Abstract
We have developed a novel assay system for systematic analysis of protein–protein interactions (PPIs) that is characteristic of a PCR-mediated rapid sample preparation and a high-throughput assay system based on the mammalian two-hybrid method. Using gene-specific primers, we successfully constructed the assay samples by two rounds of PCR with up to 3.6 kb from the first-round PCR fragments. In the assay system, we designed all the steps to be performed by adding only samples, reagents, and cells into 384-well assay plates using two types of semiautomatic multiple dispensers. The system enabled us examine more than 20,000 assay wells per day. We detected 145 interactions in our pilot study using 3500 samples derived from mouse full-length enriched cDNAs. Analysis of the interaction data showed both several significant interaction clusters and predicted functions of a few uncharacterized proteins. In combination with our comprehensive mouse full-length cDNA clone bank covering a large part of the whole genes, our high-throughput assay system will discover many interactions to facilitate understanding of the function of uncharacterized proteins and the molecular mechanism of crucial biological processes, and also enable completion of a rough draft of the entire PPI panel in certain cell types or tissues of mouse within a short time.
As in the case of Saccharomyces cerevisiae (budding yeast), Caenorhabditis elegans, Drosophila, and Arabidopsis (Mewes et al. 1997; The C. elegans Sequencing Consortium 1998; Adams et al. 2000; The Arabidopsis Genome Initiative 2000), large-scale genome sequencing and cDNA libraries brought us a rough draft of whole genes in higher organisms such as human and mouse, wherein many of the genes were novel ones of unknown function (International Human Genome Sequencing Consortium 2001; The RIKEN Genome Exploration Research Group Phase II Team and the FANTOM Consortium 2001; Venter et al. 2001). To uncover the function of each gene, systematic examination of protein–protein interactions (PPIs) covering entire genes is very important. PPIs play pivotal roles in the network of cellular biological processes (Oliver 2000; Pawson and Nash 2000) and they also should be potential targets for drug development (Cochran 2000).
Although there are many approaches to examine PPIs, the two-hybrid method has contributed excellently to the genome-wide systematic analysis of PPI. The PPI search using the two-hybrid method can be divided into two types of approaches, the so-called matrix approach and library screening (Legrain and Selig 2000). The matrix method has been used favorably in the genome-wide analysis because all possible combinations can be screened one by one using sets of predefined open reading frames (or protein coding sequences). A large-scale comprehensive analysis of PPIs in budding yeast has been performed (Uetz et al. 2000; Ito et al. 2001) and a systematic analysis of PPIs in C. elegans has been started (Walhout et al. 2000); both of these have been performed by the matrix method using the interaction mating-mediated yeast two-hybrid system (Colas and Brent 1998). In organisms with several ten-thousands of genes, however, it seems less easy to establish entire PPI panels (matrix) because the total number of examinations in human or mouse is estimated to be far larger than those for budding yeast or C. elegans. Here we report two key developments to address this difficulty, PCR-mediated sample preparation and a high-throughput PPI assay system, that allowed us to obtain interaction data very rapidly.
RESULTS
PCR-Mediated Rapid Sample Preparation
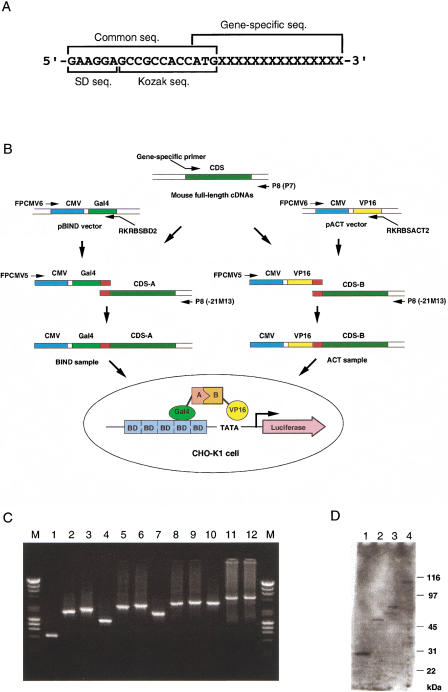
We have prepared the samples for PPI assay by PCR without any cloning steps (Fig. 1). First, we have synthesized each gene-specific forward primer possessing an 18-base common sequence followed by the gene-specific sequence (Fig. 1A). We constructed the samples by two rounds of PCR (Fig. 1B and Methods). In the first PCR, we amplified each cDNA using the gene-specific primer and the M13 universal primer to make the protein coding sequence (CDS) fragment with the common 18-base sequence at the 5′ terminus. We also amplified DNA fragments for human cytomegalovirus (CMV) immediate early promoter followed by the Gal4 DNA-binding domain or herpes virus VP16 transcriptional activation domain, in which both DNA fragments have a common sequence at the 3′ termini. We used the common sequence as a margin to connect the first PCR products with the Gal4 or VP16 fragments. In the second PCR (the overlapping PCR [Higuchi et al. 1988; Ho et al. 1989]) we amplified the first PCR products and the Gal4 or VP16 fragments to make the PCR products, in which protein derived from each cDNA was designed to be expressed as fusion proteins with the Gal4 DNA-binding domain or the VP16 transcriptional activation domain and was under the control of the CMV promoter (BIND and ACT samples, respectively). We successfully constructed assay samples with up to 3.6 kb from the first-round PCR fragments with a success rate of more than 95% (Fig. 1C). Further, we transfected the BIND samples into CHO-K1 cells and detected the expressed fusion proteins with almost reasonable size by Western blotting analysis using a monoclonal antibody against the Gal4 DNA-binding domain (Fig. 1D). To confirm that the samples were applicable to the PPI assay, we applied the PCR-mediated positive control samples, BIND-inhibitor of differentiation (BIND-ID), and ACT-myogenic regulatory protein (ACT-MyoD), to the standard mammalian two-hybrid method according to the manufacturer's method. We observed significant positive signals in the assay; the activity of the luciferase reporter gene (count) was 54,009 whereas those from the assay using either BIND-ID or ACT-MyoD were 3454 and 974, respectively.
