ABSTRACT
Simian virus 40 (SV40) is a nonenveloped DNA virus that traffics through the endoplasmic reticulum (ER) en route to the nucleus, but the mechanisms of capsid disassembly and ER exit are poorly understood. We conducted an unbiased RNA interference screen to identify cellular genes required for SV40 infection. SV40 infection was specifically inhibited by up to 50-fold by knockdown of four different DNAJ molecular cochaperones or by inhibition of BiP, the Hsp70 partner of DNAJB11. These proteins were not required for the initiation of capsid disassembly, but knockdown markedly inhibited SV40 exit from the ER. In addition, BiP formed a complex with SV40 capsids in the ER in a DNAJB11-dependent fashion. These experiments identify five new cellular proteins required for SV40 infection and suggest that the binding of BiP to the capsid is required for ER exit. Further studies of these proteins will provide insight into the molecular mechanisms of polyomavirus infection and ER function.
IMPORTANCE
The polyomaviruses, including simian virus 40 (SV40), are important human pathogens and model systems for exploring the general features of virus replication and cell biology. We used a genetic system to interrogate the role of cellular genes in SV40 infection. Based on the results of this unbiased genetic screen and analysis of proteins related to the strongest hit from the screen, we identified five new cellular proteins required for the entry of SV40 into cells. These proteins physically associate with SV40 in the endoplasmic reticulum (ER) during virus entry and are required for exit of the partially disassembled virus from this organelle. These results demonstrate that the polyomaviruses have coopted an ER-localized protein quality control process to initiate disassembly and transit through the cell on their way to the nuclear site of virus replication.
Introduction
A virus capsid leads a double life. In the extracellular environment, it must serve as a stable shell that protects the viral genome, but once the capsid has entered cells, it must release the genome at the correct site. An effective strategy to convert a stable capsid into a device that efficiently releases the genome inside cells is to rely on intracellular machinery for disassembly. In addition, before depositing its cargo in the nucleus, nonenveloped DNA viruses must cross multiple membrane barriers (1). The highly ordered processes by which viruses disassemble and cross membranes presumably involve the sequential action of cellular proteins that carry out vital cell functions. Thus, the identification of cellular proteins that mediate virus entry will provide new insights into basic cellular processes.
The polyomaviruses (PyVs) are small DNA tumor viruses of great scientific and medical interest. Study of the monkey polyomavirus simian virus 40 (SV40) has provided seminal insights into many aspects of biology, and two viruses closely related to SV40, BK virus (BKV) and JC virus (JCV), are widespread in the human population and can cause serious disease. The Merkel cell polyomavirus is likely to be a new human tumor virus (2).
The nonenveloped, icosahedral polyomavirus capsid is composed primarily of 360 molecules of the major capsid protein VP1 arranged in 72 pentamers linked by disulfide bonds (3, 4). The internal face of each pentamer binds to one of the minor capsid proteins, VP2 or VP3. After binding to the cell surface, polyomavirus particles traffic in cell-derived vesicles to the endoplasmic reticulum (ER), where they are deposited into the lumen as naked capsids (5–7). Capsids undergo disassembly and cross the ER membrane before they can access the nucleus, where viral gene expression and DNA replication occur. The mechanisms utilized by the polyomaviruses for disassembly, ER exit, and nuclear entry are poorly understood.
In the ER lumen, protein disulfide isomerase (PDI), ERp57, and ERp29 catalyze the rearrangement of VP1 disulfide bonds as an initial step in the uncoating of SV40 and murine polyomavirus (PyV) (5, 8, 9). Disulfide bond rearrangement detaches some VP1 pentamers, which initially remain associated with the rest of the capsid by strong hydrophobic interactions and calcium bridges (3, 4, 9). Disulfide bond isomerization causes local unfolding of the VP1 C terminus and exposes VP2, allowing membrane binding, perhaps by insertion of a hydrophobic segment of VP2/3 into the ER membrane (8, 10, 11).
Partially disassembled capsids are thought to enter the cytoplasm before reaching the nucleus (10, 12, 13). The exit of PyV particles from the ER is decreased by inhibition of the ER membrane protein Derlin-2, which is implicated in ER-associated degradation (ERAD), a quality control process that removes misfolded proteins from the ER into the cytoplasm, where proteosomal degradation occurs (5, 14, 15). Efficient SV40 and BKV infection requires Derlin-1 and Sel1L, which are also involved in ERAD (9, 15–19). Repression of these ER components typically resulted in only a ~2- to 5-fold reduction in infection, implying that important factors in ER exit remain to be identified. Infection is also inhibited by chemicals that block trafficking to the ER or induce the unfolded protein response (UPR) (9, 20).
A major molecular chaperone in the ER lumen is BiP, an Hsp70 family member that plays a critical role in ERAD (21). Although BiP has not been linked to infection by the polyomaviruses, it is implicated in the life cycles of other viruses (22). The DNAJ proteins belong to the Hsp40 class of cochaperones and associate specifically with Hsp70 ATPases (also known as DNAK proteins) to stimulate protein folding and remodeling reactions. DNAJB11, also known as ERj3, binds to BiP and participates in ERAD (23, 24).
DNAJ/K proteins have been previously implicated in infection by the polyomaviruses. As assessed with a broadly specific Hsc70/Hsp70 antibody, input PyV VP1 associates with Hsc70 proteins in mouse cells, but the specific chaperones engaged by VP1 were not identified (25). In addition, DNAJ/K preparations induced partial in vitro disassembly of PyV, implying that this family of chaperones catalyzes capsid rearrangement during infection (25). However, bacterial DNAJ/K preparations displayed this activity, and specific DNAJ/K proteins involved in the early phase of polyomavirus infection have not been identified. Cellular chaperones may also play a role in polyomavirus assembly (26, 27). In addition, SV40 large T antigen contains a DNAJ domain required for viral replication and cell transformation (28).
Unbiased RNA interference screens provide a powerful approach to identify cellular genes required for virus replication. We developed a cell-based assay to screen a short hairpin RNA (shRNA) library to identify cellular genes required for SV40 infection and identified DNAJB12 as the strongest hit. In follow-up experiments, we discovered that four DNAJ proteins and BiP are required for the exit of SV40 capsids from the ER. In addition, BiP associates with the viral capsid in the ER, suggesting that DNAJ/K complexes directly mediate capsid disassembly or membrane transport.
RESULTS
Design of an RNA interference screen for cellular genes required for SV40 infection.
We designed a cell-based assay to screen an shRNA library for cellular genes required for infection by SV40. Human HeLa cervical carcinoma cells continuously express the human papillomavirus type 18 (HPV18) E6 and E7 oncogenes, which inactivate the cellular p53 and retinoblastoma (RB) tumor suppressor pathways, respectively. Expression of E6 and E7 can be repressed by the bovine papillomavirus (BPV) E2 protein, which binds to the HPV E6/7 promoter (29). HeLa/E6 cells constitutively express an E2-resistant HPV16 E6 gene, which maintains the p53 pathway in an inactive state even after E2 expression (30). To study SV40 infection, we used a replication-defective recombinant virus (designated Pava), in which the SV40 early region is replaced with the BPV E2 gene (29). Transfecting Pava DNA into permissive cells that express large T antigen in trans allows its replication and packaging into SV40 capsids. Pava infection represses the HPV18 E7 gene in HeLa/E6 cells, causing them to undergo RB-dependent growth arrest (30). Agents that inhibit Pava infection and expression of the E2 protein allow ongoing cell proliferation despite exposure to Pava, thus providing a positive selection for genes that block SV40 infection. Constitutive expression of the E6 gene reduces the chance that an agent affecting the p53 pathway would score in this assay.
DNAJB12 is required for efficient infection by SV40.
We infected HeLa/E6 cells at a low multiplicity of infection (MOI) with a pooled lentiviral library expressing shRNAs that target in aggregate all human genes (Fig. 1A). We then challenged the cells three times with Pava at a high MOI. Although most of the cells ceased proliferation, a small number of colonies grew among a background of growth-arrested cells. shRNA genes cloned from the growing cells were reintroduced individually into HeLa/E6 cells and tested for biological activity. An shRNA predicted to target the DNAJB12 gene reproducibly caused the most severe inhibition of Pava-mediated growth suppression (data not shown). For subsequent experiments, we designed and used an shRNA that repressed DNAJB12 mRNA levels to a greater extent than the original one from the library and conferred greater resistance to growth suppression.
