Abstract
In an attempt to understand the origin of CpG islands (CGIs) in mammalian genomes, we have studied their location and structure according to the expression pattern of genes and to the G + C content of isochores in which they are embedded. We show that CGIs located over the transcription start site (named start CGIs) are very different structurally from the others (named no-start CGIs): (1) 61.6% of the no-start CGIs are due to repeated sequences (79 % are due to Alus), whereas only 5.6% of the start CGIs are due to such repeats; (2) start CGIs are longer and display a higher CpGo/e ratio and G + C level than no-start CGIs. The frequency of tissue-specific genes associated to a start CGI varies according to the genomic G + C content, from 25% in G + C-poor isochores to 64% in G + C-rich isochores. Conversely, the frequency of housekeeping genes associated to a start CGI (90%) is independent of the isochore context. Interestingly, the structure of start CGIs is very similar for tissue-specific and housekeeping genes. Moreover, 93% of genes expressed in early embryo are found to exhibit a CpG island over their transcription start point. These observations are consistent with the hypothesis that the occurrence of these CGIs is the consequence of gene expression at this stage, when the methylation pattern is installed.
In mammalian genomes, CpG dinucleotides are present at ∼25% of their expected frequency. This deficiency is thought to be due to the following two factors: (1) in these genomes, 60% to 90% of cytosines at CpG dinucleotides are methylated (Bird and Tagart 1980), and (2) methylated cytosines mutate to thymines at a very high rate (Coulondre et al. 1978). This high mutation rate from CpG to TpG (or CpA on the complementary strand) has been confirmed by many studies on DNA polymorphism or genetic diseases (Cargill et al. 1999; Giannelli et al. 1999; Halushka et al. 1999), and simulation studies have shown that the CpG deficiency observed in mammalian genomes correspond exactly to what would be expected with such mutation rates (Duret and Galtier 2000).
Experiments of DNA digestion with restriction enzymes sensitive to cytosines methylation have revealed the existence of regions (making up 1% to 2% of the genome) that escape methylation (Bird 1986). The analysis of these DNA fragments (originally called HTF, for HpaII tiny fragments) revealed that they are relatively G + C rich and are less depleted in CpG dinucleotides than the rest of the genome, and hence, in contrast, appear as relatively CpG rich (Bird 1986; Antequera and Bird 1999). These so-called CpG islands (CGIs) are typically several hundred bases to several kilobases long and are dispersed throughout the genome. The estimated number of CGIs in the human haploid genome is between 26,000 and 45,000 (Antequera and Bird 1993; Ewing and Green 2000). It is generally admitted that the relative CpG richness of CGIs directly reflects the absence of methylation. More precisely, the CpG depletion observed in the genome should reflect the methylation status in the germ line (methylation in somatic cells may induce mutations, but such mutations are not transmitted to the offspring and, hence, do not participate in genome evolution). The compositional features of CGIs (CpG frequency and G + C content) allow their direct identification in DNA sequences by computer analysis. However, it should be stressed that the identification of a region that has the properties of a CGI is not sufficient to prove that the region actually escapes methylation.
CGIs are often overlapping with promoters and many studies have shown that the presence of CGIs in the promoter region is correlated with particular gene expression patterns. According to the data from Larsen et al. (1992), all housekeeping genes exhibit a CGI covering the transcription start site (TSS), whereas only 25% of tissue-specific genes have a CGI over the TSS. Furthermore, CGIs have an open chromatin structure and may be the site of interactions between transcription factors and promoters (Tazi and Bird 1990). In vivo, some CGIs are methylated on the inactive X chromosome and in immortalized cell lines (Antequera et al. 1990; Pieper et al. 1999). Methylation of CGIs appears to be one route by which genes are epigenetically silenced in cancer (for review, see Schmutte and Jones 1998). In vitro, artificial methylation or demethylation of gene sequences result in repression or activation of gene expression, respectively (Razin and Cedar 1991). However, present knowledge does not allow us determine whether CGIs are the cause or a consequence of the regulation of expression.
Further hypotheses were proposed to explain the methylation-free status and therefore the maintenance of CGIs. Franck et al. (1991) and MacLeod et al. (1994) proposed that CGIs could be protected from methylation by the binding of transcription factors at the promoter sequences, which are colocalized with promoters active in the germ line. Furthermore, the CGIs could be associated with replication origins occurring at active promoters during early development (Antequera and Bird 1999). A more general hypothesis suggests that CGIs are the direct consequence of interactions between some DNA-binding proteins (transcription factors or others proteins) and a demethylase enzyme (Lin et al. 2000).
