Abstract
By using sensitive homology-search and gene-finding programs, we have found that a genomic region from the tip of the short arm of human chromosome 16 (16p13.3) encodes a putative secreted protein consisting of a domain related to the whey acidic protein (WAP) domain, a domain homologous with follistatin modules of the Kazal-domain family (FS module), an immunoglobulin-related domain (Ig domain), two tandem domains related to Kunitz-type protease inhibitor modules (KU domains), and a domain belonging to the recently defined NTR-module family (NTR domain). The gene encoding these WAP, FS, Ig, KU, and NTR modules (hereafter referred to as the WFIKKN gene) is intron-depleted—its single 1,157-bp intron splits the WAP module. The validity of our gene prediction was confirmed by sequencing a WFIKKN cDNA cloned from a lung cDNA library. Studies on the tissue-expression pattern of the WFIKKN gene have shown that the gene is expressed primarily in pancreas, kidney, liver, placenta, and lung. As to the function of the WFIKKN protein, it is noteworthy that it contains FS, WAP, and KU modules, i.e., three different module types homologous with domains frequently involved in inhibition of serine proteases. The protein also contains an NTR module, a domain type implicated in inhibition of zinc metalloproteinases of the metzincin family. On the basis of its intriguing homologies, we suggest that the WFIKKN protein is a multivalent protease inhibitor that may control the action of multiple types of serine proteases as well as metalloproteinase(s).
Keywords: multidomain protease inhibitors, zinc metalloproteinases, serine and cysteine proteinases, NTR modules, netrins
The NTR module has been defined recently as a domain type (1) present in netrins; complement proteins C3, C4, C5; secreted frizzled-related proteins; type I procollagen C-proteinase enhancer proteins (PCOLCE); and tissue inhibitors of metalloproteinases (TIMPs). Because the NTR domains of TIMPs are responsible for their metalloprotease inhibitory activity, we have suggested that NTR modules might be involved in controlling the activity of zinc metalloproteases (1). Our prediction that the NTR module of PCOLCE may be a metalloprotease inhibitor has been confirmed by recent experiments (2).
Studies on the netrin–netrin receptor system also have provided evidence that netrins may serve to inhibit metalloprotease activity. The axonal chemoattractant netrin-1 guides spinal commissural axons by activating the netrin receptor deleted in colorectal cancer (DCC). Galko and Tessier-Lavigne (3) have found that chemical inhibitors of metalloproteases potentiate netrin-mediated axon outgrowth in vitro. Their experiments suggest that potentiation of netrin activity by metalloprotease inhibitors results from stabilization of DCC on the axons against proteolysis. The synergistic action of chemical metalloprotease inhibitors and netrins raises the possibility that netrins may protect DCC from metalloproteases via NTR module-mediated inhibition of the metalloprotease (4). Consistent with this prediction, in Caenorhabditis elegans there is genetic evidence for an interaction between a metalloproteinase of the ADAM family (MIG-17) and the nematode netrin, UNC-6 (5).
In view of the emerging role of NTR modules as inhibitors of metalloproteases, we have subjected human genomic sequences to sensitive homology searches to identify additional members of this module family. In the present work we have identified a human gene containing an NTR module. The gene encoding this NTR module also encodes a whey acidic protein (WAP)-type protease inhibitory module, a follistatin/Kazal-type module, an Ig module, and two Kunitz-type protease inhibitory modules.
Our finding that an NTR module is present in a multidomain protein containing multiple types of domains that most frequently serve as protease inhibitors adds new weight to our contention that the NTR module is a protease-inhibitory module. The data presented in this work raise the possibility that this multidomain protein serves to control the action of diverse serine proteases as well as metalloproteinase(s).
Materials and Methods
Sequence Analyses.