Figure 1.
Strategy for the high-throughput in vivo assay. (A) Design of the gene-specific forward primers. Each gene-specific forward primer was designed to anneal just downstream of the predicted initiation ATG of the gene. Each gene-specific forward primer has a common sequence that is used as a margin to connect the cDNA with other DNA sequences. The common sequence consists of the Shine-Dalgarno (SD) sequence for a prokaryotic ribosome-binding site, GAAGGA, and the Kozak consensus sequence for a eukaryotic translation initiation site, GCCGCCACCATG. (B) Schematic representation of the sample preparation and assay methods. (Thin arrows) PCR primers used; (red boxes) the common sequence region. The assay was performed based on the mammalian two-hybrid system. The pG5luc vector contains five Gal4 binding sites (BD) and a minimal TATA box, both of which are upstream of the luciferase gene; interaction between the BIND and ACT fusion proteins increases luciferase expression. (CDS) Protein coding sequence; (CMV) human cytomegalo virus immediate early promoter; (Gal4) yeast Gal4 DNA-binding domain; (VP16) herpes virus VP16 transcriptional-activation domain. (C) Agarose gel electrophoresis of the PCR-mediated constructs from various lengths of cDNAs. The constructs were prepared by two steps of PCR as described in the Methods. Two microliters of the first PCR products, BIND samples, and ACT samples were subjected to the 1% agarose gel in this order. A mixture of 250 ng of λ-HindIII and 250 ng of φX174-HaeIII was used as the size marker (M). Clone ID of each cDNA and the size of the first PCR product calculated from the nucleotide sequences were as follows: (lanes 1–3) 2010004E10, 0.6 kb; (lanes 4–6) 2310016E22, 1.2 kb; (lanes 7–9) 2310009C19, 1.9 kb; (lanes 10–12) 4931412A05, 3.3 kb. (D) Expression of the fusion proteins from the PCR-mediated samples. The fusion proteins expressed from the BIND samples in C were detected by Western blotting analysis using a monoclonal antibody against the Gal4 DNA-binding domain. Clone ID of each BIND sample and the size of the fusion proteins calculated from the deduced amino acid sequences were as follows: (lane 1) 2010004E10, 30 kD; (lane 2) 2310016E22, 57 kD; (lane 3) 2310009C19, 72 kD; (lane 4) 4931412A05, 101 kD. The size of the fusion protein in lane 4 seemed to be slightly larger than the calculated size. It is unclear whether it may be because of the posttranslational modifications.
High-Throughput Assay System
We established an assay system using 384-well assay plates that is based on the mammalian two-hybrid method (Dang et al. 1991; Fearon et al. 1992). There is a difficulty for efficient assay in the standard mammalian two-hybrid method because the cultured cells must be prepared in each well of tissue culture dishes before the assay. To facilitate high throughput, we designed all the assay steps to be performed by adding only samples, reagents, and cells into the assay plates using two types of semiautomatic multiple dispensers and computerized sample tracking (see Methods). The ACT samples were prepared in 96-well plates by mixing them with the culture medium supplemented with a plasmid for the luciferase gene. The BIND samples were prepared in the same way. All combinations consisting of 96 ACT samples and 4 BIND samples were prepared in each 384-well assay plate. The ACT and BIND samples were added into the plates by 96-channel dispensers and 8-channel workstations, respectively. After adding the ACT and BIND samples, the transfection reagent and suspended CHO-K1 cells were added into each well in this order by 384-channel dispensers and were suspended by pipetting several times. If the expressed proteins interact, transcription of the luciferase gene is activated (Fig. 1B). We measured luciferase activity by the detection reagent after incubation of the assay samples in a CO2 incubator overnight.
Figure 2 shows the sequence of procedures of the assay system. Because some BIND samples (Gal4 fusion proteins) increase luciferase gene expression without interaction with any ACT samples, we first performed a preassay to remove the BIND samples with high background. We found that ∼2% of BIND samples had high background values, and these samples were excluded from further analysis. In the first assay we used a mixture of BIND samples and ACT samples to increase efficiency. In a test experiment shown in Table 1, the interaction was detectable using 1/16 BIND-ID and 1/4 or 1/8 ACT-MyoD, although the luciferase activity decreased drastically depending on the dilution of positive control samples. To examine whether these dilution ratios were applicable to the actual sample mixture, the other test experiment was performed using 16-mixture (16-mix) of BIND samples and 6-mix of ACT mixture samples in which BIND-ID and ACT-MyoD were involved in one of the BIND and ACT samples, respectively. The interaction between the positive control samples was significantly detected; the luciferase activity of the positive combination was 2032, where mean and standard deviation (SD) for the BIND sample with BIND-ID were 800 and 419 (n = 24), respectively, and those for the ACT sample with ACT-MyoD were 1296 and 349 (n = 12), respectively. Similar results were also obtained using another positive control pair, BIND-SV40 large T-antigen and ACT-p53 (data not shown). Thus, we performed the first assay using a 16-mix of BIND samples and a 6-mix of ACT samples at one time in a well of the assay plates.
Figure 2.
Flow chart of the high-throughput assay system. (BKG, background).
Table 1.
Result of a Dilution Experiment Using the Positive Control Samples
| BIND-ID | 1 | ACT-MyoD | 0 | |
|---|---|---|---|---|
| 1/4 | 1/8 | |||
| 1 | 53708 | 4200 | 2015 | 1004 |
| 1/16 | 3510 | 1003 | 639 | 388 |
| 1/32 | 1627 | 652 | 493 | 359 |
| 0 | 482 | 385 | 358 | 311 |
The positive control samples, BIND-ID and ACT-MyoD, were diluted as shown in the table with negative control samples, BIND-Fos and ACT-SV40 large T-antigen, respectively. Combinations of the diluted BIND- and ACT-samples were assayed in duplicate and the mean luciferase activity (count) is shown.