Other studies have shown that the distribution of CGIs varies widely with G + C content along chromosomes; in G + C-rich isochores (H3), the density of CGIs is 15 times higher than in G + C-poor isochores (L1 + L2) (Aissani and Bernardi 1991; Jabbari and Bernardi 1998). This difference is partly explained by the higher gene density in G + C-rich compared with G + C-poor isochores (Mouchiroud et al. 1991). Taking this effect into account, Aissani and Bernardi (1991) estimated that there were four times more genes associated with CpG islands in H3 compared with L1 + L2. Because housekeeping genes are always associated with CGIs, it has been proposed that housekeeping genes might be concentrated in G + C-rich isochores (Bernardi 1993, 1995). In contradiction with this hypothesis, however, we have shown recently,that housekeeping genes were not G + C rich (Gonçalves et al. 2000). The reason why CGIs are more frequent in G + C-rich isochores, therefore, remained an open question.
Recently, expressed sequence tag (EST) projects have been initiated in different species with an aim to make an inventory of all of the mRNAs that they express. More than 4,000,000 ESTs from ∼40 different tissues have been sampled in human and mouse. These sequences are generally partial (typically, sequences are 300–500 nucleotides long) and with a relatively high rate of sequencing errors (∼1%–3%). However, these ESTs are accurate and long enough to unambiguously identify their corresponding genes. Thus, these data can be used to estimate the tissue distribution breadth of human or mouse genes.
We took advantage of these data to re-evaluate the relationship between gene expression, isochore G + C content, and the presence of CpG islands in 1593 human genes for which the complete genomic sequence was available. Notably, we tested the hypothesis that CGIs are associated with genes that are active during early development. Our analyses bring new insights into the factors that determine the presence and the structure of CGIs throughout the genome.
RESULTS
CGIs were systematically searched in the DNA sequence of human protein-coding genes for which the TSS is annotated. CGIs are defined here as sequences longer than 500 bp, with a ratio observed over the expected number of CpG (CpGo/e) >0.6 and a G + C frequency >0.5. CGIs located over the TSS were classified as start CGIs, whereas other islands were classified as no-start CGIs.
It should be noted that previous publications did not always use the same length criteria to define CGIs; in some analyses, the threshold was set to 200 bp (Gardiner-Garden and Frommer 1987; Larsen et al., 1992; Ioshikhes and Zhang, 2000), whereas other authors used 500 bp (Matsuo et al. 1993; Jabbari and Bernardi 1998). There was no clear justification for using one threshold rather than another. In a first approach, we compared the 200- and 500-bp thresholds to identify CGIs. The number of no-start CGIs detected varies from 7717 (with the 200-bp threshold) to 1848 (with the 500-bp threshold). We noticed that most (71.3%) of short (<500 bp) CGIs corresponded to repeated sequences (essentially Alu, see below). If repeated sequences are excluded, the number of no-start CGIs detected varies from 2714 (with the 200-bp threshold) to 762 (with the 500-bp threshold). Thus, independent of repeated sequences, the number of no-start CGIs detected depends strongly on the length of the threshold. Conversely, 96.9% of start CGIs detected with the 200-bp threshold can also be detected with the threshold of 500 bp. The origin and potential function (if any) of short no-start CGIs is not known, but they clearly differ in structure from start-CGIs (see below). Because we are interested primarily in CGIs that potentially correspond to promoter regions (i.e., start CGIs), we decided to consider short CGIs as false-positives. Thus, hereafter, we will only analyze CGIs longer than 500 bp.
The complete data set is composed of 2429 CGIs, among which 1846 are complete (the 583 partial CGIs correspond to CGIs that overlap one extremity of the sequence). The data set exhibits a density of 6.8 CGIs per 100 kb, which is higher than the frequency observed for the whole human genome (2.2/100 kb; from data of Lander et al. 2001). This difference can be explained partly by a higher density of genes in our dataset (2.7 genes/100 kb vs. 1.3 genes/100 kb; from data of Lander et al. 2001) and association of CGIs with promoters. Statistical analyses of the structure and composition of CGIs were based only on complete CGIs to avoid potential biases caused by the truncated ones.
The tissue distribution of human genes was estimated by comparing their protein coding sequences (CDS) to a database of ESTs representing 24 tissues. Hereafter, genes that are expressed in at least 17 tissues are considered as housekeeping, whereas those that are detected in 0 or 1 tissue are considered as tissue specific. Housekeeping and tissue-specific genes make up 6% and 30%, respectively, of the data set. As mentioned in the Methods section, the DNA sequences were classified into the three isochore classes of increasing G + C levels, L1–L2, H1–H2, and H3 (Mouchiroud et al. 1991).