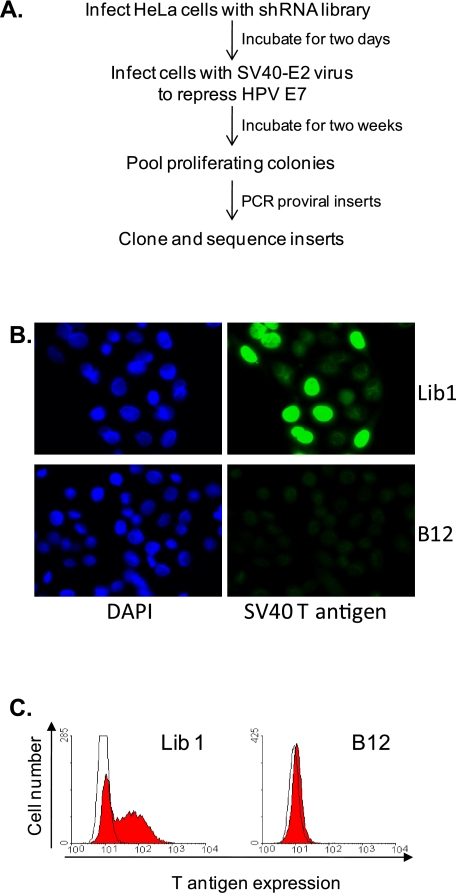
FIG 1 .
Specific DNAJ proteins are required for SV40 infection. (A) Scheme to identify genes required for SV40 infection. See text for details. (B) Large T antigen was detected in Lib1 and DNAJB12 knockdown HeLa/E6 cells by immunofluorescence 20 hours after SV40 infection. The images show the same fields stained for nuclei (left) and T antigen (right). DAPI, 4′,6-diamidino-2-phenylindole. (C) HeLa/E6 cells expressing either control Lib1 or DNAJB12 shRNA were infected with SV40 at an MOI of ~1. Twenty hours later, the cells were stained for SV40 large T antigen and analyzed by flow cytometry. Open and filled histograms show staining of uninfected and infected cells, respectively.
HeLa cells contain the machinery to support the early phase of SV40 infection up to expression of large T antigen, a viral early gene product. To test whether DNAJB12 knockdown inhibited SV40 infection, HeLa/E6 cells expressing a random shRNA from the library (Lib1) or the optimized DNAJB12 shRNA were infected with SV40, and after 20 h, immunofluorescence microscopy was used to assess expression of SV40 large T antigen. As shown in Fig. 1B, DNAJB12 knockdown dramatically inhibited nuclear large T antigen expression. To objectively assess the level of inhibition, infected cells were immunostained for large T antigen and analyzed by flow cytometry. As shown in Fig. 1C, DNAJB12 knockdown caused a dramatic reduction in the fraction of cells expressing large T antigen.
Multiple DNAJ genes are required for efficient infection by SV40 in human and monkey cells.
Because DNAJB12 was the strongest hit in the unbiased screen, we focused on the DNAJ family and associated proteins for further experiments. We tested shRNAs targeting DNAJB14 and DNAJC18 to determine whether these genes, whose sequences are closely related to DNAJB12 (31), were also required for SV40 infection. These shRNAs specifically knocked down the expression of the targeted genes (see Fig. S1A in the supplemental material). Knockdown of DNAJB12 or DNAJB14 was highly effective at blocking infection by SV40 (causing an approximately 50-fold reduction in the fraction of cells expressing T antigen), and DNAJC18 knockdown caused a 4-fold reduction (Fig. 2A).
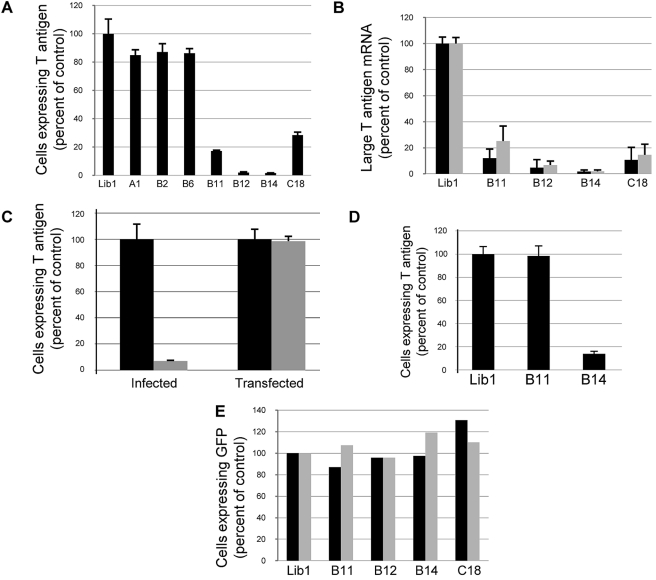
FIG 2 .
Specific effect of DNAJ knockdowns. (A) HeLa/E6 cells expressing the indicated shRNA were analyzed for T antigen expression by flow cytometry 20 h after infection as described in the legend to Fig. 1C. The percentage of positive cells was corrected for multiple infections and normalized to the results from control Lib1 cells. Averaged results from triplicate experiments are shown. Each error bar represents one standard deviation. (B) HeLa/E6 cells expressing the indicated shRNA were infected with SV40 for 20 h (black bars) or BKV for 3 days (gray bars), and levels of large T antigen mRNA were determined by qRT-PCR and presented as the percentage of expression in control Lib1 cells. Averaged results from triplicate experiments are shown. (C) Lib1 (black bars) or DNAJB14 knockdown (gray bars) HeLa/E6 cells were infected with SV40 or transfected with SV40 DNA, and after 30 hours, the proportion of cells expressing large T antigen was determined, as described in panel A. Averaged results from independent experiments are shown. (D) CV1 cells expressing the indicated shRNAs were infected with SV40, and after 24 hours, the fraction of cells expressing large T antigen was determined, as described in panel A. Averaged results from independent experiments are shown. (E) HeLa/E6 cells expressing the indicated shRNAs were infected with adenovirus-GFP for 20 hours (black bars) or HPV16-GFP for 48 hours (gray bars). GFP expression was determined by flow cytometry and normalized to expression in Lib1 cells.
We also tested shRNAs that targeted DNAJ family members more distantly related to DNAJB12 (see Fig. S1A and B in the supplemental material). Knockdown of DNAJB11 caused an approximately 6-fold reduction in the fraction of SV40-infected cells that expressed SV40 large T antigen, whereas knockdown of several other DNAJ proteins had little if any effect (Fig. 2A and data not shown). The inhibition caused by knockdown of individual genes implied that the required DNAJ proteins do not carry out redundant functions during infection of HeLa/E6 cells. To determine the basis for the decreased expression of T antigen, we used quantitative reverse transcription-PCR (qRT-PCR) to measure the level of T antigen mRNA in SV40-infected cells at 20 h after infection. As shown in Fig. 2B, knockdown of DNAJB12 and DNAJB14 caused a >20-fold reduction in T antigen mRNA, and knockdown of DNAJB11 and DNAJC18 resulted in a ~10-fold reduction. Furthermore, knockdown of these DNAJ proteins did not induce ER stress, as assessed by the lack of upregulation of standard markers of the UPR, or affect the ability of thapsigargin to induce the UPR (Fig. S1C).
The ability of DNAJ knockdown to inhibit large T antigen expression indicated that infection is inhibited during the early phase of infection prior to the onset of viral DNA replication. To test if DNAJ knockdown affected an early step of infection, we assayed the ability of transfected SV40 DNA to initiate infection in DNAJB14 knockdown cells, which displayed the most stable knockdown. Transfection delivers DNA to the nucleus and is predicted to bypass barriers to virus entry. Circular SV40 DNA was transfected into control and DNAJB14 knockdown HeLa/E6 cells, which were also infected in parallel with SV40, and T antigen expression was determined 30 h later. The fraction of cells expressing T antigen was dramatically reduced after infection of the DNAJB14 knockdown cells, but the fraction of cells expressing T antigen from transfected SV40 DNA was not affected by knockdown (Fig. 2C). Thus, transfection bypassed the block to SV40 infection, confirming that DNAJB14 is required prior to the delivery of functional viral DNA into the nucleus.