After measurement of the luciferase activity in the first assay, we analyzed these results statistically. Positive candidates were selected as those values revealing SD values both more than 3.0 for each 16-mix BIND sample and more than 2.0 for each 6-mix ACT sample (see Methods). The positive control wells containing BIND-ID and ACT-MyoD were detected as positives in 93% of the first assays in this condition. The mean SD values of the positive control wells were 7.14 and 4.11 for the BIND and ACT samples, respectively. Although the first assay is a rate-limiting step in our assay flow, our system enabled us to evaluate more than 20,000 assay wells per day (60 384-well assay plates per day). In the second assay, we examined the positive combinations in the first assay by the combination of a single BIND sample and a 6-mix ACT sample, and a 16-mix BIND sample and a single ACT sample, to identify the interaction pair candidates. Finally, we examined the positive assay with a single BIND sample and a single ACT sample to confirm the interaction pairs.
Results of the Pilot Study
In the pilot study, examinations were performed using 3500 BIND samples and 3400 ACT samples, which were derived from mouse full-length enriched cDNAs with sequence data available for the primer design (The RIKEN Genome Exploration Research Group Phase II Team and the FANTOM Consortium 2001; http://genome.gsc.riken.go.jp/). As shown in Table 2, we detected 145 PPIs of which 27 were self-interactions (all interaction data are available at http://www.genome.org/ and http://genome.gsc.riken.go.jp/). Judging from the annotation for each interacting gene, the number of interactions between genes of known function, between genes of known and unknown function, and between genes of unknown function were 77, 39, and 29, respectively.
Table 2.
Protein-Protein Interaction Pairs Identified in the High-Throughput in vivo Assay
| PPI No. | BIND clone ID | annotation | ACT clone ID | annotation |
|---|---|---|---|---|
| 1 | 0610006G13 | (J04716) ferritin light chain [M] | 0610006G13 | (J04716) ferrin light chain [M] |
| 2 | 0610006G13 | (J04716) ferritin light chain [M] | 1500001P18 | (J03941) ferritin heavy chain [M] |
| 3 | 0610007F07 | (K01515) hypoxanthine phosphoribosyltransferase [M] | 0610007F07 | (K01515) hypoxanthine phosphoribosyltransferase [M] |
| 4 | 0610007L03 | (U46837) SRB7 [H] | 2010309C18 | (U94662) TFG protein [M] |
| 5 | 0610007L13 | (X13752) delta-aminolevulinate dehydratase (AA 1-330) [M] | 0610007L13 | (X13752) delta-aminolevulinate dehydratase [M] |
| 6 | 0610009D16 | (D14811) hypothetical protein KIAA0110 [H] e-119 | 2410002G23 | (U96131) HPV16 E1 protein binding protein [H] |
| 7 | 0610009O09 | (M13955) type II mesothelial keratin K7 [H] | 1200007G13 | (U13921) cytokeratin 13 [M] |
| 8 | 0610009O09 | (M13955) type II mesothelial keratin K7 [H] | 2210407G07 | (AB013608) cytokeratin 17 [M] |
| 9 | 0610009O09 | (M13955) type II mesothelial keratin K7 [H] | 4631426H08 | (AB013607) c29 [M] e-165 |
| 10 | 0610010C03 | (M28723) antioxidant protein 1 [M] | 0610010C03 | (M28723) antioxidant protein 1 [M] |
| 11 | 0610010F19 | (AB013360) DPM2 [M] | 130015L18 | (AF050157) MHC class II beta chain [M] |
| 12 | 0610011M09 | (AL049610) transcription elongation factor A (SII)-like 1 [H] | 1700023B02 | (AF098297) CBF1 interacting corepressor CIR [H] |
| 13 | 0610012G03 | novel protein | 0610009O08 | (U24223) alpha-complex proein 1 [H] |
| 14 | 0610012H09 | (M36429) transducin beta-2 subunit [H] | 0610009O08 | (U24223) alpha-complex protein 1 [H] |
| 15 | 0610012K15 | (AF043225) 6-pyruvoyl-tetrahydropterin synthase [M] | 0610012K15 | (AF043225) 6-pyruvoyl-tetrahydropterin synthase [M] |
| 16 | 0610027F08 | (AL023859) putative tRNA splicing endonuclease γ subunit [Sc] 9e-12 | 110001K11 | (M81086) beta-tropomyosin [M] |
| 17 | 0610030M18 | (AF022813) tetraspan [H] | 1500006F05 | (U19582) claudin-11 [M] |
| 18 | 0610037N03 | (D63902) estrogen-responsive finger protein [M] 7e-10 | 2010309C18 | (U94662) TFG protein [M] |
| 19 | 0610042A16 | (AF068179) calcium modulating cyclophilin ligand CAMLG [H] | 2010107G23 | novel protein |
| 20 | 0610042H17 | (Z67995) pyrroline-5-carboxylate reductase [C] 1e-64 | 0610042H17 | (Z67995) pyrroline-5-carboxylate reductase [C] 1e-64 |
| 21 | 0910001B06 | (AF182293) U6 snRNA-associated Sm-like protein LSm7 [H] | 2310034K10 | (AJ238097) Lsm5 protein [H] |
| 22 | 0910001F03 | (AF151884) CGI-126 protein [H] | 311001N19 | (AC000098) Similar to unknown protein [C] e-105 |
| 23 | 1010001P06 | (AF047659) No definition line found [C] 9e-92 | 2010323J02 | (U49112) ALG-2 [M] |
| 24 | 1020013A21 | (D87438) Similar to a C. elegans protein [H] | 2310069P03 | (AC004839) similar to AL031532 [H] |
| 25 | 1100001A17 | (D28557) RYB-a [R] | 0610042H17 | (Z67995) pyrroline-5-carboxylate reductase [C] 1e-64 |