CpG Islands and Repeated Sequences
As some repeated sequences present a base composition similar to CGIs (i.e., high CpGo/e and G + C frequency), we searched for repeated sequences over the 1846 complete CGIs. The parameters of the CGIs (length, %G + C, and CpGo/e) were recalculated after elimination of the repeated sequences, and the CGIs with at least one parameter under the thresholds (500 bp, G + C ≥0.5 and CpGo/e ≥0.6) were considered as being the consequence of the presence of the repeated sequences. These CGIs are hereafter called repeat-CGIs. The result shows that 50.3% of the CGIs detected previously are due to repeated sequences, and 79% of these repeat-CGIs are due to Alu elements. Among them, 74.7% are due to young Alus (Alu Y, 1–30 Myr; Lander et al. 2001), whereas only 51% are due to middle-age Alus (Alu S, 25–60 Myr) and 11.0% are due to the oldest Alus (Alu J, 60–100 Myr). The sum of these percentages is >100% as some CGIs contain several Alus and can be the result of Alu elements from different families. Alus Y, S, and J represent, respectively, 13.3%, 60.6%, and 26.1% of the Alu elements present in the human genome (data from Lander et al. 2001) and this indicates that young Alus are strongly over-represented in the repeat-CGIs.
Only 5.6% of start CGIs correspond to repeat-CGIs, against 61.6% for no-start CGIs. In contrast to no-start CGIs, the start CGIs due to the repeated sequences, were not considered separately because of their low frequency.
Structure of CpG Islands
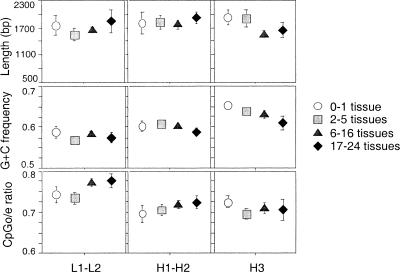
We compared the structure of the 908 repeat-CGIs, the 565 other no-start CGIs, and the 373 start CGIs according to the tissue-distribution breadth of the associated gene and to the isochore G + C content. The first observation is that the characteristics of the 373 complete start CGIs are similar between housekeeping and tissue-specific genes (Fig. 1). The length, the G + C frequency, and the CpGo/e ratio do not vary significantly according to the tissue breadth of expression (Kruskal-Wallis test, P >0.05).
Figure 1.
The length, the CpGo/e ratio, and the (G + C) frequency of start CGIs according to the tissue breadth of expression and the isochore classes of the associated genes. (○) 0–1 tissue; (▪) 2–5 tissues; (▴) 6–16 tissues; (♦) 17–24 tissues. Error bars, SE. n = 373.
We noticed that the average G + C level of CGIs increases from L1–L2 to H3 isochore classes (Kruskal-Wallis test, P <0.0001) (Fig. 1). This result, which is also observed for the others CGIs (data not shown), was expected as it has been shown that the effect of isochore context on G + C content affects all positions along the gene (exons, introns, and flanking sequences) (D'Onofrio et al. 1991).
Secondly, the structure of complete CGIs shows a sharp difference between the three classes of CGIs as follows: repeat-CGIs, no-start-CGIs, and start-CGIs (Fig. 2). Repeat-CGIs are smaller (659 bp; P <0.0001) and exhibit a lower G + C level (0.53; P <0.0001) and CpGo/e ratio (0.62; P <0.0001) than other CGIs. Moreover, no-start CGIs are very different structurally from start CGIs. On average, start CGIs are longer than no-start CGIs (1753 bp vs. 1267 bp; P <0.0001), they have a higher CpGo/e ratio (0.73 vs. 0.67; P <0.0001) and they have a higher G + C content (0.61 vs. 0.59; P <0.0001). The average size and composition of CGIs are different from that published recently by Ioshikhes and Zhang (2000) because we used a more stringent length criteria to identify CGIs (500 instead of 200 bp) and we considered the repeat-CGIs separately. However, our analyses totally confirm their observation that start CGIs are structurally very different from no-start CGIs. We show further that this difference remains detectable even when repeat-CGIs are considered separately. This difference between start and no-start is probably underestimated because the data set of no-start CGIs may contain some unidentified TSS.
Figure 2.