Detection of distant homologies was carried out with the consensus sequence method (6, 7). The principle of this procedure is that, in the case of distant homologues, scores come primarily from positions that are conserved characteristically in the family. Accordingly, in this procedure the evolutionary information characterizing the given family is condensed first into consensus sequences in which low-scoring positions are suppressed, thereby sharpening key features of the family. When searching databases with such consensus sequences, “bad hits” with variable regions are eliminated, thereby increasing the discriminating power of the consensus sequence. In the present work, multiple alignments of the NTR-module family (1) were constructed by using clustal w (8). Consensus sequences were defined for each subgroup of this module family and were used iteratively to identify further members of the family (6). Database searches were performed with the fasta and tfasta programs of the GCG Wisconsin Package (Version 10.0) and E() scores were calculated to evaluate the significance of the similarities.
The codonpreference program of the GCG Wisconsin Package (Version 10.0) was used as a frame-specific gene finder program. This program recognizes protein-coding sequences by virtue of the similarity of their codon usage to codon frequency tables, and by the bias of their composition in the third position of codons. codonpreference is useful for locating protein-coding regions and estimating the level of expression of a gene. In the present work, the codon-frequency table that was derived from highly expressed human genes was used as a reference.
The spscan program of the GCG Wisconsin Package (Version 10.0) was used to predict secretory signal peptides.
Proteins containing combinations of different protease-inhibitory modules were identified by using the SMART [(9, 10); http://smart.embl-heidelberg.de/)] and Pfam [(11); http://www.sanger.ac.uk/Software/Pfam/)] databases.
Cloning and Sequencing of the cDNA of the Predicted WFIKKN Gene.
PCR primers (sense, 5′-GCGGAATTCATGCCCGCCCTACGTCC-3′ and antisense, 5′-GCGAAGCTTGGAGTGCGTTTATTCACCAGG-3′) corresponding to positions 26593–26609 and 29424–29444 of cosmid 398G5 from human chromosome 16 (GenBank accession no. Z84479) were used to amplify the translated region of the predicted cDNA of the WFIKKN gene from a human lung cDNA library (CLONTECH). The PCR products were cloned into the EcoRI/HindIII site of the M13mp18 vector for sequencing.
Restriction enzymes were purchased from New England Biolabs. The M13 sequencing kit used for dideoxy sequencing of cloned DNA was obtained from New England Biolabs. Escherichia coli strain JM-109 was used to propagate the M13 phage.
Tissue Expression Pattern of the WFIKKN Gene.
The tissue-expression pattern of the WFIKKN gene was assessed with Multiple Tissue cDNA panels from CLONTECH by using WFIKKN-specific PCR primers corresponding to positions 28984–29006 and 29424–29444 (sense, 5′-GAGTCGACGCGCACACCGCCCTGCCGCGCC-3′ and antisense, 5′-GCGAAGCTTGGAGTGCGTTTATTCACCAGG-3′) of cosmid 398G5 from human chromosome 16. The amplified segment encodes the NTR domain of the human WFIKKN gene.
Results
Detection of Human Genomic Sequences Encoding NTR Modules.
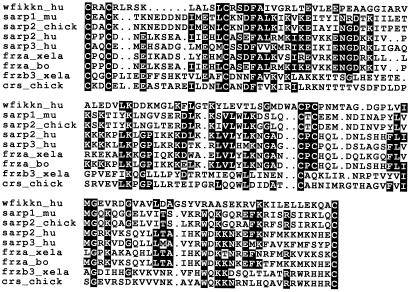
We have performed iterative homology searches with consensus sequences of different subfamilies of the NTR-domain superfamily (1) as described in Materials and Methods. The tfasta search of nucleic acid databases with the consensus sequence for the NTR domains of secreted frizzled-related proteins (SFRP) has revealed first [E() score 3.3 × 10−7] that a genomic region (GenBank accession no. Z84479) from the tip of the short arm of human chromosome 16 (16p13.3) encodes a protein domain homologous with the recently identified NTR modules. The amino acid sequence of this NTR module has all of the features typical of NTR modules (Fig. 1). Evolutionary analyses (not shown) indicate that it is related most closely to the SFRP subfamily of NTR modules.
Figure 1.