| 26 | 1110001H08 | (M59293) Id2 protein [M] | 3300001C01 | (U16321) MITF-2A protein [M] |
| 27 | 1110001H08 | (M69293) Id2 protein [M] | 5730435I22 | (X54549) Pan-2 [R] |
| 28 | 1110003H09 | (Z96932) nuclear autoantigen of 14 kDa [H] | 1110003H09 | (Z96932) nuclear autoantigen of 14 kDa [H] |
| 29 | 1110004E04 | novel protein | 1110004E04 | novel protein |
| 30 | 1110004E04 | novel protein | 1110018O07 | novel protein |
| 31 | 1110008E06 | novel protein | 2010309C18 | (U94662) TFG protein [M] |
| 32 | 1110013A16 | (AF063937) squamous cell carcinoma antigen 2 [M] | 2900011O08 | (U95740) Unknown gene product [H] |
| 33 | 1110014J03 | (AL110500) hypothetical protein [C] | 2310047L21 | (AC006465) supported by mouse EST AA277724 [H] 6e-31 |
| 34 | 1110020E15 | novel protein | 1810038N03 | novel protein |
| 35 | 1110033F04 | (X80035) hair keratin associated protein [O] 5e-06 | 1110004E04 | novel protein |
| 36 | 1110033F04 | (X80035) hair keratin associated protein [O] 5e-06 | 1110018O07 | novel protein |
| 37 | 1110054P19 | novel protein | 1110004E04 | novel protein |
| 38 | 1110054P19 | novel protein | 1110007B04 | novel protein |
| 39 | 1190002C06 | novel protein | 1190002C06 | novel protein |
| 40 | 1200005I04 | (AF095193) BAG-family molecualr chaperone regulator-3; BAG-3 [H] | 1300018P04 | (AF133207) protein kinase [H] |
| 41 | 1200007G13 | (U13921) cytokeratin 13 [M] | 0610009O09 | (M13955) type II mesothelial keratin K7 [H] |
| 42 | 1200015A19 | novel protein | 1200015A19 | novel protein |
| 43 | 1300007C18 | (U66900) acid labile subunit [M] | 2010323J02 | (U49112) ALG-2 [M] |
| 44 | 1300008O09 | novel protein | 0610012K15 | (AF043225) 6-pyruvoyl-tetrahydropterin synthase [M] |
| 45 | 1500001L03 | novel protein | 0610012K15 | (AF043225) 6-pyruvoyl-tetrahydropterin synthase [M] |
| 46 | 1500001P18 | (J03941) ferritin heavy chain [M] | 0610006G13 | (J04716) ferritin light chain [M] |
| 47 | 1500001P18 | (J03941) ferritin heavy chain [M] | 1500001P18 | (J03941) ferritin heavy chain [M] |
| 48 | 1500003N18 | (AF061346) Edp 1 protein [M] e-103 | 1500003N18 | (AF061346) Edp 1 protein [M] e-103 |
| 49 | 1500006O17 | (J04716) ferritin light chain [M] | 1500031L05 | (AF026465) putative cell adhesion molecule [M] |
| 50 | 1500012F11 | (AF074723) RNA polymerase transcriptional regulation mediator [H] | 1200015A19 | novel protein |
| 51 | 1500016H19 | (AB001740) p27 [H] | 1500016H19 | (AB001740) p27 [H] |
| 52 | 1500016H19 | (AB001740) p27 [H] | 2700002C15 | (Z31399) CCTeta protein eta chain [M] |
| 53 | 1500037O10 | (U28068) neurogenic differentiation factor (neuroD) [M] | 3300001C01 | (U16321) MITF-2A protein [M] |
| 54 | 1500040C19 | (X93357) SYT [M] | 0610043D20 | (Z85979) histone H3.3A [M] |
| 55 | 1600014M03 | (U06755) acidic calponin [R] | 2310007K12 | (D37837) 65-kDa macrophage protein [M] |
| 56 | 1700003P11 | (AF151883) CGI-125 protein [H] | 1110001O11 | (AF119676) RAB25 [M] |
| 57 | 1700022I15 | (X75959) polyA binding protein [M] | 1700021C22 | (NM_011517) synaptonemal complex protein 3 [M] |
| 58 | 1700025D04 | (U81002) TRAF4 associated factor 1 [H] 2e-74 | 2310047M10 | novel protein |
| 59 | 1700026B03 | (L32752) GTPase (Ran) [M] | 2400006H24 | (Z49574) ORF YJR074w [Sc] 1e-10 |
| 60 | 1700029P02 | (AF019926) protein kinase [M] | 2010309C18 | (U94662) TFG protein [M] |
| 62 | 1810014B23 | (X63469) TFIIE-beta [H] | 2810048P05 | novel protein |
| 65 | 1810043E06 | (M60523) Id3 protein [M] | 3300001C01 | (U16321) MTF-2A protein [M] |
| 66 | 1810043E06 | (M60523) Id3 protein [M] | 3110001I23 | (D32007) CBFA2T1(Mtg8a) [M] |
| 72 | 2010016A14 | (U43884) Id1B protein [M] | 3110001I23 | (D32007) CBFA2T1(Mtg8a) [M] |
| 73 | 2010016A14 | (U43884) Id1B protein [M] | 3300001C01 | (U16321) MITF-2A protein [M] |
| 74 | 2010016A14 | (U43884) Id1B protein [M] | 5730435I22 | (X54549) Pan-2 [R] |
| 85 | 2210407G07 | (AB013608) cytokeratin 17 [M] | 0610009O09 | (M13955) type II mesothelial keratin K7 [H] |
| 91 | 2310015J09 | (M27734) keratin type I [M] | 330001P10 | (M19723) keratin K5 [H] |
| 119 | 2610027O10 | (AF029753) basic helix-loop-helix factor Cor1 [M] | 5730435I22 | (X54549) Pan-2 [R] |
| 133 | 2900002E16 | (D83999) RNA polymerase II 3 (Rpo2-3) [M] | 2810048P05 | novel protein |
| 141 | 3300001C01 | (U16321) MITF-2A proein [M] | 1110001H08 | (M69293) Id2 protein [M] |
Clone IDs and annotations of the participants in part of 145 interaction pairs are shown. The complete table is available as an on-line supplement at http://www.genome.org/ and also available at our Web site, http://genome.gsc.riken.go.jp/. Results of BLASTX (2.0.11) search were used for the annotation. Where similar genes with an E-value of <e-5 are specified, their accession number in parenthesis, gene name, species (in brackets), and E-value are shown. In the case of identical or orthlogous genes, their E-values are omitted. Other clones are described as novel proteins.
