to one of the senses of the ambiguous term. This task is not straightforward since assigning a collocation to one of the ambiguous senses requires some notion of disambiguation. Figure 2 shows the two processes we use to assign a collocation to one of the senses. The first process is strictly for left side collocations using because of their relevant word composition properties which can be exploited in the assignment to one of the candidate senses. The second process is used for the remainder of the collocations and those left side collocations that were not assigned a sense using LSC processing. This process assigns senses to the collocations based on the k-NN algorithm. We discuss each of these processes below.
Figure 2.
Collocation assignment diagram
LSC processing
In the case of left side collocations, we use the Metathesaurus to do a preliminary assignment of the ambiguous word based on UMLS semantic types. In refinement or adaptation of existing lexical and ontological resources, head and modifier heuristics are often used to identify new hyponyms. In our work, as the head noun is an ambiguous term, we need a different way to perform this assignment. As each UMLS concept is assigned one or more semantic types, we propose to classify these terms into one of these categories.
Then, we look for the term in the UMLS Metathesaurus and, if the term already exists, use the semantic type already assigned to the term to assign the sense of the ambiguous term. In addition, this might be used to identify relations between existing terms in the Metathesaurus which are not already related.
We have found that some related terms have similar semantic types but cannot be identified just by looking at a flat structure of semantic types. For instance, cerebrospinal fluid is assigned to Body Substance while the related ambiguous sense of fluid is assigned to Substance. In this work, the taxonomy of the UMLS Semantic Network is used to identify these cases. This is an improvement on [12], where only the semantic group derived from the semantic type is used without considering the taxonomy provided by the semantic network. If the same semantic type is assigned to more than one of the senses of the ambiguous term, then we discard this collocation term since we rely in the semantic type to do the term categorization.
k-NN approach
For the other collocation types, we used a k-NN (k-Nearest Neighbor) approach. This approach is used, as well, if the LSC processing cannot assign one of the candidate senses. In the k-NN approach, examples of the collocation with the ambiguous term are collected retrieving 100 documents from PUBMED. We give more relevance to precision, so we avoid taking any categorization where the number of neighbors is lower than 66 out of 100 votes. We choose a large number of examples and a large number of neighbors, over half of the examples, to discard collocations which might be used in combination with any of the candidate senses of the ambiguous word.
The assignment of a candidate sense is done using one of the following methods. The first method simply uses the Naïve Bayes algorithm to train a model using the AEC corpus. The model is then used to assign a sense to each of the collocations.
The second method performs categorization of the examples into one of the semantic groups derived from the concept metadata. In cases where the concepts in the Metathesaurus are assigned to the same semantic group this method cannot be applied. The following section explains how these sets are built. As we have seen in the discussion of the approaches above, we can make use of categorization of terms or citations. Unfortunately, we have no manually annotated terms or citations with semantic groups in MEDLINE to train a classifier, therefore, we build profile vectors for UMLS semantic types and groups based on MEDLINE and monosemous terms.
For each semantic type, a profile vector is built as follows. Monosemous terms are selected randomly from the UMLS. MEDLINE citations containing these monosemous terms are retrieved using PUBMED. Sentence boundaries are detected and sentences containing the monosemous terms are selected. This corpus is tokenized and lowercased, and stopwords are removed. Dimensions of the vector are the extracted tokens. Each dimension in the vector is assigned a weight with the frequency in the corpus multiplied by the inverse document frequency obtained from MEDLINE. As explained above, profile vectors for terms and citations are obtained in a similar way.
In Table 4, top terms in the profile vectors are shown for selected semantic types. We find that semantic types T046 (Pathologic Function) and T047 (Disease or Syndrome) are quite similar; so it is difficult to provide a proper classification into semantic types given a disorder. The same thing happens with semantic types T116 (Amino Acid, Peptide, or Protein) and T126 (Enzyme).
Table 4.