We also tested whether DNAJ proteins were required for efficient infection of monkey cells permissive for the complete SV40 life cycle. We tested the ability of SV40 to infect CV1 cells expressing shRNAs that repressed DNAJB14 or DNAJB11 (see Fig. S1B in the supplemental material). (We were not able to establish monkey cells stably expressing DNAJB12 or DNAJC18 shRNA, presumably because these genes are essential for CV1 cell growth.) As measured by flow cytometry, DNAJB14 knockdown caused a 7-fold reduction in infection (Fig. 2D). Thus, DNAJB14 knockdown inhibited SV40 infection in both semipermissive human cells and permissive monkey cells. In contrast, knockdown of DNAJB11 in CV1 cells did not inhibit SV40 infection (Fig. 2D).
Cellular DNAJ proteins are specifically required for SV40 and BKV infection.
We tested whether the genes identified above were also required for infection by BKV, a pathogenic human polyomavirus. HeLa/E6 knockdown cells were infected with BKV, and three days later, the relative amounts of BKV T antigen mRNA were determined. As was the case for SV40, the level of BKV T antigen mRNA was dramatically reduced by knockdown of DNAJB12 or DNAJB14 or, to a lesser extent, DNAJB11 or DNAJC18 (Fig. 2B).
To determine whether these DNAJ proteins were required for infection with unrelated DNA viruses, we tested the ability of HeLa/E6 knockdown cells to support infection by adenovirus or HPV16, neither of which traffics through the ER. As assessed by green fluorescent protein (GFP) expression from recombinant viruses, adenovirus and HPV16 efficiently infected the cells despite knockdown of any of the DNAJ genes (Fig. 2E). These results indicated that repression of the tested DNAJ genes caused a specific defect in infection by the polyomaviruses.
Rescue of DNAJ knockdown restores SV40 infectivity.
To determine if the block to SV40 infection caused by DNAJ knockdown was due to repression of the target genes, we conducted rescue experiments. We first constructed silent mutations in the shRNA binding sites in cloned human DNAJ genes to generate DNAJB11*, DNAJB12*, and DNAJB14*, which were predicted to be resistant to shRNA-mediated silencing, and stably introduced these genes or empty vector into the cognate HeLa/E6 knockdown cells. The cells were then infected with SV40, and the fraction of cells expressing large T antigen was measured. When the shRNA-resistant gene was expressed, the ability of SV40 to infect the knockdown cells was largely restored compared to that of cells that received the empty retrovirus vector (Fig. 3A). The cells, particularly DNAJB12 knockdown cells, tended to lose knockdown with passage (data not shown), which accounts for their decreased resistance to SV40 infection after the empty vector was introduced. Thus, the ability of DNAJB11, DNAJB12, and DNAJB14 shRNAs to inhibit SV40 infection was due to the repression of the target gene and not to off-target or nonspecific effects.
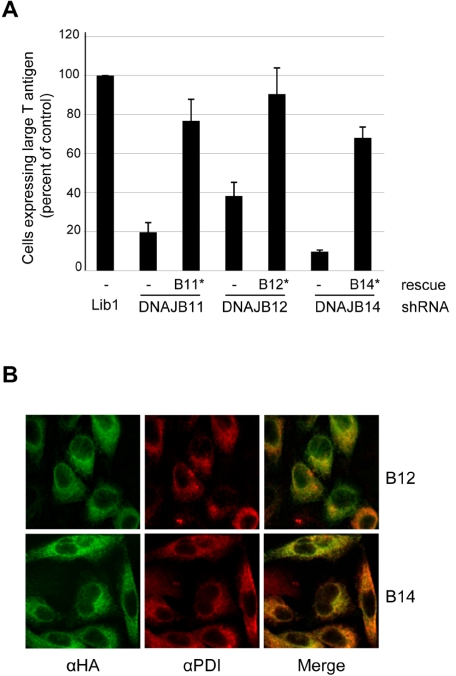
FIG 3 .
Rescue of SV40 infection and localization of DNAJ proteins. (A) Lib1, DNAJB11, DNAJB12, and DNAJB14 knockdown HeLa/E6 cells were stably transduced with a DNAJB11*, DNAJB12*, or DNAJB14* gene containing a mutation in the shRNA binding site, as indicated, or empty vector (–). Forty-eight hours after SV40 infection, flow cytometry was used as described in the legend to Fig. 2A to measure the fraction of cells expressing large T antigen. Averaged results from independent experiments are shown. (B) DNAJB12*-HA or DNAJB14*-HA was transduced into HeLa/E6 cells expressing DNAJB12 or DNAJB14 shRNA, respectively. The cells were stained for the HA tag (green) and the ER marker PDI (red). Colocalization is shown in yellow in the merged images. The same confocal slice is shown in each field.
DNAJB12 and DNAJB14 proteins reside in the ER.
DNAB12 and DNAJB14 are each predicted to contain a C-terminal transmembrane domain. Recently, mammalian DNAJB12 was localized to the ER membrane (32, 33). To localize the DNAJB14 protein and confirm the localization of DNAJB12, we added the influenza virus hemagglutinin (HA) tag to the carboxy terminus of these proteins in the context of the silent mutations that prevent shRNA recognition. The HA-tagged versions of these genes restored SV40 infection in the cognate HeLa/E6 knockdown cells, indicating that the tag did not disrupt biological activity (see example shown in Fig. S2A in the supplemental material).
We used immunofluorescence microscopy to localize the HA-tagged proteins. Cells expressing DNAJB12*-HA and DNAJB14*-HA showed reticular staining that is excluded from the nuclei, whereas nontransduced cells did not stain (Fig. 3B and data not shown). Staining for the ER marker protein disulfide isomerase (PDI) showed a similar pattern, and the merged images exhibited a high degree of overlap, indicating that these DNAJ proteins reside primarily in the ER. Finally, detergent partitioning studies demonstrated that DNAJB12-HA and DNAJB14-HA were integral membrane proteins (see Fig. S2B in the supplemental material).
BiP is required for SV40 infection in human and monkey cells.
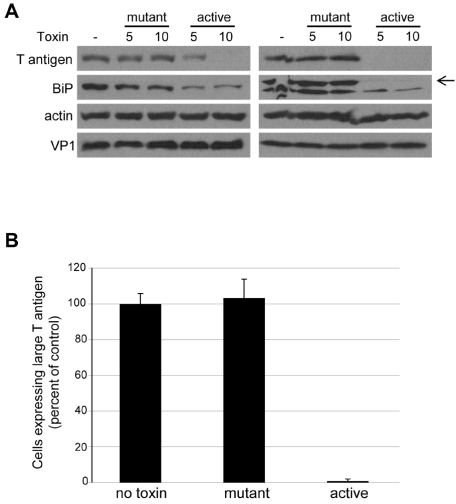
DNAJ proteins affect protein folding by activating their partner Hsp70 ATPases. Because BiP is the Hsp70 partner of DNAJB11, we examined the role of BiP in SV40 infection. For these experiments, we used the subtilase cytotoxin, a protease that specifically cleaves and inactivates BiP (34, 35). We measured infection in HeLa/E6 or CV1 cells treated with either the active toxin (SubAB) or a mutant lacking catalytic activity (SubAA272B). As shown in Fig. 4A, BiP levels were downregulated by treatment with the active toxin but not the mutant toxin. Toxin treatment had no effect on virus uptake, as assessed by levels of cell-associated VP1 at early times after infection. Western blotting demonstrated that SV40 large T antigen levels were markedly decreased in cells treated with the wild-type toxin, but treatment with the inactive mutant toxin had no effect (Fig. 4A). Depletion of BiP also caused a ~50-fold reduction in the number of CV1 cells expressing large T antigen, as measured by flow cytometry (Fig. 4B), but did not inhibit adenovirus or HPV16 (see Fig. S3 in the supplemental material). These results indicated that BiP, like its cochaperone, DNAJB11, was required for efficient infection by SV40. Similarly, BiP cleavage inhibited BKV infection (data not shown). Unlike DNAJ knockdown, BiP cleavage induced the UPR (data not shown). Therefore, it is possible that BiP plays an indirect role in infection.
FIG 4 .
BiP is required for efficient infection. (A) HeLa/E6 (left) or CV1 (right) cells were treated with 5 or 10 ng/ml of mutant or active subtilase cytotoxin and infected with SV40 at an MOI of 2 for 20 hours. Cells were then lysed and analyzed by Western blotting for expression of SV40 large T antigen, BiP, and β-actin. The arrow on the right indicates BiP from CV1 cells. VP1 levels were assessed 8 hours after infection. (B) CV1 cells were infected with SV40 and treated with 10 ng/ml active subtilase toxin. Twenty hours later, the fraction of cells expressing large T antigen was measured by flow cytometry. Averaged results from independent experiments are shown.
BiP and DNAJ proteins are not required for VP1 bond isomerization or exposure of VP2/3.