Abbreviations of species are as follows: A, Arabidopsis thaliana; C, Caenorhabditis elegans; G, Gallus gallus; H, Homo sapiens; M, Mus musculus; Mt, Mycobacterium tuberculosis; O, Oryctolagus cuniculus; R, Rattus norvegicus; Sc, Saccharomyces cerevisiae; Sp, Schizosaccharomyces pombe; X, Xenopus laevis.
All interaction data in this report have been submitted to the public database BIND (http://www.bind.ca/).
We analyzed the network of interactions by our software called PPI network viewer to confirm whether the detected interactions were biologically significant. We found several predictable protein network clusters. For example, we detected a quite large protein cluster with a basic helix-loop-helix (bHLH) motif (PPI numbers 26, 27, 53, 65, 66, 72, 73, 74, 119, and 141 in Table 2). It is well known that this motif appears often in some transcription factors and their regulatory proteins and is responsible for heterodimerization of the bHLH-containing proteins (Norton et al. 1998). We compared the interaction data of this cluster with those deposited in the ProNet PPI database (http://pronet.doubletwist.com/). We found four out of six interactions of the ProNet data (MITF-2A–Id1B, MITF-2A–Id2, MITF-2A–Id3, and Pan-2–Id2) were detected in our study. The remaining two interactions were LYL1–MITF-2A and LYL1–Pan-2, which have been established by the yeast two-hybrid method and/or immunoprecipitation (Miyamoto et al. 1996). We have also detected interactions among keratin family proteins and keratin-related protein (PPI numbers 7, 8, 9, 41, 85, and 91 in Table 2).
Recently, several groups tried to predict the function of uncharacterized proteins using yeast global PPI data (Schwikowski et al. 2000; Uetz et al. 2000; Walhout et al. 2000; Ito et al. 2001). The main concept behind such analyses is that known proteins interacting with uncharacterized proteins provide a valuable clue to the function of the uncharacterized proteins because many proteins play a role in the network of cellular biological processes by associating with other related proteins (guilt-by-association [Oliver 2000]). Considering the confidence of the functional prediction using the two-hybrid analysis data, we searched our pilot experiment data for uncharacterized proteins that interact with more than two known proteins of similar function. We found that Clone 2810048P05 is a good example of this situation; it interacts with the third largest subunit of RNA polymerase II (PPI number 133) and the β-subunit of transcription factor IIE (PPI number 62), indicating that 2810048P05 is involved in the transcription process. Another example is the CBFA2T1/MTG8 gene (3110001I23). CBFA2T1/MTG8 is a component of the nuclear receptor corepressor (Lutterbach et al. 1998; Wang et al. 1998). Our result showing CBFA2T1/MTG8 interacting with two isoforms of Id proteins (PPI numbers 66 and 72) indicates that Ids may play a role in the regulation of the nuclear receptor corepressor.
DISCUSSION
To construct assay samples efficiently from each cDNA, we have synthesized each gene-specific forward primer. The primer has a common sequence and we used it as the margin to connect two DNA fragments by overlapping PCR for the fusion proteins. The gene-specific primers are also useful for simple expression of the proteins both in vitro and in vivo, because the common sequence consists of the Shine-Dalgarno sequence for a prokaryotic ribosome-binding site, GAAGGA, and the Kozak consensus sequence for a eukaryotic translation initiation site, GCCGCCACCATG (Fig. 1A). We have expressed the proteins successfully in vitro using constructs in which the T7 promoter sequence was added upstream of the first-round PCR products by extension PCR (data not shown).
The main characteristic of our strategy for PPI search is quickness through all the steps from sample preparation through assay. The PCR-mediated sample could be prepared very quickly within 1 d because it is not necessary to recover the clones as plasmids from bacteria. The assay is based on the mammalian two-hybrid method in which the assay could be performed using transiently expressed proteins. Therefore, the PCR-mediated samples were applied directly to the assay. Further, incubation time necessary for the assay is only 20 h, which is faster than that for the yeast two-hybrid method, which takes at least several days. The results of the assay are quantified by measuring luciferase activity. These values are also suitable for quick judgment of positives (or positive candidates). In addition, the values for interaction pairs will be useful for evaluating the strength of each interaction because the luciferase activity roughly parallels the strength of interaction.
There are advantages and disadvantages in the PCR-mediated sample preparation. In addition to the rapid sample preparation described above, the direct use of the PCR products as samples could minimize the problem of mutations incorporated into the samples by thermostable DNA polymerase. Actually, we confirmed the expressed fusion proteins of reasonable size using the PCR-mediated samples (Fig. 1D). We showed successful construction of assay samples with up to 3.6 kb from the first-round PCR fragments (Fig. 1C). However, the concentration of assay samples had a tendency to decrease in first-round PCR fragments of >3 kb. Because the intensity of luciferase activity was affected strongly by the concentration of the assay samples, as shown in Table 1, it is plausible that the less-amplified assay samples may not be screened effectively.