The length, the CpGo/e ratio, and the (G + C) frequency of the different types of CGIs, start CGIs (n = 373), no-start CGIs (n = 565), and repeat-CGIs (n = 908). (○) Repeat CGIs; (▪) no-start CGIs; (▴); start CGIs. Error bars, SE.
Distribution of Start CpG Islands Along the Genes
To ensure that all start CGIs could be detected, human sequences with <500 nucleotides upstream the TSS were excluded from this analysis. Overall, 58.0% of the 864 coding genes analyzed exhibit a start CGI. As shown in Table 1, there is a strong positive correlation between the frequency of genes with a start CGI and tissue-distribution breadth, from 42% in tissue-specific to 90% in housekeeping genes (χ2 test, P <0.0001). However, contrary to previous studies (Larsen et al. 1992), we found that a significant fraction of housekeeping genes (10%) do not possess a start CGI. It should be noted that this previous study was conducted with a much smaller data set and that they used different criteria to classify their gene in housekeeping or tissue-specific classes (see Discussion).
Table 1.
Frequency of Genes With a Start CGI According to Their Tissue Distribution Breadth and the Isochore G + C Content
| No. of genes | Frequency of genes with a start CGI (%) | ||
|---|---|---|---|
| Tissue | 0–1 | 274 | 41.6 |
| distribution | 2–5 | 276 | 52.2 |
| 6–16 | 263 | 74.9 | |
| 17–24 | 51 | 90.2 | |
| Isochores | L1–L2 | 272 | 55.1 |
| H1–H2 | 343 | 51.9 | |
| H3 | 249 | 69.5 | |
| Total | 864 | 58.0 | |
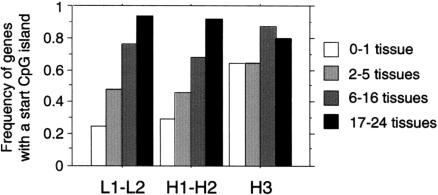
The frequency of genes with a start CGI varies according to the isochore classes also. Genes localized in H3 isochore are significantly associated more frequently with a start CGI than genes localized in L1–L2 isochores (70% vs. 55%, χ2 test: P <0.0001) (Table 1). Interestingly, this link between isochore context and the presence of start CGI depends on the tissue breadth of expression (Fig. 3). The percentage of tissue-specific genes with a start CGI is 2.5 greater in H3 isochore than in L1–L2 isochores (25%, 29%, and 64% for L1–L2, H1–H2, and H3, respectively; χ2 test: P <0.0001), whereas 90% of housekeeping genes are associated to a start CGI, independent of the isochore structure (94%, 92%, and 80% for L1–L2, H1–H2, and H3, respectively; χ2 test: P >0.52). Thus, the presence of CGI over the TSS appears to be independent of the isochore context for housekeeping genes, whereas the frequency of tissue-specific genes with start CGI is correlated positively with isochores G + C content.
Figure 3.
Frequency of genes with a start CGI according to their tissue breadth of expression and the isochore G + C content. n = 864 genes. (○) 0–1 tissue; (▪) 2–5 tissues; (▴) 6–16 tissues; (♦) 17–24 tissues.
Start CpG Islands and Expression in Early Embryo
Some authors have suggested that start CGIs of tissue-specific or housekeeping genes are localized on promoters that are active during early development (Choi and Chae 1991; MacLeod et al. 1998) and more particularly in totipotent cells (Antequera and Bird 1999). To test this hypothesis, we analyzed genes expressed in early embryo. EST data from early embryo cDNA libraries are available for mouse but not for human. Unfortunately, there are presently not enough mouse genomic sequences with identified TSS to conduct a statistical analysis. We therefore used an indirect approach based on the assumption that human and mouse orthologous genes have similar expression patterns. First, we took our data set of human genes with identified TSS, searched for their orthologs in mouse, and then compared these orthologs with mouse early embryo ESTs. Early embryo corresponds to the stages from the fertilized egg to the blastocyste. From the data set, 367 human genes have an orthologous gene in mouse, among which 102 were detected in early embryo (Table 2). As expected, most (30/33) of the genes classified here as housekeeping are found to be expressed in early embryo, compared with only 9% for tissue-specific genes. Globally, 93% of the early embryo genes are associated with a start CGI compared with 56% of other genes. The most noteworthy result is that this high frequency of start CGI among genes expressed in early embryo is observed both for housekeeping and tissue-specific genes. Notably, all the tissue-specific genes detected in early embryo exhibit a start CGI (Table 2). Thus, nearly all genes expressed in early embryo at the totipotent cell stage or in the blastocyste are associated to a start CGI independent of their tissue-distribution breadth in somatic cells.