During the initial stages of capsid disassembly in the ER, disulfide bonds that cross-link a subset of VP1 pentamers with neighboring pentamers undergo isomerization. These pentamers can be released from the residual capsid and dissociated into monomeric VP1 by treatment with SDS in vitro (9). To test whether DNAJ proteins were required for disulfide bond isomerization in the ER, SV40-infected HeLa/E6 cells were lysed in radioimmunoprecipitation assay (RIPA) buffer containing an alkylating agent to prevent further disulfide bond rearrangement. Proteins in the lysates were separated by nonreducing SDS-PAGE and analyzed by Western blotting with a VP1 antibody (see Fig. S4A in the supplemental material). As expected, only low levels of monomeric VP1 were detected at 2 h, prior to the time of ER entry and pentamer detachment. Levels of released VP1 were substantially higher by 8 h of infection of control Lib1 cells, demonstrating that SV40 had reached the ER and disulfide bond isomerization had occurred. Similar levels of VP1 were released in extracts of the knockdown cells, indicating that SV40 entry into the ER and isomerization of VP1 disulfide bonds occurred despite DNAJ knockdown. The levels of total, intracellular VP1 in all samples were comparable, demonstrating that DNAJ knockdown did not affect virus uptake. Similarly, virus uptake and VP1 release were not affected by treating cells with subtilase toxin (Fig. S4B). These results demonstrated that neither the tested DNAJ proteins nor BiP was required for disulfide bond isomerization in the ER or for any of the preceding steps.
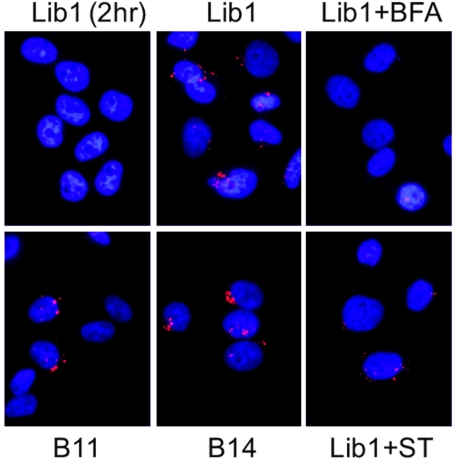
In the intact capsid, the internal minor capsid proteins are not accessible to antibody, but capsid rearrangement following disulfide bond isomerization exposes VP2/3, allowing the visualization of capsids that had proceeded farther along the disassembly process (20). We infected HeLa/E6 cells with SV40 at an MOI of 200 and at various times performed immunofluorescence microscopy with the anti-VP2/3 antibody. Anti-VP2/3 did not stain infected control cells for the first 6 h of infection, but by 8 h, prior to the de novo synthesis of VP2/3, cells showed intense staining in relatively few perinuclear spots (Fig. 5; see also Fig. S5A in the supplemental material). VP2/3 exposure was blocked by brefeldin A (Fig. 5), which prevents SV40 trafficking to the ER (20). Thus, exposure of VP2/3 required ER entry and was consistent with the known timing of SV40 disassembly. All VP2/3 spots colocalized with PDI (Fig. S5B), but PDI staining was much more widespread than VP2/3 staining, indicating that VP2/3 is not exposed throughout the ER. In contrast, antibody to VP1, which stains the bulk of SV40 that enters the cells, not just capsids that have initiated disassembly, showed a wide distribution throughout the cell, with only a small fraction of spots colocalizing with PDI (Fig. S5C). It is possible that VP1 may show a more restricted localization at lower MOIs.
FIG 5 .
DNAJ proteins and BiP are not required for VP2/3 exposure. HeLa/E6 cells expressing Lib1, DNAJB11, or DNAJB14 shRNA were treated with 5 µg/ml brefeldin A (BFA) or 50 ng/ml wild-type subtilase toxin (ST) or left untreated. Cells were then infected with SV40 at an MOI of 200 for 10 hours (or for 2 h [top left]). Cells were then immunostained for VP2/3 (red) and visualized by confocal microscopy. Nuclei are stained blue. Individual slices were compressed along the z axis to generate these images.
To determine whether DNAJ activity was required for exposure of VP2/3, we stained the DNAJ knockdown cells at 10 h postinfection. The pattern and intensity of VP2/3 staining were not affected by knockdown of any of the four tested DNAJ family members (Fig. 5 and data not shown). These results demonstrated that these DNAJ proteins are not required for VP2/3 exposure during SV40 disassembly in the ER. Similarly, subtilase toxin did not prevent VP2/3 staining, indicating that BiP activity was not required for VP2/3 exposure (Fig. 5). However, we note that the VP2/3 signal intensity is reduced in toxin-treated cells, possibly as a consequence of UPR induction.
BiP and DNAJ proteins are required for SV40 exit from the ER.
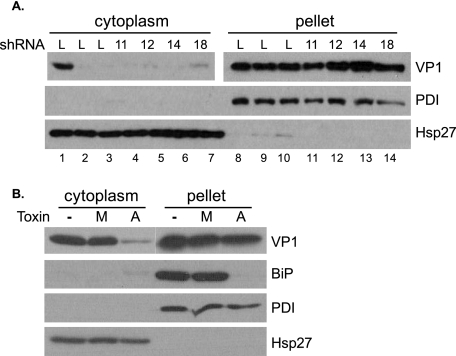
The results presented above indicated that the tested DNAJ proteins and BiP were not required for entry of SV40 into the ER or the initiation of disassembly. We used a fractionation-based assay to test whether exit from the ER was blocked by DNAJ knockdown or BiP inactivation (12). Control and knockdown HeLa/E6 cells were infected with SV40 at an MOI of 30 for 2 or 14 h and harvested in buffer containing 0.03% digitonin, which permeabilizes the plasma membrane but not organellar membranes. The lysates were fractionated by centrifugation into a supernatant fraction containing the cytoplasm and a pellet containing the plasma membrane and cellular organelles and their contents. Thus, incoming virus contained within vesicular organelles, including the ER, was pelleted, whereas virus that had exited the ER and entered the cytoplasm was in the supernatant. Blotting for PDI and cytoplasmic Hsp27 demonstrated the proper fractionation of the lysates (Fig. 6A). VP1 in each fraction was assayed by SDS-PAGE and Western blotting (Fig. 6A). At 2 h after infection, prior to SV40 entry into the ER, there was only a small amount of VP1 in the cytoplasm of either control or knockdown cells (data not shown). In contrast, by 14 h, prior to de novo synthesis of VP1, levels of cytoplasmic VP1 increased significantly in control Lib1 cells, indicating the exit of capsids from the ER into the cytoplasm (Fig. 6A, lane 1) (12). As expected, treatment with brefeldin A or thapsigargin prevented exit from the ER (Fig. 6A, lanes 2 and 3, respectively). Strikingly, the levels of cytoplasmic VP1 remained low in the DNAJ knockdown cells, indicating that these DNAJ proteins were required for efficient export of SV40 from the ER. Blotting for total VP1 in the pellet fraction documented that all cells took up similar levels of SV40. Similarly, the levels of cytoplasmic VP1 in HeLa/E6 (see Fig. S6 in the supplemental material) and CV1 (Fig. 6B) cells treated with active subtilase toxin were low compared to those in cells treated with the mutant toxin. As expected, BiP was present in the pellet fraction and was cleaved by the wild-type toxin only. These results indicated that export of SV40 from the ER requires BiP and its cochaperone DNAJB11, as well as several other DNAJ proteins.
FIG 6 .
DNAJ proteins and BiP are required for ER exit. (A) HeLa/E6 cells expressing the indicated DNAJ shRNA were infected with SV40 at an MOI of 30 for 14 hours. Digitonin lysates were fractionated by centrifugation, and VP1 was detected by immunoblotting in the cytoplasmic and pellet fractions under nonreducing and reducing conditions, respectively. At the time of infection, cells were treated with thapsigargin (lanes 2 and 9) or brefeldin A, (lanes 3 and 10). A shorter exposure of the pellet VP1 blot is shown to allow comparison with the supernatant samples. L, cells expressing Lib1 control shRNA. (B) CV1 cells were infected with SV40 at an MOI of 30 for 14 hours in the presence of 2 ng/ml mutant (M) or active (A) subtilase cytotoxin or in the absence of toxin. The cells were analyzed as described in the legend to panel A.
BiP binds to the SV40 capsid in a DNAJB11-dependent fashion.