Generally, the two-hybrid method is not completely reliable because the results usually contain many false positives. Actually, very little overlap of the yeast PPI data from two independent research groups using the yeast two-hybrid method was observed, even though part of the reason may be explained by the different conditions of the experiments such as the sample construction procedure, the selection strategy, and the depth of the examinations (Uetz et al. 2000; Ito et al. 2001). The false-positive interactions in the two-hybrid method could be classified into two types: less-reproducible interactions and physiologically less-significant interactions. Our assay strategy may have several advantages to minimize the false positives. First, because we have three examinations (the first, second, and final assays) before determining each interaction pair, such repeated assays should be expected to decrease less-reproducible interactions. This assertion was confirmed by the results of reexamination of some of the assays: almost 80% of the interaction pairs could be found again in the reexamination (data not shown). Second, we applied the results of the first assay to the statistical analysis because the luciferase activity was quantitative. The statistical selection of the positive candidates must be effective in reducing the false positives because it enabled exclusion of most of the pseudopositive values caused by high background in some BIND or ACT samples. Finally, because the assay system is performed in mammalian cells, expressed proteins are more likely to be in their native conformation with appropriate posttranslational modifications (Fagan et al. 1994). Thus, there may be fewer false interactions in the mammalian two-hybrid system than would occur in the yeast two-hybrid system. Nonetheless, this does not mean that all the interactions detected in our assay are physiologically significant. It is clear that we should examine the significance of each interaction by additional experiments such as immunoprecipitation.
We found 145 interactions using 3500 BIND samples and 3400 ACT samples. When we suppose that all possible combinations (1.2 × 107 combinations) were tested significantly, this means that interactions were detected approximately once every 80,000 combinations. Although it has not been well-known how often PPIs are found in mammals, the frequency seems to be relatively lower compared with the value of once per 30,000 combinations obtained by systematic analysis of PPIs in yeast (Uetz et al. 2000). The major reason is that we may overlook PPIs with moderate or weak interaction affinity because we used the positive control samples with relatively high interaction affinity in the establishment of the assay parameters. This was confirmed by a trial of the assay without making the sample mixture using the 192 BIND samples and 192 ACT samples in which any interaction pairs have not been detected using the 16-mix BIND samples and the 6-mix ACT samples; judging from the luciferase activity, this procedure detected five interaction candidates with moderate affinity and five interaction candidates with weak affinity. Actually, one of the interaction candidates with moderate affinity was an established interaction: translation initiation factor 4E and its binding protein 2 (Lin and Lawrence 1996). Therefore, if we suppose that the interaction candidates with moderate affinity should be detected, we overlook many interactions. Generally, the false negatives increase in any high-throughput assay systems in proportion to the increase in the complexity level handling at one time. Thus, it should be necessary to reduce the complexity level further in our assay system for the detailed analysis of PPI with various interaction affinities. The other reason for the observed low frequency of interactions with our PPI systems is that as a first attempt we did not examine the assay without selecting genes coexpressed together. Unlike yeast, higher organisms consist of many types of cells with various gene expression spectrums, in which each cell may express only some of the genes. Because proteins could not compose biologically significant interactions without a chance to meet, the appropriate selection of genes for PPI search may enhance PPI discovery more efficiently. Actually, genes involving similar functions or the same pathways, some of which are likely to associate with each other, could be clustered together according to their expression profiles (Eisen et al. 1998; Miki et al. 2001).
Viewing PPIs globally through a network is valuable because it increases the confidence level for each interaction and because it suggests the function of uncharacterized proteins (Mayer and Hieter 2000). Thus, global analysis has been performed in yeast PPIs, which showed that most of the interactions were involved in a complicated large network; the results were used to reasonably predict the function of 29 previously uncharacterized proteins (Schwikowski et al. 2000). We have also predicted the function of a few previously uncharacterized proteins. Thus, computer analysis of the PPI data seems to be a very powerful tool to explore the functions of proteins and the molecular mechanisms of biological processes. However, it should be stressed again that predictions made using such a bioinformatics approach must be confirmed by additional experiments.
The RIKEN mouse genome project has already collected a large number of mouse full-length enriched cDNAs (The RIKEN Genome Exploration Research Group Phase II Team and the FANTOM Consortium 2001; http://genome.gsc.riken.go.jp/). Applying these cDNAs to the assay systems described here paved the way for us toward completion of the rough draft of a PPI panel covering most genes in certain cell types or tissues of a higher organism within a short time.
METHODS
Primers
Each gene-specific forward primer was designed manually as shown in Figure 1. In some cDNAs with partial coding sequence (CDS), we designed the primer at the 5′ end of the CDS. Other primers used in this work were as follows: FPCMV6, 5′-CCAATATGACCGCCATGTTGGC-3′ and its nested primer FPCMV5, 5′-GCCATGTTGGCATTGATTATT GAC-3′; RKRBSBD2, 5′-CATGGTGGCGGCTCCTTCCGGC GATACAGTCAACTG-3′; RKRBSACT2, 5′-CATGGTGGCG GCTCCTTCAAGTCGACGGATCCCTGGC-3′; M13 universal forward primer P7, 5′-CGCCAGGGTTTTCCCAGTCACGAC-3′ and its nested primer −21M13, 5′-TGTAAAACGACGGC CAGT-3′; and universal reverse primer P8, 5′-AGCGGATAACAATTTCACACAGGAAA-3′.