Table 2.
Start CGIs and Expression at Early Embryo Stage
| Tissue distribution | Early embryo genes? | No. of genes | Genes with a start CGI (%) | χ2P value |
|---|---|---|---|---|
| 0–1 | yes | 9 | 100.00 | 0.026* |
| no | 88 | 45.45 | ||
| 2–5 | yes | 15 | 80.00 | 0.056* |
| no | 98 | 53.06 | ||
| 6–16 | yes | 48 | 95.83 | 0.0014* |
| no | 76 | 73.68 | ||
| 17–24 | yes | 30 | 93.33 | 0.033* |
| no | 3 | 33.33 | ||
| Total | yes | 102 | 93.14 | |
| no | 265 | 56.23 |
The number and percentage of genes with a start CGI are indicated according to their overall tissue breadth of expression and to their expression at early embryo stage. The table indicates the P value for the comparison of start CGI frequencies between the genes detected and not detected in early embryo.
DISCUSSION
In previous works, CGIs distribution has been studied according to the isochore's structure (Jabbari and Bernardi 1998) or according to the expression pattern of the genes (Larsen et al. 1992). The former studies concluded that CGIs are concentrated in G + C-richer isochore, whereas the latter showed that all housekeeping genes exhibit a CGI on the TSS (start CGI). In the present study, we analyzed the start CGIs according to the two preceding factors taken simultaneously.
At Least Three Classes of CpG Islands
Our analyses with a larger data set confirm the results of Ioshikhes and Zhang (2000). These authors have shown that there exist two classes of CGIs characterized by different structural properties, the start CGIs located over the TSS and the no-start CGIs located upstream or downstream of this point. However, with the length criteria that they used to identify CGIs (>200 bp), it is likely that their data set included many CGIs composed of repeat sequences such as Alus. In our analysis, we took into account the presence of repeated sequences and found that >60% of the no-start CGIs are due to such repeated elements (repeat-CGIs) and correspond essentially to young Alus. It is unlikely that repeat-CGIs correspond to true CGIs (i.e., regions that escape methylation). Only a very small number of replicatively competent Alu elements is responsible for the insertion of new copies in the human genome (Quentin, 1988 Deininger et al. 1992). Whereas these master copies are active (and presumably unmethylated), the large majority of Alu elements are transcriptionnaly silent (Deininger et al. 1992). Moreover, it has been shown that Alu sequences are heavily methylated and that their CpG dinucleotides are not evolutionary stable (Hellmann-Blumberg et al. 1993; Rubin et al. 1994). Recently inserted Alu elements still have the features of unmethylated sequences (because there was not enough time to accumulate mutations at CpG dinucleotides), and hence, by sequence analysis, they are identified as CGIs. However, a majority of these repeat-CGIs are probably methylated and should be considered as false-CGIs.
Even after the elimination of repeat-CGIs, the start CGIs are longer and exhibit higher CpGo/e ratio and G + C level than no-start CGIs. All start CGIs tested so far have been shown to be methylation free in the germ line, even when they were methylated in some tissue (Choi and Chae 1991; Frank et al. 1991; MacLeod et al. 1998). Their methylation-free status is associated with the fixation of transcription factors (Sp1) over the promoter. On the other hand, the status of no-start CGIs is not well known. Their relatively low CpGo/e ratio suggests that these CGIs are not as protected from methylation as are the start CGIs. Furthermore, this difference between start and no-start CGIs may be underestimated because some CGIs qualified here as no-start may correspond to unknown TSS (MacLeod et al. 1998).
Start CpG Islands and Isochore G + C Content
An unexpected result is that tissue-specific genes are more frequently associated with a start CGI in G + C-rich than in G + C-poor isochore (64% vs. 25%), whereas for housekeeping genes, the presence of start CGI is independent of the isochore context. It could be argued that start CGIs associated with these tissue-specific genes in the G + C-rich isochore are simply the consequence of local base composition and are not really nonmethylated sequences. However, we found no significant structural difference between start CGIs of housekeeping and tissue-specific genes (Fig. 1), which suggests that these latter CGIs really correspond to unmethylated islands.