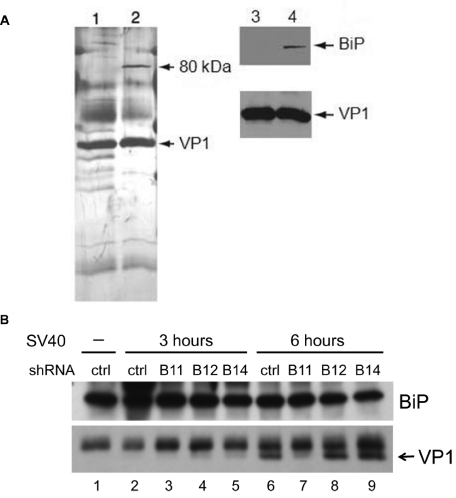
To determine whether ER proteins formed a stable complex with SV40 during disassembly, we performed mass spectrometry. CV1 cells were infected with SV40 at an MOI of 30. After 12 h, cells were lysed in digitonin buffer and fractionated to isolate SV40 particles that were present in the ER lumen (12). These virus particles were immunoprecipitated with antibodies against VP1 and analyzed by SDS-PAGE and silver staining. A prominent band of approximately 80 kDa was detected in the sample from SV40-infected CV1 cells but absent from an immunoprecipitate of an uninfected ER preparation mixed with purified SV40 (Fig. 7A, lane 1 versus lane 2). The 80-kDa band was subjected to mass spectrometry. The sequences of numerous peptides generated from this band matched the sequence of BiP, identifying BiP as a binding partner of ER-localized SV40 capsids (see Fig. S7A in the supplemental material). This assignment was confirmed by immunoblotting (Fig. 7A, lanes 3 and 4). These results demonstrated that BiP and the SV40 capsid formed a stable complex in the ER.
FIG 7 .
BiP forms a complex with SV40. (A) CV1 cells were infected with SV40 at an MOI of 30 for 12 hours. ER-localized SV40 particles were precipitated with VP1 antibodies and analyzed by SDS-PAGE, followed by silver staining (lane 2) or Western blotting for BiP and VP1 (lane 4). VP1 was also immunoprecipitated from a control preparation from the ERs of uninfected cells mixed with SV40 (lanes 1 and 3). (B) HeLa/E6 cells expressing a Lib1 (ctrl) or the indicated DNAJ shRNA were mock infected (–) or infected with SV40 at an MOI of 10 for 3 or 6 hours. After lysis in the presence of 1 mM DSP, samples were immunoprecipitated with a BiP antibody and analyzed by Western blotting for BiP and VP1.
To determine whether the BiP-VP1 complex could be detected in unfractionated cell extracts, we infected HeLa/E6 cells with SV40 at an MOI of 10, lysed the cells at 3 and 6 hours postinfection in a 1% NP-40 buffer containing the chemical cross-linker DSP, and immunoprecipitated them with a BiP antibody. As shown in Fig. 7B, lane 6, VP1 was present in the immunoprecipitate by 6 h after infection. VP1 coimmunoprecipitation was blocked by a BiP peptide that competes for binding to the BiP antibody (see Fig. S7B in the supplemental material).
To test whether DNAJ proteins were required for complex formation between BiP and VP1, we performed coimmunoprecipitation in extracts from the DNAJ knockdown cells (Fig. 7B). VP1-BiP complex formation was not detectable at 3 h after infection in any of the cell lines. Strikingly, at 6 h postinfection, coimmunoprecipitated VP1 was readily detectable in control, DNAJB12, and DNAJB14 knockdown cells but not in DNAJB11 knockdown cells (Fig. 7B, lane 7). This result demonstrated that DNAJB11 was specifically required for BiP to associate with the SV40 capsid, whereas DNAJB12 and DNAJB14 were not.
DISCUSSION
Nonenveloped viruses need to penetrate various cell membranes and undergo intracellular disassembly to release the viral genome. Because small DNA viruses have limited genetic coding capacity, they rely to a great extent on cellular machinery to accomplish these tasks. We conducted a genetic screen and recovered a DNAJB12 shRNA that markedly inhibited infection by SV40. Analysis of additional DNAJ genes showed that DNAJB11, DNAJB14, and DNAJC18 were also required for efficient infection. Knockdown of DNAJB12 or DNAJB14 inhibited infection by up to 50-fold, demonstrating a stringent requirement for these genes. Infection also required the luminal ER Hsp70 protein BiP, the partner of DNAJB11. Although BiP inactivation upregulates the UPR, the physical association of BiP with incoming capsids suggests that BiP plays a direct role in SV40 infection. None of these proteins were previously implicated in infection by the polyomaviruses, and all are required for efficient SV40 exit from the ER. Thus, we have discovered five new cellular proteins that play a role in this poorly understood step in SV40 entry. Three of them, BiP, DNAJB11, and DNAJB12, are involved in ERAD, demonstrating the central importance of this process in SV40 infection, and the other two, DNAJB14 and DNAJC18, were not previously characterized. Like DNAJB12, DNAJB14 localizes to ER membranes. The required DNAJ proteins are not interchangeable because knocking down single DNAJ genes inhibits infection and because knockdowns of different DNAJ proteins have different effects on BiP-VP1 complex formation. Our results imply that ER exit is a complex, rate-limiting step in infection. The reliance of SV40 on ER enzymes to catalyze capsid disassembly and membrane penetration is an effective mechanism to defer these events until the virus has reached the correct intracellular membrane.
The identified DNAJ proteins and BiP are required after ER entry and VP2/3 exposure. It is striking that the intracellular localization of exposed VP2/3 is so much more restricted than that of VP1 or PDI. This implies that VP2/3 exposure occurs in a subcompartment of the ER and affects a small minority of input capsids.
The proteins identified here may be essential components of the ER exit machine utilized by SV40, or they may prime the capsid for exit by catalyzing an obligatory capsid disassembly or membrane insertion step. The association of BiP with the SV40 capsid in a DNAJB11-dependent fashion suggests that BiP and DNAJB11 catalyze capsid remodeling events required for ER exit, a conclusion consistent with the ability of crude DNAJ/K preparations to catalyze partial PyV disassembly in vitro (25). We speculate that the DNAJB11 knockdown HeLa/E6 cells are resistant to SV40 infection because they cannot form the BiP-capsid complex. Although BiP is required for infection of CV1 cells, DNAJB11 is not, suggesting that another DNAJ protein can activate BiP in monkey cells. Consistent with this interpretation, BiP-VP1 complex formation does not depend on DNAJB11 in CV1 cells (our unpublished data).
DNAJ/K protein complexes typically engage unfolded hydrophobic peptide segments (21). Such segments are buried in intact capsids, but VP1 disulfide bond rearrangement in the ER lumen exposes a hydrophobic segment of VP2/3 (10). We propose that the DNAJB11-BiP complex recognizes this segment and escorts it to a restricted domain of the ER membrane for insertion (10, 11). According to this model, the role of BiP in SV40 infection is analogous to its role in the delivery of misfolded proteins and bacterial toxins to the ER membrane during ERAD (21, 24). The viral capsid may then be transferred through the membrane by the transmembrane DNAJ proteins, analogous to the role of DNAJB12 in mediating ERAD of transmembrane proteins (32, 33). We also note that DNAJB12 is in a complex with Derlin-1, which like Sel1L, is also implicated in SV40 entry and involved in ERAD (9, 32, 36). Similarly, Derlin-2 is an ERAD component required for PyV infection (5). Thus, different polyomaviruses have hijacked multiple components of this complex pathway to mediate ER exit.
Topologically, ER exit involves the transfer of the capsid from the ER lumen to the cytoplasm. DNAJB11 and BiP are luminal, whereas the J domain of DNAJB12 resides in the cytoplasm and associates with cytosolic Hsc70 (21, 32, 33). Thus, SV40 disassembly and exit appear to require the action of J domains in the luminal and cytoplasmic compartments together with distinct Hsp70 partners. Because the partially disassembled capsid is much larger than typical ERAD substrates, exit from the ER may entail membrane rupture or clustering of smaller structures to form a channel that can accommodate the capsid.
Cellular DNAJ proteins evolved to carry out cellular functions, not to mediate virus infection. Our analysis of SV40 suggests that these proteins, including the previously uncharacterized DNAJB14 and DNAJC18, normally act together to catalyze the transfer of proteins from the ER into the cytoplasm. Further analysis of SV40 entry will shed new light on the complex activities of cellular ER chaperones. Finally, pathogenic polyomaviruses infect billions of people worldwide, but there are no specific therapies for polyomavirus diseases. The cellular DNAJ proteins identified here and their DNAK partners, including BiP, are potential antiviral targets. Conversely, agents identified because they inhibit or stimulate SV40 infection may be useful modulators of these important cellular processes.