Example top terms for profile vectors for semantic types
| Type: T046 | Type: T047 | Type: T116 | Type: T126 |
|---|---|---|---|
| patients | patients | activity | activity |
| management | case | delta | ec |
| case | hypoxic | rat | delta |
| cases | raeb | human | liver |
| diagnosis | management | liver | human |
| acute | diagnosis | ec | rat |
| treatment | treatment | deficiency | mitochondrial |
| spontaneous | allergic | mitochondrial | activities |
| massive | patient | alpha | enzyme |
| chronic | cases | enzyme | inhibition |
Fortunately, there is a higher-level semantic categorization which clusters semantic types into semantic groups. In this categorization, T046 and T047 belong to the group DISO (Disorders) and T116 and T126 to the group CHEM (Chemicals & Drugs). Semantic group profile vectors are built on the semantic type profiles. Semantic types are assigned to one semantic group. So retrieved sentences belonging to a semantic type are assigned to its semantic group. This corpus is processed as explained above to produce the profile vectors. Top terms for selected semantic groups are shown in Table 5.
Table 5.
Example top terms for profile vectors for semantic groups
| Grp: DISO | Grp: CHEM | Grp: CONC | Grp: ANAT |
|---|---|---|---|
| patients | human | health | human |
| case | activity | patients | rat |
| treatment | acid | based | cells |
| cases | effects | study | function |
| diagnosis | effect | children | anatomy |
| management | rat | inter | normal |
| children | alpha | care | patients |
| congenital | synthesis | medical | case |
| patient | mg | data | left |
| syndrome | treatment | evaluation | neurons |
Cosine similarity is used to compare the profile vector of a given semantic group (c) from the set (C) with the profile vectors of terms and citations (cx) used above; as shown in Equation 1.
| (1) |
Categories like CONC (Concepts & Ideas) or ANAT (Anatomy) do not seem to behave coherently in a manual assessment and are not considered in any of the approaches presented in this study. The CONC group is very generic and its profile seems to always rank higher than any other group profile. The ANAT group is never assigned since the different body parts are linked to a disorder, which is always ranked higher.
Method 2: Second-order features
Second-order co-occurrence vectors were first described by Schütze [18] and extended by Purandare and Pedersen [19] and Patwardhan and Pedersen [20] for the task of word sense discrimination. Later, McInnes [8] adapted these vectors for the task of disambiguation rather than discrimination.
We extract second-order features for our purposes by first creating a co-occurrence matrix in which rows represent the words surrounding the ambiguous term, and the columns represent words that co-occur in a corpus with those words. Each cell in this matrix contains the frequency in which the word found in the row occurs with the word in the column. We use the second-order features that occur in the matrix at least five times but no more than 150. The second-order features are combined with the first-order features when performing disambiguation.
Modification of WSD algorithms
Once we have obtained the collocations, we have to adapt each one of the disambiguation algorithms to combine these new features with the ones already obtained from the UMLS Metathesaurus.
Method 1: modification of MRD
This disambiguation method collects terms from the Metathesaurus for each candidate sense to produce a profile used to compare to the context of the ambiguous word. To generate this profile, the terms extracted from the Metathesaurus are put into a list of words. The integration of the collocations and second-order features is done by simply adding the terms within this list.
Method 2: modification of AEC
This disambiguation method builds a query using terms from the Metathesaurus for each candidate concept. As mentioned above, terms are collected from the synonyms and the related terms. AEC makes the distinction between synonyms and related terms [5]. Synonyms are added as alternative wordings of the ambiguous word. Related terms are combined with the ambiguous word assuming one sense per collocation. AEC is modified as follows in order to accomodate the extracted collocations and second-order features into the list of terms obtained from the Metathesaurus for each one of the senses.
For collocations, the left side collocations are used, in order to generate the query, as synonyms since they are hyponyms of the ambiguous word and as alternative wording of the ambiguous word once they are combined with the ambiguous word. This means that if dna is a left side collocation for one of the candidate senses of repair, it is added in the list of synonyms as dna repair.
The other collocation types and second-order features are considered as related terms and added to the list of related terms for each one of the candidate senses to which the feature has been assigned.
Evaluation data sets
An evaluation has been performed on two available data sets which have been annotated with Metathesaurus concept identifiers. These data sets are based on examples from MEDLINE but they have been developed using different approaches.