Alignment of the sequence of the NTR module of the WFIKKN protein (wfikkn_hu) with those of murine-, chicken-, and human-secreted apoptosis-related proteins (sarp1_mu, AC O35297; sarp2_chick, AC AF218056; sarp-2_hu, AC O14779 and sarp-3_hu, AC O14780), bullfrog and bovine frza proteins (frza_xela, AC Q9YI24 and frza_bo, AC O19116), bullfrog frzb3 protein (frzb3_xela, AF136184), and chicken crescent protein (crs_chick, AF006508). Residues conserved in more than 50% of the sequences are highlighted in bold.
A tfasta search of nucleic acid databases with this human NTR domain has revealed that an identical amino acid sequence [E() score 6.9 × 10−57] is encoded by segments derived from human chromosome 2 by using low-pass sequence sampling (GenBank accession no. AC068332). The corresponding segments from human chromosomes 16 and 2 are also identical at the nucleotide sequence level. The tfasta search with the NTR sequence from chromosome 16 also has detected the presence of related NTR domains encoded by segments derived through low-pass sequence sampling from chromosomes 17 [GenBank accession no. AC015841; E() score 1.7 × 10−28] and 15 [GenBank accession no. AC068831; E() score 3.2 × 10−13]. Comparison of the NTR sequence from chromosome 16 with that from chromosome 17 revealed 68% identity at the nucleotide sequence level and 55.6% identity at the amino acid sequence level. Note that the sequences originating from chromosomes 17 and 15 are 98.6% identical at the nucleotide sequence level and 97.2% identical at the amino acid sequence level. The slight difference is caused by sequence ambiguities in these clones. Thus, it seems likely that the sequences of the NTR modules from chromosomes 15 and 17 are identical.
The NTR module encoded by the 2-Mb cosmid 398G5 from chromosome 16 (GenBank accession no. Z84479) comes from the C terminus of a long ORF (1,227 bp, 409 amino acids) that is likely to encode a protein. This suggestion is also supported by data obtained with the codonpreference/gene-finding program of the GCG Wisconsin Package. Analysis of the nucleotide sequence of cosmid 398G5 with the codonpreference program has revealed that this long ORF (corresponding to nucleotides 28170–29396 of Z84479) shows a codon usage and very strong third position GC bias expected for translated regions of human genes.
Prediction of the Structure of the Gene Encoding the NTR Module.
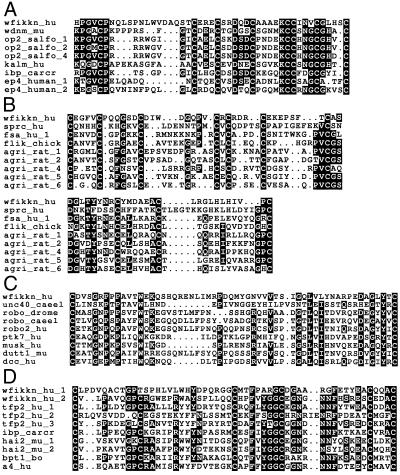
The putative amino acid sequence encoded by the ORF encoding the NTR domain was subjected to a homology search to identify additional domains. These iterative homology searches have revealed that upstream of the NTR module there are two tandem Kunitz- type protease inhibitor modules (Fig. 2). fasta searches using the first Kunitz-type module gave E() scores lower than 1 × 10−3 with several Kunitz domain-containing proteins. The lowest E() score was obtained with type 2 hepatocyte growth factor activator inhibitor 2. In the case of the second Kunitz domain of the putative protein, E() scores lower than 1 × 10−9 were obtained with various members of the Kunitz-domain family. The score with the Kunitz-inhibitor domain of Red Sea turtle protease inhibitor (chelonianin) was 7.6 × 10−10.
Figure 2.