Bernardi (1993, 1995) suggested that housekeeping genes were concentrated in G + C-rich isochore. However, we did not find this pattern; the frequency of housekeeping was rather slightly higher in L1 + L2 (6%) than in H3 (4%), which confirms previous observations (Gonçalves et al. 2000). Bernardi's proposition was based on the positive correlation between the CGIs frequency and isochores G + C content (Aissani and Bernardi 1991) and on the association between housekeeping genes and start CGIs (Larsen et al. 1992). In fact, we confirm the finding that start CGIs are more frequent in G + C-rich than in G + C-poor isochores, but we show that this difference is due to CGIs in tissue-specific genes and not to housekeeping genes.
Start CpG Islands and Tissue Breadth of Expression
The distribution of start CGIs according to expression and isochore's G + C content shows that housekeeping genes are almost always associated with a start CGI, independent of the isochore structure. Larsen et al. (1992) found that 100% of housekeeping genes were associated with start CGIs. In our analyses, a significant fraction of housekeeping genes (5/51) do not contain any start CGI. Several hypotheses could explain this discrepancy. The CGIs definition used here is more stringent (minimal length, 500 vs. 200 nucleotides) in order to limit the detection of repeat-CGIs. However, using the 200-bp criteria does not radically change our results, only one more start CGI is found among these five housekeeping genes. In fact, the 500-bp threshold does not appear to be a limitation for the identification of start CGIs that are generally much larger (1753 bp on average). The percentages of genes identified with a start CGI in the two studies are similar (58% vs. 56%). The difference between our results and previous results is probably due to the estimation of tissue distribution breadth. We used EST data, whereas Larsen and colleagues extracted expression data from the work referred to in the sequence databases. The advantage of our approach is that the same procedure and the same tissue samples were used to estimate the expression patterns of all genes, whereas Larsen et al. (1992) used data obtained from different methods and different tissue samples. For example, in agreement with Larsen and colleagues, we found that galectin 1 and E-apolipoprotein genes do not contain CGI on their TSS. However, whereas these two genes were considered as restrictedly expressed by Larsen and colleagues, we detected matching ESTs in 19/24 and 16/24 tissues, respectively. Data from the literature also confirmed that these two genes are widely expressed (Mahley 1988; Perillo et al. 1998). Thus, some housekeeping genes seem to lack start CGI. However, some of the three other genes might, in fact, be tissue specific because of possible contamination artefact in EST sequences. It is also possible that some TSS annotations were incomplete because of unknown alternative promoters. Moreover, an alternative explanation could be that these genes are widely expressed in somatic tissues, but not in the germ line (see below).
The structure (length and base composition) of start CGIs of tissue-specific genes is very similar to that of housekeeping genes. This result disagrees with the conclusions of Edwards (1990) on the basis of the analysis of 44 vertebrate genes, which indicated that CGIs of housekeeping genes were more efficiently protected from CpG depletion than CGIs of tissue-specific genes. In fact, our results, based on a much larger data set, suggest that the lack of CpG depletion (i.e., absence of methylation) at start CGIs is independent of the tissue distribution breadth in the whole organism. Therefore, this result suggests that there is a common characteristic between 90% of housekeeping genes and 42% of tissue-specific genes that is responsible for the presence of CpG island over their promoter. It has been proposed that start CGIs correspond to promoters of genes that are active in the germ line, and notably during early development (Franck et al. 1991; MacLeod et al. 1994; Antequera and Bird 1999). During development, the methylation pattern is totally removed at the totipotent stage and is installed de novo at the blastocyste stage (Razin and Shemer 1995). Specific demethylation of CGIs is observed in embryonic cells (Frank et al. 1991) and could protect CGIs against methylation. Consistent with this hypothesis, we found that nearly all genes expressed in early embryo are associated with a start CGI, not only housekeeping, but also tissue-specific genes. However, further work is necessary to test whether all CGIs correspond to promoters that are active in the germ line. The most surprising observation is the strong correlation between isochores G + C content and the frequency of tissue-specific genes with a start CGI. One possible hypothesis would be that tissue-specific genes expressed in the germ line are preferentially located in G + C-rich isochores. However, in mouse (in which we have EST data from early embryo), we found no significant difference in G + C content between tissue-specific genes expressed in early embryo (n = 63 G + C = 59.4%) and other tissue-specific genes (n = 1341 G + C = 57.5%; t-test P = 0.15). The correlation between start-CGI frequency in tissue-specific genes and isochores G + C content therefore remains an open question.
METHODS
Sequence Data
CDS of human genes were selected from HOVERGEN (release 114, October 1999, Duret et al. 1994) by use of the ACNUC retrieval system (Gouy et al. 1985). All CDS were compared with each other using BLASTN2 (Altschul et al. 1997) to remove redundant sequences and splice variants. Two sequences were considered as redundant if BLASTN2 alignment showed at least 97% of identity with 200 nucleotides or more.