MATERIALS AND METHODS
The experimental procedures used are described in detail in the supplemental material. Oligonucleotide sequences are presented in Table S1 in the supplemental material.
Cells and viruses.
CV1 cells, 293T cells, BKV, and SV40 DNA were purchased from the American Type Culture Collection (ATCC). HeLa/E6 cells are a clonal line of HeLa cells constitutively expressing an exogenous HPV16 E6 gene (30). All cells were cultured in standard medium containing 10% fetal bovine serum (FBS). A packaged, pooled feline leukemia virus (FeLV)-based lentiviral shRNA library expressing 200,000 unique shRNAs targeting 47,400 human transcripts was purchased from System Biosciences (SBI) (SI 202B-1; Mountain View, CA). Standard molecular biology techniques were used to insert silent mutations and the HA tag into DNAJ genes. Pava was produced in CMT4 cells as described previously (29). SV40 was produced and its titers were determined in CV1 cells from cloned DNA. Adenovirus type 5 expressing GFP (Ad5-GFP) and HPV16-GFP were obtained from Vector Biolabs and C. Buck (NCI), respectively.
SV40 infection screen.
A total of 4 × 106 HeLa/E6 cells were infected with the shRNA library at an MOI of 0.5. Two, five, and eight days later, cells were infected with Pava at an MOI of 20. At day 14, RNA was prepared from pooled cells, and shRNA sequences were recovered by PCR amplification. Amplification products were cloned and introduced individually into HeLa/E6 cells for confirmatory testing. ShRNAs targeting genes of interest were designed by using the Invitrogen BLOCK-iT RNA interference (RNAi) designer, inserted into the pSiren vector (Clontech, Mountain View, CA), and packaged as retroviruses in 293T cells. Knockdown of the targeted mRNAs in stably transduced cells was confirmed by qRT-PCR.
Microscopy.
To detect large T antigen, HeLa/E6 cells were infected with SV40 at an MOI of 1, and the cells were stained with large T antigen antibodies 20 h later. Anti-HA antibody was used to stain uninfected cells. To detect capsid proteins, HeLa/E6 cells were infected with SV40 at an MOI of 200. After 2 h, fresh medium was added, and the infection proceeded for the indicated times. The cells were stained with rabbit VP1 or VP2/3 antibody or mouse PDI antibody. After incubation with the appropriate secondary antibodies, images were obtained by confocal microscopy.
Flow cytometry.
To detect large T antigen, cells were stained with mouse anti-T antigen monoclonal antibodies and incubated with Alexa Fluor 488-conjugated donkey anti-mouse IgG. GFP-positive cells were analyzed without antibody staining. Flow cytometry was performed with a FACSCalibur flow cytometer (Becton Dickinson).
qRT-PCR.
Cells were infected with SV40 at an MOI of ~0.5 or with 100 µl of crude BKV. Cells were harvested 20 (SV40) or 72 (BKV) hours after infection, and total RNA was prepared and analyzed in triplicate by qRT-PCR using a single-color real-time PCR detection system (Bio-Rad) and specific DNA primers. The same method was used to measure mRNA knockdown in uninfected cells.
Transfection assay.
HeLa/E6 cells expressing Lib1 or DNAJB14 shRNA were transfected with circular SV40 DNA by using Lipofectamine LTX or infected in parallel with SV40 at an MOI of 1. The cells were harvested 30 h later and subjected to flow cytometry for large T antigen expression.
Subtilase treatment.
HeLa/E6 or CV1 cells infected with SV40 at an MOI of 2 were left untreated or treated with the mutant or active toxin for 20 h, lysed in sample buffer containing SDS and reducing agents, and analyzed by SDS-PAGE and immunoblotting or harvested for flow cytometry.
ER exit assay.
Cells were infected with SV40 at an MOI of 30 for 2 h at 4°C and then incubated at 37°C for 2 or 14 h. In some cases, cells were treated with thapsigargin, brefeldin A, or subtilase toxin. Cells were lysed with cold 0.03% digitonin buffer supplemented with N-ethylmaleimide (NEM) and protease inhibitors. After centrifugation, the resulting supernatant and pellet fractions were assayed by SDS-PAGE under nonreducing and reducing conditions, respectively, and by immunoblotting.
VP1 and BiP immunoprecipitation and mass spectrometry.
A total of 1 × 107 CV1 cells were mock infected or infected with SV40 at an MOI of 30 for 12 h, and then SV40 capsids in the ER were isolated as described in the supplemental material. The viral particle fraction was immunoprecipitated with a mixture of SV40 monoclonal antibodies. Purified SV40 was added to the uninfected cell material prior to immunoprecipitation as a control. Bound proteins were eluted from the beads with glycine, separated by SDS-PAGE, and subjected to silver staining or immunoblotting for VP1 and BiP. The 80-kDa coimmunoprecipitated band was excised from the stained gel and analyzed by mass spectroscopy at the Taplin Biological Mass Spectrometry Facility (Harvard Medical School). HeLa/E6 cells were infected with SV40 at an MOI of 10 for 3 or 6 h and lysed in 1% NP-40 in phosphate-buffered saline (PBS) containing protease inhibitors and the cross-linker DSP dithiobis[succinimidyl propionate]. Extracts were immunoprecipitated with BiP antibody in the presence or absence of competing immunogenic peptide. Precipitates were analyzed with SDS-PAGE and immunoblotting.
SUPPLEMENTAL MATERIAL
Supplemental methods. Download Text S1 , DOC file, 0.071 MB.
Specific knockdown of DNAJ family members and analysis of UPR. (A) HeLa/E6 cells stably expressing Lib1, DNAJB11, DNAJB12, DNAJB14, or DNAJC18 shRNA were assayed by qRT-PCR with primers specific for human DNAJ transcripts to determine mRNA levels. Black bars, DNAJB11 mRNA; dark gray bars, DNAJB12 mRNA; light gray bars, DNAJB14 mRNA; white bars, DNAJC18 mRNA. Results are expressed as the fold repression of the indicated mRNA relative to that of cells expressing control Lib1 shRNA. (B) HeLa/E6 cells stably expressing DNAJA1, DNAJB2, DNAJB6, or DNAJB12 shRNA and CV1 cells stably expressing DNAJB11 or DNAJB14 shRNA were assayed by qRT-PCR for target mRNA knockdown. Results are expressed as described in the legend to panel A. (C) HeLa/E6 cells stably expressing Lib1, DNAJB11, or DNAJB14 shRNA were either left untreated or treated with 100 nM thapsigargin for 24 hours. Levels of spliced XBP-1, CHOP, or calreticulin mRNA were then assayed by qRT-PCR and expressed as the fold induction of mRNA relative to that of untreated shLib1 cells. Download Figure S1, PDF file, 0.083 MB.
Rescue of infection defect by HA-tagged DNAJB12 and membrane localization of DNAJ proteins. (A) HeLa/E6 cells expressing Lib1 or DNAJB12 shRNA were stably transduced with the empty vector or mutant DNAJB12-HA* gene, as indicated. Forty-eight hours after SV40 infection, flow cytometry was used to measure the fraction of cells expressing large T antigen, which is expressed relative to that of control cells expressing the vector. (B) Extracts from cells expressing HA-tagged DNAJB12 or DNAJB14 were subjected to detergent partitioning as described in the supplemental material. The soluble supernatant (S) and detergent-rich membrane (M) fractions were subjected to immunoblotting against the HA tag (left two panels). The integral membrane ER protein HRD1 and the cytoplasmic p97 protein from cells expressing HA-tagged DNAJB14 are also shown to document proper fractionation. Download Figure S2, PDF file, 0.048 MB.
BiP inactivation does not inhibit infection by adenovirus or HPV16. HeLa/E6 cells were treated with increasing doses of the active subtilase cytotoxin and infected with HPV16-GFP (gray) or Ad5-GFP (black) for 48 or 20 hours, respectively. Cells were fixed and analyzed by flow cytometry to measure the fraction of cells expressing GFP, expressed relative to that expressed by untreated cells. Download Figure S3, PDF file, 0.032 MB.