Multiple alignments of the sequences of the WAP-, follistatin-, Ig- and Kunitz-type modules of WFIKKN with those of representative members of the given module families. (A) Multiple alignment of WAP modules of the human WFIKKN protein (wfikkn_hu), murine wdnm1 protein (wdnm_mu, AC Q62477), trout ovulatory protein 2 (op2_salfo_1, op2_salfo_2, and op2_salfo_3, AC Q98988), human Kallmann syndrome protein (kalm_hu, AC P23352), chelonianin of Red Sea turtle (ibp_carcr, AC P00993), and human major epididymis-specific protein e4 precursor (ep4_hu_1 and ep4_hu_2, AC Q14508). (B) Multiple alignment of follistatin domains of the human WFIKKN protein (wfikkn_hu), human SPARC protein (sprc_hu, AC P09486), human follistatin (fsa_hu_1, AC P19883), chicken FLIK protein (flik_chick(AC Q9W600), and rat agrin (agri_rat_1, agri_rat_2, agri_rat_4, agri_rat_5, and agri_rat_6, AC P25304). (C) Multiple alignment of Ig domains of the human WFIKKN protein (wfikkn_hu), unc40 of C. elegans (unc40_caeel, AC U70618), robo proteins of C. elegans, Drosophila melanogaster, and human (robo_caeel, AC T42405; robo_drome, AC Q9W213; robo2_hu, AC O43608), human protein tyrosine kinase 7 (ptk7_hu, AC Q13308), human muscle-specific kinase (musk_hu, AC O15146), murine dutt1 protein (dutt1_mu, AC O89026), and human deleted in colorectal carcinoma protein (dcc_hu, AC P43146). (D) Multiple alignment of Kunitz domains of human WFIKKN protein (wfikkn_hu_1, wfikkn_hu_2), human tissue factor pathway inhibitor 2 (tfp2_hu_1, tfp2_hu_2, tfp2_hu_3, AC P48307), chelonianin of Red Sea turtle (ibp_carcr, AC P00993), murine hepatocyte growth factor activator-inhibitor type 2 (hai2_mu_1, hai2_mu_2, AC Q9WU03), bovine pancreatic trypsin inhibitor (bpt1_bo, AC P00974), and human Alzheimer's disease amyloid a4 protein precursor (a4_hu, AC P05067). Residues conserved in more than 50% of the aligned sequences are highlighted in bold.
Adjacent to the N-terminus of the first Kunitz-type module, the ORF was found to contain an Ig domain (Fig. 2). A fasta search with this domain gave the lowest E() scores (1.5 × 10−3) with Ig domains of human roundabout 2 and human netrin receptor.
Upstream of the Ig domain, the same reading frame (but extending beyond the methionine of the ORF) was found to contain a cysteine-rich domain (Fig. 2) related to the follistatin/Kazal-module family (12). fasta searches with the follistatin module of the putative protein gave E() scores lower than 1 × 10−3 with follistatin modules of agrin and several other follistatin-related proteins. That the follistatin module extends beyond the methionine of the predicted ORF indicated that translation of the putative protein may be initiated from an upstream exon.
A 5′ extension of the reading frame until the nearest stop codon (position 27720) predicted a protein segment that showed weak sequence similarity with the four-disulfide core or WAP-module family. The lowest score was obtained with the WAP domain of the Kallman syndrome protein (Fig. 2). Sequence similarity was limited to the C-terminal half of WAP domains (a segment that contains five of the eight conserved cysteines of this module family), raising the possibility that the missing N-terminal part of the WAP domain may be encoded by an upstream exon.
To find the exon containing the N-terminal part of the predicted WAP domain, we have used iterative gene-finding and homology search procedures. As a result of these searches we have identified an ORF between nucleotides 26593 and 27687 as a candidate region. The 5′ part of this region (between nucleotides 26593 and 26820) showed a codon usage and strong third position GC bias as expected for translated regions of human genes. Analysis of the putative protein product of this region has shown homology with the N-terminal half of WAP modules. On the basis of these findings, we assumed that the region lying between the segments encoding the N- and C-terminal parts of a WAP domain may correspond to an intron. By using intron splice-site consensus sequences of 5′ and 3′ splice sites (13), we have found that a donor splice site, AG/GTGAGT, conforming to the AG/GTRAGT consensus sequence is present between nucleotides 26763 and 26764, and a CAG/G acceptor site conforming to the acceptor consensus sequence is present between positions 27920 and 27921. Because both predicted splice sites are phase 0, splicing of this predicted intron would join the two parts of the predicted WAP domain in the same reading frame. It is noteworthy that the predicted intron contains a mammalian-wide interspersed repeat at nucleotides 27609–27724 (compare annotation of GenBank accession no. Z84479).