The NLM WSD data set [21,22] contains 50 ambiguous terms which have been annotated with a sense number. Each sense number has been mapped to a UMLS concept identifier. 100 manually disambiguated cases are provided for each term. In case no UMLS concept is appropriate, None of the above has been assigned in the NLM WSD. The selection of the 50 ambiguous words was based on an ambiguity study of 409,337 citations added to the database in 1998. MetaMap was used to annotate UMLS concepts to the titles and abstracts based on the 1999 version of the UMLS. 50 highly frequent ambiguous strings were selected for inclusion in the test collection. Out of 4,051,445 ambiguous cases found in these citations, 552,153 cases are represented by these 50 terms. This means that a large number of ambiguous cases can be solved dealing with these highly frequent cases. A team of 11 individuals annotated the ambiguous cases with Metathesaurus concept identifier. The data set is available from [23]. No CUIs were provided with the set, but there is a mapping to UMLS CUIs for the 1999 version of the UMLS Metathesaurus. In addition, from the same site [22] it is possible to obtain the version of the UMLS used for the development of the NLM WSD data set which we have used in our work. We have considered the same setup as Humphrey et al. [24] and discarded the None of the above category. Since the ambiguous term association has been assigned entirely to None of the above, it has been discarded. This means that we will present results for 49 out of the 50 ambiguous terms.
In addition, we used a second WSD test set, referred to as the MSH WSD set, developed automatically using MeSH indexing from MEDLINE [6]. This automatically developed set is based on the 2009AB version of the Metathesaurus and MEDLINE up to May 2010 using PUBMED to recover the documents. The Metathesaurus is screened to identify ambiguous terms which contain MeSH headings. Then, each ambiguous term and the MeSH headings linked to it are used to recover MEDLINE citations using PUBMED where the term and only one of the MeSH headings co-occur. The term found in the MEDLINE citation is assigned the UMLS concept identifier linked to the MeSH heading. Because this initial set is noisy, we filtered out some of the ambiguous terms to enhance precision of the set. The filtering process targeted cases where at least 15 examples are available for each sense, filtered out noisy examples and ensured that each ambiguous word has more than 1 character. This filtered set has 203 ambiguous terms and includes not only words but abbreviations which, in some cases, are used as terms. In addition, it covers a larger set of semantic types compared to the NLM WSD set.
Results
In this section, we present the comparison of the performance of the disambiguation methods before (Initial system) and after using the different collocation types. Comparisons of the results with different values of the different configurations are presented. Accuracy is used to compare the approaches and is defined in Equation 2.
| (2) |
Statistical significance of the results is done using a randomization version of the two sample t-test [25]. In the result tables, · indicates p < 0.1, † indicates p < 0.05 and ‡ indicates p < 0.01.
Words occurring in the citation text where the ambiguous terms appear are used as the context of the ambiguous word. Two baselines are used to compare the approaches. The first one is the Maximum Frequency Sense (MFS) baseline, where the counts are obtained from the benchmark. This baseline is standard in WSD evaluation. The results are also compared against a Naïve Bayes (NB) approach. NB is trained and tested using the evaluation sets sampled based on 10-fold cross-validation.
Tables 6 and 7 compare the baseline results and the initial configuration without collocations to the results after adding the collocations, where LSC stands for left side collocations, Coll stands for co-occurrence collocations and CollParser stands for syntactic dependent collocations. These tables contain the highest performance for each approach, where different parameters have been tested. We find that the semantic group profiles used to assign collocations to candidate senses obtain a high accuracy in the NLM WSD set, but add noise to the MSH WSD set. Second-order features have two results per method. In the first one (2-MRD), all the features which appear more than five times are used while in the second one (2-MRDFilter) only the collocations which, in addition, are statistically significant are considered. This allows us to use these features with the AEC method which otherwise could not cope with a large set of features. Second-order features after filtering provide the largest improvement to the MRD method with the NLM WSD data set but it adds noise to the queries built by the AEC approach.
Table 6.
NLM WSD results comparing the baselines and the proposed methods
| AEC | MRD | |
|---|---|---|
| Initial | 0.7007 | 0.6362 |
| LSC | 0.7226† | 0.6368 |
| Coll | 0.7163 | 0.6365 |
| CollParser | 0.7233† | 0.6406 |
| 2-MRD | - | 0.7158‡ |
| 2-MRDFilter | 0.6295 | 0.6825‡ |
| MFS | 0.8550 | 0.8550 |
| NB | 0.8830 | 0.8830 |
Accuracy results of the different methods using the NLM WSD set. The Initial system consists of the knowledge-based method being evaluated and off the shelf UMLS.