G + C content of CDS was computed with the Java application JaDis (Gonçalves et al. 1999). CDS were classified in three isochores classes according to their G + C content at the third codon position. L1–L2, <0.57, H1–H2, between 0.57 and 0.75, and H3 >0.75 (Mouchiroud et al. 1991).
For each sequence, the position of the TSS, when available, were extracted from the annotations of the database. Moreover, for each gene with a known TSS, the genomic sequences were aligned with ESTs matching the CDS to identify potential alternative start sites.
Expression Profile
We selected from GenBank (release 115, December, 1999) 1,775,942 ESTs from 24 human tissues as follows: aorta, brain (adult, fetal, and infant), breast, colon, endothelial cells, eye, fetal heart, lung (adult and fetal), fibroblasts, germinal center of B cells, kidney, liver, muscle, neuroepithelium, pancreas, placenta, prostate, testis, uterus, bone and bone marrow, and lymph-associated tissues. cDNA libraries from cell culture, tumors, pooled organs, or unidentified tissues were excluded. To limit stochastic variations in expression measures, we only retained cDNA libraries that had been sampled with at least 10,000 ESTs. Selected CDS were first filtered with XBLAST program (Claverie and States 1993) to mask repetitive elements. CDS were then compared with the EST data set by using BLASTN2. BLASTN2 alignments showing at least 95% identity over 100 nucleotides or more were counted as a sequence match. This criteria was chosen to be low enough to allow the detection of most ESTs despite sequencing error (the average sequence accuracy of ESTs is ∼97%) (Hillier et al. 1996), but stringent enough to distinguish — in most cases — different members of highly conserved gene families (e.g., β- and γ-actins, proteins are 98% identical, CDS are 91% identical; cardiac and skeletal α-actins, proteins are 99% identical, CDS are 85% identical; histones H3.3A and H3.3B, proteins are 100% identical, CDS are 79% identical) (Duret and Mouchiroud 2000). Genes were classified in four classes as follows: 0–1 tissues, 2–5 tissues, 6–16 tissues, and 17–24 tissues. Each of the three first classes represents ∼30% of the data set, whereas the last class represents 6% of genes.
Expression in Early Embryo
ESTs from early embryo are available in mouse but not in human. Therefore, we used an indirect approach to identify human genes expressed at this stage. First, we used the HOVERGEN database to identify in our data set the genes for which a mouse ortholog was available. By use of the same procedure as described above, these mouse orthologs were compared with a data set of 13,457 ESTs from mouse early embryo (selected from GenBank release 115, December, 1999). In this way, we identified 367 human genes for which a mouse ortholog was available, among which 102 have at least one matching EST in early embryo.
CpG Islands
CGIs were defined as DNA regions longer than 500 nucleotides, with a moving average G + C frequency above 0.5 and a moving average ratio of observed over expected CpG (CpGo/e) >0.6. The ratio CpGo/e was calculated according to Gardiner-Garden and Frommer (1987) as follows: CpGo/e = (CpG × L)/(C × G), in which CpG is the number of CpG, L the length of the sequence, C the number of C ,and G the number of G. Moving average values for %G + C and for CpGo/e were calculated for each sequence by use of a 500-nucleotide window moving along the sequence in steps of 1 nucleotide. Overlapping windows with a G + C frequency higher than 0.5 and a CpGo/e ratio higher than 0.6 were grouped to form CGIs. CGIs were characterized by their length, global G + C frequency, and global CpGo/e ratio. They were classified according to their location along the sequence. Thus, two classes were identified as follows: the CGI located over the initiation site of transcription, named start CGIs, and the other CGIs, named no-start CGIs (Larsen et al. 1992). CGIs overlapping the 5′ or 3′ end of the sequences were considered as partial, and were not used for the statistical analysis of the structure of the CGIs.
The repeated sequences (Alus, satellites…) located over the CGIs were identified and masked using RepeatMasker (A. Smit and P. Green, unpubl., http://ftp.genome.washington.edu/RM/RepeatMasker.html). The data of Alu elements in the human genome was extracted from RepeatMasker analysis of the Human Genome Project annotation (October 7, 2000; Lander et al. 2001) available at http://genome.cse.ucsc.edu/.
All of the data files and source files of the software used to search CGIs are available at http://pbil.univ-lyon1.fr/datasets/Ponger2001/data.html.
Acknowledgments
This work is supported by the CNRS (Centre National de la Recherche Scientifique).