Homology searches with the predicted WAP domain have yielded the lowest E() scores (1.3 × 10−3) with WAP domains of trout ovulatory protein 2 (Fig. 2).
Upstream of the WAP domain, the spscan program predicted a secretory signal peptide with a high score (10.6), the cleavage site being predicted between residues 19 and 20, following the predicted initiating Met1 residue.
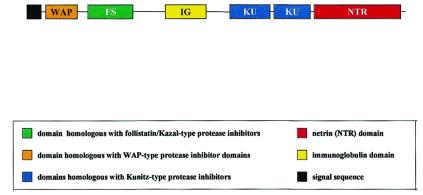
On the basis of homology searches and gene-finding algorithms, we predict that two exons on chromosome 16 encode a secreted multidomain protein of 548 residues, containing a WAP module, a follistatin module, an Ig module, two Kunitz modules, and an NTR module (Fig. 3). The putative protein consisting of a WAP, an FS, an Ig, two KU, and an NTR module is referred to hereafter as the WFIKKN protein.
Figure 3.
Schematic representation of the modular architecture of the predicted WFIKKN protein. The boxes representing domains are drawn to scale.
As discussed above, chromosomes 2, 15, and 17 encode proteins related closely to the WFIKKN protein encoded by chromosome 16. tfasta searches of the (unordered) genomic regions derived from chromosome 17 have identified significant matches not only with the NTR module of WFIKKN but also with its two Kunitz- inhibitor, Ig, follistatin, and WAP modules. On the basis of the near identity of the NTR modules of the WFIKKN-related genes on chromosomes 2/16 and 15/17, it seems likely that there are (at least) four WFIKKN-related genes in the human genome.
Evidence that the WFIKKN Gene Is Expressed.
Searching expressed sequence tag (EST) databases with the sequence of the predicted cDNA sequence of the WFIKKN gene has shown that a 453-bp human EST clone from a cervical tumor (gbAW182905) and a 314-bp human EST clone from squamous cell carcinoma of lung (gbAI819096) correspond to nucleotides 27895–28347 and 28071–28385 of the WFIKKN gene, respectively. Both ESTs are transcribed from the predicted exon encoding the NTR module, suggesting that the WFIKKN gene is expressed.
That codon usage of the predicted WFIKKN mRNA (Table 1) shows a very strong GC bias in synonymous positions, similar to that observed for highly expressed human genes, suggests that the WFIKKN gene may be expressed at a high level.
Table 1.
Comparison of the codon usage of the WFIKKN gene (WFIKKN) with that of highly expressed human genes (HIGH)
| Amino acid | Codon | WFIKKN fraction | HIGH fraction | Amino acid | Codon | WFIKKN fraction | HIGH fraction |
|---|---|---|---|---|---|---|---|
| Gly | GGG | 0.26 | 0.24 | Trp | TGG | 1.00 | 1.00 |
| Gly | GGA | 0.02 | 0.14 | End | TGA | 0.00 | 0.55 |
| Gly | GGT | 0.07 | 0.12 | Cys | TGT | 0.18 | 0.32 |
| Gly | GGC | 0.65 | 0.50 | Cys | TGC | 0.82 | 0.68 |
| Glu | GAG | 0.97 | 0.75 | End | TAG | 1.00 | 0.21 |
| Glu | GAA | 0.03 | 0.25 | End | TAA | 0.00 | 0.23 |
| Asp | GAT | 0.21 | 0.25 | Tyr | TAT | 0.15 | 0.26 |