DNAJ proteins and BiP are not required for initiation of disassembly. (A) Total cellular protein was extracted from control and knockdown HeLa/E6 cells at 2 hours or 8 hours postinfection with SV40 at an MOI of 5 (left) or 10 (right) and subjected to SDS-PAGE in the absence (top) or presence (bottom) of reducing agents. VP1 was detected by immunoblotting. The four sets of panels on the right were taken from a single gel at the same exposure, but the order of the lanes was changed. (B) HeLa/E6 cells were infected with SV40 at an MOI of 10 for 2 or 8 hours in the absence of subtilase toxin (−) or in the presence of 50 ng/ml mutant (M) or active (A) toxin. Cell lysates were analyzed as described in the legend to panel A. Download Figure S4, PDF file, 0.115 MB.
Exposure of SV40 VP2/3 and colocalization of VP2/3 and VP1 with an ER marker. (A) HeLa/E6 cells expressing Lib1 shRNA were infected with SV40 at an MOI of 200 for the indicated times. Cells were then immunostained for VP2/3 (red) and visualized by confocal microscopy. Nuclei are stained blue. Individual slices were compressed along the z axis to generate these images. (B) SV40-infected HeLa/E6 Lib1 cells were fixed 10 hours postinfection. The same field shows VP2/3 staining in red, ER marker PDI in green, and DNA in blue. The ImageJ fast Fourier transform FFT band-pass filter was used to visualize a single confocal slice to reduce the complex ER staining pattern, allowing for better determination of overlap (Qian M, Cai D, Verhey KJ, Tsai B, PLoS Pathog. 5:e1000465, 2009). Arrows in the merged image indicated yellow areas of overlap between VP2/3 and PDI. (C) As in panel B, except VP1 staining is shown in red. The lower set of panels shows a magnified view of the upper panels. Download Figure S5, PDF file, 0.397 MB.
BiP is required for ER exit in HeLa cells. HeLa/E6 cells were infected with SV40 at an MOI of 30 for 14 hours in the presence of 50 ng/ml mutant (M) or active (A) subtilase cytotoxin. The cells were fractionated into cytoplasmic and pellet fractions and assayed by Western blotting for VP1, PDI, and BiP. Equal exposures of the PDI and BiP blots are shown; a shorter exposure of the pellet VP1 blot is shown to allow for comparison with the cytoplasmic samples. Download Figure S6, PDF file, 0.047 MB.
Complex formation between BiP and VP1. (A) The full-length amino acid sequence of human BiP is shown. Underlined sequences correspond to peptides derived from the 80-kDa protein bound to intact SV40 capsids (Fig. 7A), as identified by mass spectrometry. (B) HeLa/E6 cells were infected with SV40 at an MOI of 10 for 8 hours. After lysis in 1% NP-40 buffer containing the cross-linker DSP, equal amounts of the protein lysates were incubated overnight with a BiP antibody in the presence or absence of 3.3 µg/ml immunogenic peptide. (Left) Complexes were then precipitated and analyzed by Western blotting for BiP and VP1. (Right) Ten percent of lysates without immunoprecipitation were also analyzed. Download Figure S7, PDF file, 0.115 MB.
Primers.
ACKNOWLEDGMENTS
This paper is dedicated to the memory of William H. Prusoff.
We thank J. Alwine for providing essential reagents and J. Zulkeski for assistance in preparing the manuscript. Flow cytometry was carried out in the Flow Cytometry Shared Resource of the Yale Cancer Center.
A.L. was supported by a training grant from the NIH. This work was supported by a grant from the NCI to D.D. (CA016038).
Footnotes
Citation Goodwin EC, et al. 2011. BiP and multiple DNAJ molecular chaperones in the endoplasmic reticulum are required for efficient simian virus 40 infection. mBio 2(3):e00101-11. doi:10.1128/mBio.00101-11.
REFERENCES
- 1. Tsai B, Qian M. 2010. Cellular entry of polyomaviruses. Curr. Top. Microbiol. Immunol. 343:177–194 [DOI] [PubMed] [Google Scholar]
- 2. Feng H, Shuda M, Chang Y, Moore PS. 2008. Clonal integration of a polyomavirus in human Merkel cell carcinoma. Science 319:1096–1100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Liddington RC, et al. 1991. Structure of simian virus 40 at 3.8-A resolution. Nature 354:278–284 [DOI] [PubMed] [Google Scholar]
- 4. Stehle T, Gamblin SJ, Yan Y, Harrison SC. 1996. The structure of simian virus 40 refined at 3.1 A resolution. Structure 4:165–182 [DOI] [PubMed] [Google Scholar]
- 5. Gilbert J, Ou W, Silver J, Benjamin T. 2006. Downregulation of protein disulfide isomerase inhibits infection by the mouse polyomavirus. J. Virol. 80:10868–10870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Kartenbeck J, Stukenbrok H, Helenius A. 1989. Endocytosis of simian virus 40 into the endoplasmic reticulum. J. Cell Biol. 109:2721–2729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Pelkmans L, Kartenbeck J, Helenius A. 2001. Caveolar endocytosis of simian virus 40 reveals a new two-step vesicular-transport pathway to the ER. Nat. Cell Biol. 3:473–483 [DOI] [PubMed] [Google Scholar]
- 8. Magnuson B, et al. 2005. ERp29 triggers a conformational change in polyomavirus to stimulate membrane binding. Mol. Cell 20:289–300 [DOI] [PubMed] [Google Scholar]
- 9. Schelhaas M, et al. 2007. Simian virus 40 depends on ER protein folding and quality control factors for entry into host cells. Cell 131:516–529 [DOI] [PubMed] [Google Scholar]
- 10. Rainey-Barger EK, Magnuson B, Tsai B. 2007. A chaperone-activated nonenveloped virus perforates the physiologically relevant endoplasmic reticulum membrane. J. Virol. 81:12996–13004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Daniels R, Rusan NM, Wadsworth P, Hebert DN. 2006. SV40 VP2 and VP3 insertion into ER membranes is controlled by the capsid protein VP1: implications for DNA translocation out of the ER. Mol. Cell 24:955–966 [DOI] [PubMed] [Google Scholar]
- 12. Inoue T, Tsai B. 2011. A large and intact viral particle penetrates the endoplasmic reticulum membrane to reach the cytosol. PLoS Pathog. 7:e1002037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Nakanishi A, Clever J, Yamada M, Li PP, Kasamatsu H. 1996. Association with capsid proteins promotes nuclear targeting of simian virus 40 DNA. Proc. Natl. Acad. Sci. U. S. A. 93:96–100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Vembar SS, Brodsky JL. 2008. One step at a time: endoplasmic reticulum-associated degradation. Nat. Rev. Mol. Cell Biol. 9:944–957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Lilley BN, Gilbert JM, Ploegh HL, Benjamin TL. 2006. Murine polyomavirus requires the endoplasmic reticulum protein Derlin-2 to initiate infection. J. Virol. 80:8739–8744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Oda Y, et al. 2006. Derlin-2 and Derlin-3 are regulated by the mammalian unfolded protein response and are required for ER-associated degradation. J. Cell Biol. 172:383–393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Jiang M, Abend JR, Tsai B, Imperiale MJ. 2009. Early events during BK virus entry and disassembly. J. Virol. 83:1350–1358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Mueller B, Lilley BN, Ploegh HL. 2006. SEL1L, the homologue of yeast Hrd3p, is involved in protein dislocation from the mammalian ER. J. Cell Biol. 175:261–270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Ye Y, Shibata Y, Yun C, Ron D, Rapoport TA. 2004. A membrane protein complex mediates retro-translocation from the ER lumen into the cytosol. Nature 429:841–847 [DOI] [PubMed] [Google Scholar]
- 20. Norkin LC, Anderson HA, Wolfrom SA, Oppenheim A. 2002. Caveolar endocytosis of simian virus 40 is followed by brefeldin A-sensitive transport to the endoplasmic reticulum, where the virus disassembles. J. Virol. 76:5156–5166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Dudek J, et al. 2009. Functions and pathologies of BiP and its interaction partners. Cell. Mol. Life Sci. 66:1556–1569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Mayer MP. 2005. Recruitment of Hsp70 chaperones: a crucial part of viral survival strategies. Rev. Physiol. Biochem. Pharmacol. 153:1–46 [DOI] [PubMed] [Google Scholar]