Table 7.
MSH WSD results comparing the baselines and the proposed methods
| AEC | MRD | |
|---|---|---|
| Initial | 0.8383 | 0.8070 |
| LSC | 0.8416 | 0.8082 |
| Coll | 0.8407 | 0.8104† |
| CollParser | 0.8409 | 0.8098· |
| 2-MRD | - | 0.8069 |
| 2-MRDFilter | 0.8313 | 0.8072 |
| MFS | 0.5448 | 0.5448 |
| NB | 0.9386 | 0.9386 |
Accuracy results of the different methods using the MSH WSD set. The Initial system consists of the knowledge-based method being evaluated and off the shelf UMLS.
Results with thresholds for the k-NN method and the AEC categorization method to assign the different senses are presented in Tables 8 and 9. We find that the semantic group approach works reasonably well on the NLM WSD set but decreases in performance on the MSH WSD set, but the contrary is true for the AEC categorization. Considering the disambiguation approaches, the AEC method prefers higher threshold values compared to the MRD method. A higher value means higher confidence on the assignment to one of the candidate senses and will prefer precision to recall in the assignment. This explains as well the performance of the second-order features with these sets, where the MRD has an improvement in performance while AEC has a decrease in performance.
Table 8.
NLM WSD results at different k-NN threshold levels
| AEC | MRD | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| 66 | 75 | 85 | 95 | 66 | 75 | 85 | 95 | ||
| LSC | 0.7226 | 0.7220 | 0.7201 | 0.7082 | 0.6368 | 0.6368 | 0.6360 | 0.6360 | |
| SG | Coll | 0.7163 | 0.7038 | 0.7102 | 0.7055 | 0.6365 | 0.6365 | 0.6363 | 0.6363 |
| CollParser | 0.7120 | 0.7198 | 0.7233 | 0.7055 | 0.6362 | 0.6364 | 0.6362 | 0.6356 | |
| LSC | 0.7052 | 0.7050 | 0.7110 | 0.7053 | 0.6348 | 0.6348 | 0.6344 | 0.6352 | |
| kAEC | Coll | 0.7128 | 0.7027 | 0.6992 | 0.7004 | 0.6358 | 0.6359 | 0.6347 | 0.6347 |
| CollParser | 0.7118 | 0.7023 | 0.7079 | 0.6969 | 0.6406 | 0.6372 | 0.6356 | 0.6357 | |
Disambiguation results in terms of accuracy using the NLM WSD set. Several k-NN values are used in combination with the semantic group (SG) and the automatic extracted corpus (kAEC) methods. The disambiguation methods AEC and MRD are compared.
Table 9.
MSH WSD results at different k-NN threshold levels
| AEC | MRD | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| 66 | 75 | 85 | 95 | 66 | 75 | 85 | 95 | ||
| LSC | 0.8370 | 0.8371 | 0.8371 | 0.8377 | 0.8071 | 0.8070 | 0.8071 | 0.8071 | |
| SG | Coll | 0.8173 | 0.8214 | 0.8268 | 0.8327 | 0.8082 | 0.8077 | 0.8073 | 0.8071 |
| CollParser | 0.8284 | 0.8271 | 0.8337 | 0.8355 | 0.8076 | 0.8071 | 0.8071 | 0.8071 | |
| LSC | 0.8391 | 0.8413 | 0.8416 | 0.8400 | 0.8072 | 0.8072 | 0.8072 | 0.8071 | |
| kAEC | Coll | 0.8252 | 0.8331 | 0.8385 | 0.8407 | 0.8104 | 0.8104 | 0.8100 | 0.8092 |
| CollParser | 0.8298 | 0.8337 | 0.8396 | 0.8409 | 0.8098 | 0.8093 | 0.8090 | 0.8090 | |
Disambiguation results in terms of accuracy using the MSH WSD set. Several k-NN values are used in combination with the semantic group (SG) and the automatic extracted corpus (kAEC) methods. The disambiguation methods AEC and MRD are compared.
Discussion
Our results show that collocations improve the performance of the two knowledge-based methods used in this work. In addition, the methods had different effects on these sets which have shown a similar behavior while assigning collocations to candidate concepts. Due to this, results per disambiguation sets are presented below. Furthermore, semantic categorization based on semantic group profiles is not effective with the MSH WSD set.