| Asp | GAC | 0.79 | 0.75 | Tyr | TAC | 0.85 | 0.74 |
| Val | GTG | 0.78 | 0.64 | Leu | TTG | 0.06 | 0.06 |
| Val | GTA | 0.00 | 0.05 | Leu | TTA | 0.00 | 0.02 |
| Val | GTT | 0.03 | 0.07 | Phe | TTT | 0.18 | 0.20 |
| Val | GTC | 0.19 | 0.25 | Phe | TTC | 0.82 | 0.80 |
| Ala | GCG | 0.19 | 0.17 | Ser | TCG | 0.09 | 0.09 |
| Ala | GCA | 0.07 | 0.13 | Ser | TCA | 0.00 | 0.05 |
| Ala | GCT | 0.14 | 0.17 | Ser | TCT | 0.03 | 0.13 |
| Ala | GCC | 0.60 | 0.53 | Ser | TCC | 0.09 | 0.28 |
| Arg | AGG | 0.05 | 0.18 | Arg | CGG | 0.22 | 0.21 |
| Arg | AGA | 0.00 | 0.10 | Arg | CGA | 0.05 | 0.06 |
| Ser | AGT | 0.06 | 0.10 | Arg | CGT | 0.05 | 0.07 |
| Ser | AGC | 0.73 | 0.34 | Arg | CGC | 0.62 | 0.37 |
| Lys | AAG | 1.00 | 0.82 | Gln | CAG | 1.00 | 0.88 |
| Lys | AAA | 0.00 | 0.18 | Gln | CAA | 0.00 | 0.12 |
| Asn | AAT | 0.00 | 0.22 | His | CAT | 0.09 | 0.21 |
| Asn | AAC | 1.00 | 0.78 | His | CAC | 0.91 | 0.79 |
| Met | ATG | 1.00 | 1.00 | Leu | CTG | 0.56 | 0.58 |
| Ile | ATA | 0.00 | 0.05 | Leu | CTA | 0.02 | 0.03 |
| Ile | ATT | 0.00 | 0.18 | Leu | CTT | 0.02 | 0.05 |
| Ile | ATC | 1.00 | 0.77 | Leu | CTC | 0.35 | 0.26 |
| Thr | ACG | 0.26 | 0.15 | Pro | CCG | 0.35 | 0.17 |
| Thr | ACA | 0.13 | 0.14 | Pro | CCA | 0.15 | 0.16 |
| Thr | ACT | 0.09 | 0.14 | Pro | CCT | 0.11 | 0.19 |
| Thr | ACC | 0.52 | 0.57 | Pro | CCC | 0.40 | 0.48 |
To verify the predicted exon–intron structure and mRNA structure of the WFIKKN gene, we have designed PCR primers to amplify the predicted cDNA from human cDNA libraries. The sequence of the cDNAs amplified from the human lung cDNA library have confirmed our predictions (Fig. 4).
Figure 4.
cDNA sequence and deduced amino acid sequence of the WFIKKN gene. The signal peptide of the predicted WFIKKN protein is bold and underlined. The protein sequences corresponding to the WAP domain (WAP-domain), the follistatin domain (FS-domain), the Ig domain (Ig-domain), the two Kunitz-type inhibitor domains (KU-domains 1 and 2), and the NTR module (NTR-domain) are underlined. Nucleotides 171–172, separated by an intron in the WFIKKN gene, are highlighted by a bold double underline. The nucleotide positions matching the PCR primers used for amplification of the cDNA from a fetal lung cDNA library are underlined.
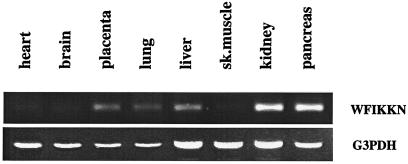
Studies on the tissue-expression pattern of the WFIKKN gene have shown that the gene is expressed widely, being most abundant in pancreas, kidney, liver, placenta, and lung, with only weak expression in brain, heart, and skeletal muscle (Fig. 5).
Figure 5.
Tissue-expression pattern of the WFIKKN gene. cDNA fragments of glyceraldehyde-3-phosphate dehydrogenase (G3PDH) and WFIKKN were amplified from human Multiple Tissue cDNA Panels (CLONTECH) by using G3PDH amplimer set no. 5406–1 (983 bp, 22 cycles) and WFIKKN primers (475 bp, 35 cycles). The PCR products were then electrophoresed on 2% (vol/vol) agarose/ethidium bromide gels.