- 23. Shen Y, Hendershot LM. 2005. ERdj3, a stress-inducible endoplasmic reticulum DnaJ homologue, serves as a cofactor for BiP’s interactions with unfolded substrates. Mol. Biol. Cell 16:40–50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Yu M, Haslam DB. 2005. Shiga toxin is transported from the endoplasmic reticulum following interaction with the luminal chaperone HEDJ/ERdj3. Infect. Immun. 73:2524–2532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Chromy LR, Oltman A, Estes PA, Garcea RL. 2006. Chaperone-mediated in vitro disassembly of polyoma- and papillomaviruses. J. Virol. 80:5086–5091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Chromy LR, Pipas JM, Garcea RL. 2003. Chaperone-mediated in vitro assembly of Polyomavirus capsids. Proc. Natl. Acad. Sci. U. S. A. 100:10477–10482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Li PP, et al. 2009. Association of simian virus 40 vp1 with 70-kilodalton heat shock proteins and viral tumor antigens. J. Virol. 83:37–46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Pipas JM. 1998. Molecular chaperone function of the SV40 large T antigen. Dev. Biol. Stand. 94:313–319 [PubMed] [Google Scholar]
- 29. Goodwin EC, DiMaio D. 2000. Repression of human papillomavirus oncogenes in HeLa cervical carcinoma cells causes the orderly reactivation of dormant tumor suppressor pathways. Proc. Natl. Acad. Sci. U. S. A. 97:12513–12518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. DeFilippis RA, Goodwin EC, Wu L, DiMaio D. 2003. Endogenous human papillomavirus E6 and E7 proteins differentially regulate proliferation, senescence, and apoptosis in HeLa cervical carcinoma cells. J. Virol. 77:1551–1563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Qiu XB, Shao YM, Miao S, Wang L. 2006. The diversity of the DnaJ/Hsp40 family, the crucial partners for Hsp70 chaperones. Cell. Mol. Life Sci. 63:2560–2570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Grove DE, Fan CY, Ren HY, Cyr DM. 2011. The endoplasmic reticulum-associated Hsp40 DNAJB12 and Hsc70 cooperate to facilitate RMA1 E3-dependent degradation of nascent CFTR{Delta}F508. Mol. Biol. Cell 22:301–314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Yamamoto YH, et al. 2010. A novel ER J-protein DNAJB12 accelerates ER-associated degradation of membrane proteins including CFTR. Cell Struct. Funct. 35:107–116 [DOI] [PubMed] [Google Scholar]
- 34. Paton AW, Srimanote P, Talbot UM, Wang H, Paton JC. 2004. A new family of potent AB(5) cytotoxins produced by Shiga toxigenic Escherichia coli. J. Exp. Med. 200:35–46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Talbot UM, Paton JC, Paton AW. 2005. Protective immunization of mice with an active-site mutant of subtilase cytotoxin of Shiga toxin-producing Escherichia coli. Infect. Immun. 73:4432–4436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Bernardi KM, Forster ML, Lencer WI, Tsai B. 2008. Derlin-1 facilitates the retro-translocation of cholera toxin. Mol. Biol. Cell 19:877–884 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental methods. Download Text S1 , DOC file, 0.071 MB.
Specific knockdown of DNAJ family members and analysis of UPR. (A) HeLa/E6 cells stably expressing Lib1, DNAJB11, DNAJB12, DNAJB14, or DNAJC18 shRNA were assayed by qRT-PCR with primers specific for human DNAJ transcripts to determine mRNA levels. Black bars, DNAJB11 mRNA; dark gray bars, DNAJB12 mRNA; light gray bars, DNAJB14 mRNA; white bars, DNAJC18 mRNA. Results are expressed as the fold repression of the indicated mRNA relative to that of cells expressing control Lib1 shRNA. (B) HeLa/E6 cells stably expressing DNAJA1, DNAJB2, DNAJB6, or DNAJB12 shRNA and CV1 cells stably expressing DNAJB11 or DNAJB14 shRNA were assayed by qRT-PCR for target mRNA knockdown. Results are expressed as described in the legend to panel A. (C) HeLa/E6 cells stably expressing Lib1, DNAJB11, or DNAJB14 shRNA were either left untreated or treated with 100 nM thapsigargin for 24 hours. Levels of spliced XBP-1, CHOP, or calreticulin mRNA were then assayed by qRT-PCR and expressed as the fold induction of mRNA relative to that of untreated shLib1 cells. Download Figure S1, PDF file, 0.083 MB.
Rescue of infection defect by HA-tagged DNAJB12 and membrane localization of DNAJ proteins. (A) HeLa/E6 cells expressing Lib1 or DNAJB12 shRNA were stably transduced with the empty vector or mutant DNAJB12-HA* gene, as indicated. Forty-eight hours after SV40 infection, flow cytometry was used to measure the fraction of cells expressing large T antigen, which is expressed relative to that of control cells expressing the vector. (B) Extracts from cells expressing HA-tagged DNAJB12 or DNAJB14 were subjected to detergent partitioning as described in the supplemental material. The soluble supernatant (S) and detergent-rich membrane (M) fractions were subjected to immunoblotting against the HA tag (left two panels). The integral membrane ER protein HRD1 and the cytoplasmic p97 protein from cells expressing HA-tagged DNAJB14 are also shown to document proper fractionation. Download Figure S2, PDF file, 0.048 MB.
BiP inactivation does not inhibit infection by adenovirus or HPV16. HeLa/E6 cells were treated with increasing doses of the active subtilase cytotoxin and infected with HPV16-GFP (gray) or Ad5-GFP (black) for 48 or 20 hours, respectively. Cells were fixed and analyzed by flow cytometry to measure the fraction of cells expressing GFP, expressed relative to that expressed by untreated cells. Download Figure S3, PDF file, 0.032 MB.
DNAJ proteins and BiP are not required for initiation of disassembly. (A) Total cellular protein was extracted from control and knockdown HeLa/E6 cells at 2 hours or 8 hours postinfection with SV40 at an MOI of 5 (left) or 10 (right) and subjected to SDS-PAGE in the absence (top) or presence (bottom) of reducing agents. VP1 was detected by immunoblotting. The four sets of panels on the right were taken from a single gel at the same exposure, but the order of the lanes was changed. (B) HeLa/E6 cells were infected with SV40 at an MOI of 10 for 2 or 8 hours in the absence of subtilase toxin (−) or in the presence of 50 ng/ml mutant (M) or active (A) toxin. Cell lysates were analyzed as described in the legend to panel A. Download Figure S4, PDF file, 0.115 MB.
Exposure of SV40 VP2/3 and colocalization of VP2/3 and VP1 with an ER marker. (A) HeLa/E6 cells expressing Lib1 shRNA were infected with SV40 at an MOI of 200 for the indicated times. Cells were then immunostained for VP2/3 (red) and visualized by confocal microscopy. Nuclei are stained blue. Individual slices were compressed along the z axis to generate these images. (B) SV40-infected HeLa/E6 Lib1 cells were fixed 10 hours postinfection. The same field shows VP2/3 staining in red, ER marker PDI in green, and DNA in blue. The ImageJ fast Fourier transform FFT band-pass filter was used to visualize a single confocal slice to reduce the complex ER staining pattern, allowing for better determination of overlap (Qian M, Cai D, Verhey KJ, Tsai B, PLoS Pathog. 5:e1000465, 2009). Arrows in the merged image indicated yellow areas of overlap between VP2/3 and PDI. (C) As in panel B, except VP1 staining is shown in red. The lower set of panels shows a magnified view of the upper panels. Download Figure S5, PDF file, 0.397 MB.
BiP is required for ER exit in HeLa cells. HeLa/E6 cells were infected with SV40 at an MOI of 30 for 14 hours in the presence of 50 ng/ml mutant (M) or active (A) subtilase cytotoxin. The cells were fractionated into cytoplasmic and pellet fractions and assayed by Western blotting for VP1, PDI, and BiP. Equal exposures of the PDI and BiP blots are shown; a shorter exposure of the pellet VP1 blot is shown to allow for comparison with the cytoplasmic samples. Download Figure S6, PDF file, 0.047 MB.
Complex formation between BiP and VP1. (A) The full-length amino acid sequence of human BiP is shown. Underlined sequences correspond to peptides derived from the 80-kDa protein bound to intact SV40 capsids (Fig. 7A), as identified by mass spectrometry. (B) HeLa/E6 cells were infected with SV40 at an MOI of 10 for 8 hours. After lysis in 1% NP-40 buffer containing the cross-linker DSP, equal amounts of the protein lysates were incubated overnight with a BiP antibody in the presence or absence of 3.3 µg/ml immunogenic peptide. (Left) Complexes were then precipitated and analyzed by Western blotting for BiP and VP1. (Right) Ten percent of lysates without immunoprecipitation were also analyzed. Download Figure S7, PDF file, 0.115 MB.
Primers.