If we compare the collocation results to Naïve Bayes, we find that statistical learning still achieves a higher performance. On the other hand, there is not enough manually annotated training data to cover all the UMLS ambiguity cases and the collocation results are the best ones available for a production system. The collection of manually annotated data is quite expensive. Manually annotating the 50 ambiguous words for the NLM WSD set spanned over several months involving 9 people. If we consider that the 2009AB has more than 24K ambiguous words, without considering morphological variations, we can see that it becomes infeasible to build a manually annotated training set. In addition, this set would need to be maintained and updated as soon as new UMLS versions become available.
NLM WSD corpus
Second-order features allow the MRD method to obtain the largest increase in performance. The ambiguous terms with the largest increase in performance are extraction, single and energy. The ambiguous terms with the largest decrease in performance are japanese and ultrasound. A largest improvement is obtained if we do not further filter the proposed features, which indicates that, in this data set, more features provide a better representation of the profile vector. On the other hand, the AEC method has lower performance after considering the second-order features. The AEC method is more sensitive to noise, so a more restricted set of features might provide better performance.
The MFS baseline achieves better performance compared to the knowledge based methods evaluated, including the methods which contain the collocations. Frequencies are obtained from the data set used for evaluation and are not available from any resource, including the UMLS. This means that senses are skewed and usually one of them has a higher chance of occurring and the proposed approaches cannot perform better than this baseline, as usually happens in the evaluation of WSD methods.
Left side collocations and dependent collocations seem to give a larger improvement. Left side collocations provide a narrower meaning of the ambiguous word; they are usually not ambiguous and seem to be assigned to the proper sense. This is partially because terms formed with these collocations and the ambiguous word found in the UMLS Metathesaurus are automatically classified into the proper semantic type. This means that the mistakes of the semantic group categorizer have a smaller impact. We find as well that using the UMLS Semantic Network taxonomy to link related types (e.g. Substance and Body Substance) improves over our previous work [12].
Collocations restricted to dependencies with the ambiguous term seem to further filter some of the spurious terms. On the other hand, we can still see some loss in accuracy compared to the original query. For example, the term nurse is assigned to the ambiguous term support.
Considering collocations within the citation text, we find that the performance increase is not that significant. This might be due to categorizer mistakes. Part of these mistakes are due to terms which could either be assigned to more than one sense of the term, or that are not related to any of the senses of the ambiguous terms. For example, terms like medicine, practice and problems are assigned to one of the senses of the ambiguous sense of pathology.
The approaches developed in our work rely on the ranking of categories provided by several categorizers. Different granularities should be considered in the categorization of entities because the coverage of the current approach is narrowed by the number of categories on which it can be applied. In addition, this process relies on the ranking of the categories, and it considers all the text in the citation so many different topics might be discussed in the document which might be similar to the topic of a different sense of the ambiguous term in the citation.
Finally, there are some ambiguous terms within the NLM WSD benchmark for which collocations could not be identified. These terms are: blood pressure, pressure, growth.
MSH WSD corpus
The AEC disambiguation method provides lower improvement compared to the results obtained with the NLM WSD set. Again, left side collocations provided an improved performance over the other types. AEC method is more sensitive to noise in the set of suggested collocations compared to the MRD method.
Simply considering the term European bat for the M2 sense (mammal) of the term BAT allows obtaining better examples considering using the AEC method. The ambiguous term cortex is another example. It refers to either the cerebral cortex or to the adrenal cortex disease. Just the added term adrenal cortex seems to identify more appropriate examples compared to the other terms in the Metathesaurus like adrenal cortex disease. On the other hand, in RBC the two candidate senses either refer to red blood cells or the counting of red blood cells. This example is similar to blood pressure in the NLM WSD set, so it is easy to add noise using the distilled collocations. Furthermore, short acronyms with a high ambiguity level like DE which stands for Delaware and Germany are prone to retrieve documents with senses not covered in the Metathesaurus. Collocations in this case contribute to the noise of the original query.
The MFS baseline achieves a lower performance compared to the MSH WSD set. Frequencies are obtained from the data set used for evaluation. In this case, the data set has been balanced to deliver the same number of examples per sense when possible. This explains why the MFS for the MSH WSD is close to 0.50, so over a random behavior.