Discussion
In the present work, we have identified a human gene on chromosome 16 that encodes WFIKKN, a secreted multidomain protein consisting of WAP, follistatin, Ig, Kunitz, and NTR modules. Homology searches also have revealed that closely related genes are present on chromosomes 2, 15, and 17.
WAP-, follistatin-, Ig-, and Kunitz-module types are all class 1–1 modules that have been spread by exon shuffling. Usually, the genes assembled from these modules have an exon–intron structure that reflects this assembly process (14, 15). The WFIKKN gene, however, lacks introns from module boundaries. In fact, it has only a single 1,157-bp intron, suggesting that the gene has been subjected to intron elimination.
A noteworthy feature of the WFIKKN protein is that it consists of domains that have been implicated frequently in inhibition of various types of proteases, suggesting that WFIKKN may be a multidomain protease inhibitor.
It is not unprecedented that different types of protease inhibitory modules are present in a multidomain protein. Homology searches and surveys of the SMART (9, 10) and Pfam (11) databases identified several proteins in which WAP-, Kunitz-, trypsin inhibitor-like cysteine-rich domain (TIL)-, thyroglobulin-, and Kazal-type protease inhibitory modules are combined (Table 2). The WAP-, Kunitz-, TIL-, and Kazal-type modules are involved most frequently in inhibition of serine proteases, whereas thyroglobulin modules are involved frequently in the inhibition of cysteine and aspartic proteinases (16–19).
Table 2.
Multidomain proteins containing combinations of WAP-, Kunitz-, thyroglobulin-, TIL-, and Kazal-type protease inhibitory modules
| Name | Accession no. | Species | Modules* | ||||
|---|---|---|---|---|---|---|---|
| C08G9.2 | 044131 | Caenorhabditis elegans | WA (9) | TY (6) | KU (1) | ||
| W01F3.3 | 045881 | Caenorhabditis elegans | WA (1) | TY (1) | KU (10) | ||
| Cosmid b0222 | Q17457 | Caenorhabditis elegans | TY (2) | KU (1) | |||
| r12a1.3 | 016701 | Caenorhabditis elegans | WA (1) | KU (1) | |||
| t01d3.3 | P90956 | Caenorhabditis elegans | TI (6) | TY (1) | |||
| Lacunin | q9u8g8 | Manduca sexta | WA (1) | KU (4) | |||
| CG5639 | Q9VB21 | Drosophila melanogaster | WA (2) | TY (6) | KU (1) | ||
| CG2264 | Q9V5F6 | Drosophila melanogaster | TY (2) | KAZ (1) | |||
| CG13830 | Q9VCR3 | Drosophila melanogaster | TY (1) | KAZ (1) | |||
| ECM 18 | Q25431 | Lytechinus variegatus | TI (4) | TY (1) | |||
| Chelonianin | P00993 | Caretta caretta | WA (1) | KU (1) | |||
| SPARC-RP | Q9WVN9 | Mus musculus | TY (2) | KAZ (1) | |||
| Testican | Q62288 | Mus musculus | TY (1) | KAZ (1) | |||
| Testican | Q08629 | Homo sapiens | TY (1) | KAZ (1) | |||
| dj461p17.1 | O95959 | Homo sapiens | WA (3) | KU (1) | |||
| dj461p17.2 | O95925 | Homo sapiens | WA (1) | KU (1) | |||
WA, WAP module; TY, thyroglobulin module; KU, Kunitz-type protease inhibitory module; KAZ, Kazal-type protease inhibitory module; TI, Trypsin inhibitor like cysteine rich domain.
The numbers in parenthesis indicate the number of domains present in the given multidomain protein.
A unique aspect of the WFIKKN protein is that it is a multidomain protease inhibitor protein that also contains an NTR module, a domain type implicated in the inhibition of zinc metalloproteinases of the metzincin family (1, 2). On the basis of its intriguing homologies, we suggest that the WFIKKN protein is a multivalent protease inhibitor that functions in controlling the action of multiple types of serine proteases as well as metalloproteinase(s). That it is expressed in a variety of tissues suggests it may play a basic biological role.