Construction of the Assay Samples
Plasmid vectors pACT, pBIND, and pG5luc were from the CheckMate mammalian two-hybrid system (Promega). The protein CDS of each mouse cDNA was amplified with the corresponding gene-specific forward primer and P8 (or P7) primers (first PCR). Because some clones were difficult to amplify because of the high GC content around the initiation ATG, PCR was performed using modified dNTPs consisting of 250 μM each of dATP, dCTP, and dTTP, 167 μM of dGTP, and 83 μM of 7-deaza dGTP at the final concentration. The modified dNTPs improved amplification of the PCR products efficiently. Fragments for the CMV promoter and the Gal4 DNA-binding domain (BIND fragment) or the VP16 transcriptional activation domain (ACT fragment) were amplified from pBIND or pACT vectors by PCR using the primer sets FPCMV6 and RKRBSBD2 or FPCMV6 and RKRBSACT2, respectively. Overlapping PCR was performed to connect the CDS fragments with the BIND or ACT fragments. One to two microliters of the first PCR products was mixed with 0.5 μL of ACT or BIND fragment and amplified in 100-μL reactions using the primer set FPCMV5 and P8 (or −21M13). All of the PCR products were checked by agarose gel electrophoresis before each subsequent step in the PCR protocol.
Western Blotting Analysis
Five microliters of the BIND samples were transfected into 2.4 × 105 of CHO-K1 cells in 12-well culture plates. After 24 h of incubation, cells were washed once with ice-cold TBS and harvested using 100 μL of Lamuli sample buffer. The samples were boiled for 2 min and mixed well. Protein in Lamuli sample buffer (10 μL) was subjected to SDS-PAGE and transferred electrically onto PVDF membrane. The membrane was blocked by TBS/0.05% Tween 20 (TBS/Tween) containing 5% skim milk for 1 h and incubated with anti-Gal4 DNA-binding domain monoclonal antibody (Santa Cruz; dilution 1:200) for 1 h. After washing with TBS/Tween, the membrane was incubated with HRP-conjugated anti-mouse IgG2a (Santa Cruz; dilution 1:1000) for 1 h and washed again with TBS/Tween. Detection of the signal was performed using the ECL plus system (Amersham Pharmacia).
Assay
The PCR-mediated BIND and ACT samples were used directly in the assay without further purification. One-quarter microliter each of the BIND and ACT samples, 30 ng of pG5luc, and 9.5 μL of Opti-MEM (Lifetech) were transferred into a well of a 384-well plate. Ten microliters of the transfection reagent LF2000 (Lifetech) that was diluted 32-fold with Opti-MEM was added to the well and suspended by pipetting several times. After 20 min of incubation, 20 μL of CHO-K1 cells that were suspended in F12 medium at 1300 cells/μL were added into the transfection mixture and suspended well. The assay samples were incubated in a CO2 incubator for 20 h. The luciferase activity after the incubation was measured with the Steady-Glo Luciferase Assay System (Promega). Two types of semiautomatic multiple dispensers, 96/384-channel dispensers (Biotec) and 8-channel workstations (Tecan), were used in the high-throughput assay.
Statistical Analysis
Positive candidates were selected statistically after the first assay. The assay data consisting of 24 16-mix BIND samples and 96 6-mix ACT samples were analyzed at one time. First, the SD of each assay point was calculated for each 16-mix BIND sample. These SD values were used for the calculation of the SD for each 6-mix ACT sample because of the large variation of the background values among the BIND samples. Positive candidates were selected as those values revealing SD both more than 3.0 for each 16-mix BIND sample and more than 2.0 for each 6-mix ACT sample. These values were determined empirically.
ACKNOWLEDGMENTS
We especially thank C. Kai for his excellent technical assistance. We also thank the members of the Laboratory for Genome Exploration Research Group. This study was supported by Special Coordination Funds and a Research Grant for the RIKEN Genome Exploration Research Project from the Science Technology Agency of the Japanese Government to Y.H. This work was also supported by a Grant-in-Aid for Scientific Research on Priority Areas and Human Genome Program, from the Ministry of Education, Science and Culture, and by a Grant-in-Aid for a Second Term Comprehensive 10-Year Strategy for Cancer Control from the Ministry of Health and Welfare to Y.H.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL rgscerg@gsc.riken.go.jp; FAX 81-45-503-9216.
Article published on-line before print: Genome Res., 10.1101/gr.180101.
Article and publication are at http://www.genome.org/cgi/doi/10.1101/gr.180101.
REFERENCES
- Adams MD, Celniker SE, Holt RA, Evans CA, Gocayne JD, Amanatides PG, Scherer SE, Li PW, Hoskins RA, Galle RF, et al. The genome sequence of Drosophila melanogaster. Science. 2000;287:2185–2195. doi: 10.1126/science.287.5461.2185. [DOI] [PubMed] [Google Scholar]
- Cochran AG. Antagonists of protein–protein interactions. Chem Biol. 2000;7:R85–R94. doi: 10.1016/s1074-5521(00)00106-x. [DOI] [PubMed] [Google Scholar]
- Colas P, Brent R. The impact of two-hybrid and related methods on biotechnology. Trends Biotechnol. 1998;16:355–363. doi: 10.1016/s0167-7799(98)01225-6. [DOI] [PubMed] [Google Scholar]
- Dang CV, Barrett J, Villa-Garcia M, Resar LM, Kato GJ, Fearon ER. Intracellular leucine zipper interactions suggest c-Myc hetero-oligomerization. Mol Cell Biol. 1991;11:954–962. doi: 10.1128/mcb.11.2.954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisen MB, Spellman PT, Brown PO, Botstein D. Cluster analysis and display of genome-wide expression patterns. Proc Natl Acad Sci. 1998;95:14863–14868. doi: 10.1073/pnas.95.25.14863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fagan R, Flint KJ, Jones N. Phosphorylation of E2F-1 modulates its interaction with the retinoblastoma gene product and the adenoviral E4 19 kDa protein. Cell. 1994;78:799–811. doi: 10.1016/s0092-8674(94)90522-3. [DOI] [PubMed] [Google Scholar]