The second-order features cause a non-significant change in performance for the MRD method. As in the NLM WSD data set, the AEC method has a lower performance. Compared to the performance with the NLM WSD data set, this might indicate that the features extracted by the method did not contribute to produce better profile vectors. An explanation could be that the terms in the NLM WSD have higher frequency in MEDLINE, and consequently a larger number of co-occurring terms in the UMLS.
We can see as well that the assignment of the collocations to the senses using the semantic group categorization degrades performance. This is not surprising if we consider the results of a similar approach called JDI [24] on this data set as shown in [6]. This means as well that the JDI approach might perform reasonably well on a limited set of semantic categories and perform poorly on the rest.
Conclusions and future work
Collocations improve the performance of knowledge-based disambiguation methods, even though conclusions differ for each set and method. In the NLM WSD set, the improvement is larger for the MRD method using second-order features followed by the AEC method using dependent and left side collocations. Assignment of collocations to a candidate sense based on UMLS semantic group profiles seems to be effective. Assignment of a collocation to a candidate sense based on knowledge-based methods is effective. Globally, the AEC method is sensitive to noisy collocations, and few selected terms provide a large improvement in disambiguation performance. The MRD method handles noisy terms better but requires a larger set of terms to improve the results. This explains the difference in performance between the AEC and the MRD methods in combination with second-order features, which have provided a larger set of features compared to other collocation extraction methods.
We envisage several directions for future work. We have found that some collocations add noise and decrease disambiguation performance. We would like to study the identification and removal of noisy terms, extending this study to terms from the knowledge source which might already contribute to a lower performance of the knowledge-based methods. Some techniques have already been suggested for query reformulation in information retrieval [26].
This might mean that determining the semantic category based on the contextual features still needs more research. One possibility to use semantic categories would be to study named entity recognition techniques. But these techniques require manually annotated data which is quite expensive to produce.
Granularity in the semantic types and groups might be another issue. The study of a different organization of the semantic categories might provide better results in disambiguation performance.
Second-order features have provided an improved performance to the MRD method in the NLM WSD set. We would like to extend the search for new terms which would improve the concept profiles based on clustering approaches.
Competing interests
The authors declare that they have no competing interests.
Authors contributions
AJ designed and carried out the experiments, participated in the development of the methods and drafted the manuscript. BM designed and carried out the experiments, participated in the development of the methods and drafted the manuscript. AA designed the experiments and reviewed the manuscript. AJ, BM and AA read, commented, and approved the final version of the manuscript.
Contributor Information
Antonio Jimeno-Yepes, Email: antonio.jimeno@gmail.com.
Bridget T Mclnnes, Email: bthomson@umn.edu.
Alan R Aronson, Email: alan@nlm.nih.gov.
Acknowledgments
This work was supported in part by the Intramural Research Program of the NIH, National Library of Medicine and by an appointment of A. Jimeno-Yepes to the NLM Research Participation Program sponsored by the National Library of Medicine and administered by the Oak Ridge Institute for Science and Education.
This article has been published as part of BMC Bioinformatics Volume 12 Supplement 3, 2011: Machine Learning for Biomedical Literature Analysis and Text Retrieval. The full contents of the supplement are available online at http://www.biomedcentral.com/1471-2105/12?issue=S3.