Acknowledgments
This work was supported by Hungarian Research Fund (Budapest) Grants OTKA T022949 and OTKA T031726.
Abbreviation
- WAP
whey acidic protein
References
- 1.Bányai L, Patthy L. Protein Sci. 1999;8:1636–1642. doi: 10.1110/ps.8.8.1636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mott J D, Thomas C L, Rosenbach M T, Takahara K, Greenspan D S, Banda M J. J Biol Chem. 2000;275:1384–1390. doi: 10.1074/jbc.275.2.1384. [DOI] [PubMed] [Google Scholar]
- 3.Galko M J, Tessier-Lavigne M. Science. 2000;289:1365–1367. doi: 10.1126/science.289.5483.1365. [DOI] [PubMed] [Google Scholar]
- 4.Pasquale E. Science. 2000;289:1308–1310. doi: 10.1126/science.289.5483.1308. [DOI] [PubMed] [Google Scholar]
- 5.Nishiwaki K, Hisamoto N, Matsumoto K. Science. 2000;288:2205–2208. doi: 10.1126/science.288.5474.2205. [DOI] [PubMed] [Google Scholar]
- 6.Patthy L. J Mol Biol. 1987;198:567–577. doi: 10.1016/0022-2836(87)90200-2. [DOI] [PubMed] [Google Scholar]
- 7.Patthy L. Methods Enzymol. 1996;266:184–198. doi: 10.1016/s0076-6879(96)66014-5. [DOI] [PubMed] [Google Scholar]
- 8.Thompson J D, Higgins D G, Gibson T. Nucleic Acids Res. 1994;12:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schultz J, Milpetz F, Bork P, Ponting C P. Proc Natl Acad Sci USA. 1998;95:5857–5864. doi: 10.1073/pnas.95.11.5857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schultz J, Copley R R, Doerks T, Ponting C P, Bork P. Nucleic Acids Res. 2000;28:231–234. doi: 10.1093/nar/28.1.231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sonnhammer E L L, Eddy S R, Durbin R. Proteins. 1997;28:405–420. doi: 10.1002/(sici)1097-0134(199707)28:3<405::aid-prot10>3.0.co;2-l. [DOI] [PubMed] [Google Scholar]
- 12.Patthy L, Nikolics K. Trends Neurosci. 1993;16:76–81. doi: 10.1016/0166-2236(93)90021-d. [DOI] [PubMed] [Google Scholar]
- 13.Zhang M Q. Hum Mol Genet. 1998;7:919–932. doi: 10.1093/hmg/7.5.919. [DOI] [PubMed] [Google Scholar]
- 14.Patthy L. Protein Evolution by Exon-Shuffling. Berlin: Springer; 1995. [Google Scholar]
- 15.Patthy L. Gene. 1999;238:103–114. doi: 10.1016/s0378-1119(99)00228-0. [DOI] [PubMed] [Google Scholar]
- 16.Lenarcic B, Ritonja A, Strukelj B, Turk B, Turk V. J Biol Chem. 1997;272:13899–13903. doi: 10.1074/jbc.272.21.13899. [DOI] [PubMed] [Google Scholar]
- 17.Lenarcic B, Krishnan G, Borukhovich R, Ruck B, Turk V, Moczydlowski E. J Biol Chem. 2000;275:15572–15577. doi: 10.1074/jbc.M001406200. [DOI] [PubMed] [Google Scholar]
- 18.Lenarcic B, Bevec T. Biol Chem Hoppe–Seyler. 1998;379:105–111. [PubMed] [Google Scholar]
- 19.Strukelj B, Lenarcic B, Gruden K, Pungercar J, Rogelj B, Turk V, Bosch D, Jongsma M A. Biochem Biophys Res Commun. 2000;269:732–736. doi: 10.1006/bbrc.2000.2356. [DOI] [PubMed] [Google Scholar]