- Fearon ER, Finkel T, Gillison ML, Kennedy SP, Casella JF, Tomaselli GF, Morrow JS, Van Dang C. Karyoplasmic interaction selection strategy: A general strategy to detect protein–protein interactions in mammalian cells. Proc Natl Acad Sci. 1992;89:7958–7962. doi: 10.1073/pnas.89.17.7958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higuchi R, Krummel B, Saiki RK. A general method of in vitro preparation and specific mutagenesis of DNA fragments: Study of protein and DNA interactions. Nucleic Acids Res. 1988;16:7351–7367. doi: 10.1093/nar/16.15.7351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho SN, Hunt HD, Horton RM, Pullen JK, Pease LR. Site-directed mutagenesis by overlap extension using the polymerase chain reaction. Gene. 1989;77:51–59. doi: 10.1016/0378-1119(89)90358-2. [DOI] [PubMed] [Google Scholar]
- International Human Genome Sequencing Consortium. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- Ito T, Chiba T, Ozawa R, Yoshida M, Hattori M, Sakaki Y. A comprehensive two-hybrid analysis to explore the yeast protein interactome. Proc Natl Acad Sci. 2001;98:4569–4574. doi: 10.1073/pnas.061034498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Legrain P, Selig L. Genome-wide protein interaction maps using two-hybrid systems. FEBS Lett. 2000;480:32–36. doi: 10.1016/s0014-5793(00)01774-9. [DOI] [PubMed] [Google Scholar]
- Lin TA, Lawrence JC., Jr Control of the translational regulators PHAS-I and PHAS-II by insulin and cAMP in 3T3-L1 adipocytes. J Biol Chem. 1996;271:30199–30204. doi: 10.1074/jbc.271.47.30199. [DOI] [PubMed] [Google Scholar]
- Lutterbach B, Westendorf JJ, Linggi B, Patten A, Moniwa M, Davie JR, Huynh KD, Bardwell VJ, Lavinsky RM, Rosenfeld MG, et al. ETO, a target of t(8;21) in acute leukemia, interacts with the N-CoR and mSin3 corepressors. Mol Cell Biol. 1998;18:7176–7184. doi: 10.1128/mcb.18.12.7176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer ML, Hieter P. Protein networks—built by association. Nat Biotechnol. 2000;18:1242–1243. doi: 10.1038/82342. [DOI] [PubMed] [Google Scholar]
- Mewes HW, Albermann K, Bahr M, Frishman D, Gleissner A, Hani J, Heumann K, Kleine K, Maierl A, Oliver SG, et al. Overview of the yeast genome. Nature. 1997;387:7–65. doi: 10.1038/42755. [DOI] [PubMed] [Google Scholar]
- Miki R, Kadota K, Bono H, Mizuno Y, Tomaru Y, Carninci P, Itoh M, Shibata K, Kawai J, Konno H, et al. Delineating developmental and metabolic pathways in vivo by expression profiling using the RIKEN set of 18,816 full-length enriched mouse cDNA arrays. Proc Natl Acad Sci. 2001;98:2199–2204. doi: 10.1073/pnas.041605498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyamoto A, Cui X, Naumovski L, Cleary ML. Helix-loop-helix proteins LYL1 and E2a form heterodimeric complexes with distinctive DNA-binding properties in hematolymphoid cells. Mol Cell Biol. 1996;16:2394–2401. doi: 10.1128/mcb.16.5.2394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norton JD, Deed RW, Craggs G, Sablitzky F. Id helix-loop-helix proteins in cell growth and differentiation. Trends Cell Biol. 1998;8:58–65. [PubMed] [Google Scholar]
- Oliver S. Guilt-by-association goes global. Nature. 2000;403:601–603. doi: 10.1038/35001165. [DOI] [PubMed] [Google Scholar]
- Pawson T, Nash P. Protein–protein interactions define specificity in signal transduction. Genes & Dev. 2000;14:1027–1047. [PubMed] [Google Scholar]
- Schwikowski B, Uetz P, Fields S. A network of protein–protein interactions in yeast. Nat Biotechnol. 2000;18:1257–1261. doi: 10.1038/82360. [DOI] [PubMed] [Google Scholar]
- The Arabidopsis Genome Initiative. Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature. 2000;408:796–815. doi: 10.1038/35048692. [DOI] [PubMed] [Google Scholar]
- The C. elegans Sequencing Consortium. Genome sequence of the nematode C. elegans: A platform for investigating biology. Science. 1998;282:2012–2018. doi: 10.1126/science.282.5396.2012. [DOI] [PubMed] [Google Scholar]
- The RIKEN Genome Exploration Research Group Phase II Team and the FANTOM Consortium. Functional annotation of a full-length mouse cDNA collection. Nature. 2001;409:685–690. doi: 10.1038/35055500. [DOI] [PubMed] [Google Scholar]
- Uetz P, Giot L, Cagney G, Mansfield TA, Judson RS, Knight JR, Lockshon D, Narayan V, Srinivasan M, Pochart P, et al. A comprehensive analysis of protein–protein interactions in Saccharomyces cerevisiae. Nature. 2000;403:623–627. doi: 10.1038/35001009. [DOI] [PubMed] [Google Scholar]
- Venter JC, Adams MD, Myers EW, Li PW, Mural RJ, Sutton GG, Smith HO, Yandell M, Evans CA, Holt RA, et al. The sequence of the human genome. Science. 2001;291:1304–1351. doi: 10.1126/science.1058040. [DOI] [PubMed] [Google Scholar]
- Walhout AJ, Sordella R, Lu X, Hartley JL, Temple GF, Brasch MA, Thierry-Mieg N, Vidal M. Protein interaction mapping in C. elegans using proteins involved in vulval development. Science. 2000;287:116–122. doi: 10.1126/science.287.5450.116. [DOI] [PubMed] [Google Scholar]
- Wang J, Hoshino T, Redner RL, Kajigaya S, Liu JM. ETO, fusion partner in t(8;21) acute myeloid leukemia, represses transcription by interaction with the human N-CoR/mSin3/HDAC1 complex. Proc Natl Acad Sci. 1998;95:10860–10865. doi: 10.1073/pnas.95.18.10860. [DOI] [PMC free article] [PubMed] [Google Scholar]