References
- Bodenreider O. The unified medical language system (UMLS): integrating biomedical terminology. Nucleic acids research. 2004;32(Database Issue):D267. doi: 10.1093/nar/gkh061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- UMLS (Unified Medical Language System) http://www.nlm.nih.gov/research/umls/
- Aronson A, Lang F. An overview of MetaMap: historical perspective and recent advances. Journal of the American Medical Informatics Association. 2010;17(3):229. doi: 10.1136/jamia.2009.002733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuemie M, Kors J, Mons B. Word sense disambiguation in the biomedical domain: an overview. Journal of Computational Biology. 2005;12(5):554–565. doi: 10.1089/cmb.2005.12.554. [DOI] [PubMed] [Google Scholar]
- Jimeno-Yepes A, Aronson A. Knowledge-based biomedical word sense disambiguation: comparison of approaches. BMC bioinformatics. 2010;11:565. doi: 10.1186/1471-2105-11-569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jimeno-Yepes A, McInnes B, Aronson A. Processing the MEDLINE MeSH indexing to generate a corpus for word sense disambiguation. 2010. http://skr.nlm.nih.gov/ [DOI] [PMC free article] [PubMed]
- Stevenson M, Guo Y, Gaizauskas R. Proceedings of the 22nd International Conference on Computational Linguistics. Vol. 1. Association for Computational Linguistics; 2008. Acquiring sense tagged examples using relevance feedback; pp. 809–816. [Google Scholar]
- McInnes B. PhD thesis. University of Minnesota, Minneapolis, MN; 2009. Supervised and Knowledge-based Methods for Disambiguating Terms in Biomedical Text using the UMLS and MetaMap. [Google Scholar]
- MEDLINE. http://www.nlm.nih.gov/databases/databases_medline.html
- PUBMED. http://www.ncbi.nlm.nih.gov/sites/entrez
- Leacock C, Miller G, Chodorow M. Using corpus statistics and WordNet relations for sense identification. Computational Linguistics. 1998;24:147–165. [Google Scholar]
- Jimeno-Yepes A, Aronson A. Improving an automatically extracted corpus for UMLS Metathesaurus word sense disambiguation. Proceedings of the SEPLN’10 Workshop on Language Technology Applied to Biomedical and Health Documents. 2010.
- McInnes B. Proceedings of the ACL-08: HLT Student Research Workshop. Columbus, Ohio: Association for Computational Linguistics; 2008. An Unsupervised Vector Approach to Biomedical Term Disambiguation: Integrating UMLS and Medline; pp. 49–54.http://www.aclweb.org/anthology/P/P08/P08-3009 [Google Scholar]
- Yarowsky D. Proceedings of the 33rd annual meeting on Association for Computational Linguistics. Association for Computational Linguistics; 1995. Unsupervised word sense disambiguation rivaling supervised methods; pp. 189–196. full_text. [Google Scholar]
- Rosario B, Hearst M, Fillmore C. Proceedings of the 40th Annual Meeting on Association for Computational Linguistics. Association for Computational Linguistics; 2002. The descent of hierarchy, and selection in relational semantics; pp. 247–254. [Google Scholar]
- Stanford Parser. http://nlp.stanford.edu/software/lex-parser.shtml
- Manning C, Schütze H. Foundations of statistical natural language processing. MIT Press; 2000. [Google Scholar]
- Schütze H. Proceedings of the 1992 ACM/IEEE conference on Supercomputing. IEEE Computer Society Press Los Alamitos, CA, USA; 1992. Dimensions of meaning; pp. 787–796. [Google Scholar]
- Purandare A, Pedersen T. Word sense discrimination by clustering contexts in vector and similarity spaces. Proceedings of the Conference on CoNLL. 2004. pp. 41–48.
- Patwardhan S, Pedersen T. Proceedings of the EACL 2006 Workshop Making Sense of Sense - Bringing Computational Linguistics and Psycholinguistics Together. Vol. 1501. Trento, Italy; 2006. Using WordNet-based Context Vectors to Estimate the Semantic Relatedness of Concepts; pp. 1–8. [Google Scholar]
- Weeber M, Mork J, Aronson A. Proceedings of the AMIA Symposium. American Medical Informatics Association; 2001. Developing a test collection for biomedical word sense disambiguation; p. 746. [PMC free article] [PubMed] [Google Scholar]
- NLM WSD site. http://wsd.nlm.nih.gov/
- NLM WSD data set (restricted) http://wsd.nlm.nih.gov/Restricted/index.shtml
- Humphrey S, Rogers W, Kilicoglu H, Demner-Fushman D, Rindflesch T. Word sense disambiguation by selecting the best semantic type based on Journal Descriptor Indexing: Preliminary experiment. Journal of the American Society for Information Science and Technology (Print) 2006;57:96. doi: 10.1002/asi.20257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen PR. Empirical methods for artificial intelligence. Cambridge, MA, USA: MIT Press; 1995. [Google Scholar]
- Jimeno-Yepes A, Berlanga-Llavori R, Rebholz-Schuhmann D. Proceedings of the WSDM’09 Workshop on Exploiting Semantic Annotations in Information Retrieval. ACM; 2009. Terminological cleansing for improved information retrieval based on ontological terms; pp. 6–14. [Google Scholar]