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL ponger@biomserv.univ-lyon1.fr; FAX 33-4-78-89-27-19.
Article published on-line before print: Genome Res., 10.1101/gr.174501.
Article and publication are at http://www.genome.org/cgi/doi/10.1101/gr.174501.
REFERENCES
- Aissani B, Bernardi G. CpG islands, genes and isochores in the genomes of vertebrates. Gene. 1991;106:185–195. doi: 10.1016/0378-1119(91)90198-k. [DOI] [PubMed] [Google Scholar]
- Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antequera F, Bird A. Number of CpG islands and genes in human and mouse. Proc Natl Acad Sci. 1993;90:11995–11999. doi: 10.1073/pnas.90.24.11995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ————— CpG islands as genomic footprints of promoters that are associated with replication origins. Curr Biol. 1999;9:661–667. doi: 10.1016/s0960-9822(99)80418-7. [DOI] [PubMed] [Google Scholar]
- Antequera F, Boyes J, Bird A. High levels of de novo methylation and altered chromatin structure at CpG islands in cell lines. Cell. 1990;62:503–514. doi: 10.1016/0092-8674(90)90015-7. [DOI] [PubMed] [Google Scholar]
- Bernardi G. The isochore organization of the human genome and its evolutionary history—a review. Gene. 1993;135:57–66. doi: 10.1016/0378-1119(93)90049-9. [DOI] [PubMed] [Google Scholar]
- ————— The human genome: Organization and evolutionary history. Annu Rev Genet. 1995;29:445–476. doi: 10.1146/annurev.ge.29.120195.002305. [DOI] [PubMed] [Google Scholar]
- Bird AP. CpG-rich islands and the function of DNA methylation. Nature. 1986;321:209–213. doi: 10.1038/321209a0. [DOI] [PubMed] [Google Scholar]
- Bird AP, Taggart MH. Variable patterns of total DNA and rDNA methylation in animals. Nucleic Acids Res. 1980;8:1485–1497. doi: 10.1093/nar/8.7.1485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cargill M, Altshuler D, Ireland J, Sklar P, Ardlie K, Patil N, Shaw N, Lane CR, Lim EP, Kalyanaraman N, et al. Characterization of single-nucleotide polymorphisms in coding regions of human genes. Nat Genet. 1999;22:231–238. doi: 10.1038/10290. [DOI] [PubMed] [Google Scholar]
- Choi YC, Chae CB. DNA hypomethylation and germ cell-specific expression of testis- specific H2B histone gene. J Biol Chem. 1991;266:20504–20511. [PubMed] [Google Scholar]
- Claverie JM, States DJ. Information enhancement methods for large scale sequence analysis. Computers and Chemistry. 1993;17:191–201. [Google Scholar]
- Coulondre C, Miller JH, Farabaugh PJ, Gilbert W. Molecular basis of base substitution hotspots in Escherichia coli. Nature. 1978;274:775–780. doi: 10.1038/274775a0. [DOI] [PubMed] [Google Scholar]
- Deininger PL, Batzer MA, 3rd, Hutchison CA, Edgell MH. Master genes in mammalian repetitive DNA amplification. Trends Genet. 1992;8:307–311. doi: 10.1016/0168-9525(92)90262-3. [DOI] [PubMed] [Google Scholar]
- D'Onofrio G, Mouchiroud D, Aissani B, Gautier C, Bernardi G. Correlations between the compositional properties of human genes, codon usage, and amino acid composition of proteins. J Mol Evol. 1991;32:504–510. doi: 10.1007/BF02102652. [DOI] [PubMed] [Google Scholar]
- Duret L, Galtier N. The covariation between TpA deficiency, CpG deficiency, and G+C content of human isochores is a mathematical artifact. Mol Biol Evol. 2000;17:1620–1625. doi: 10.1093/oxfordjournals.molbev.a026261. [DOI] [PubMed] [Google Scholar]
- Duret L, Mouchiroud D. Determinants of substitution rates in mammalian genes: Expression pattern affects selection intensity but not mutation rate. Mol Biol Evol. 2000;17:68–74. doi: 10.1093/oxfordjournals.molbev.a026239. [DOI] [PubMed] [Google Scholar]
- Duret L, Mouchiroud D, Gouy M. HOVERGEN: A database of homologous vertebrate genes. Nucleic Acids Res. 1994;22:2360–2365. doi: 10.1093/nar/22.12.2360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards YH. CpG islands in genes showing tissue-specific expression. Philos Trans R Soc Lond B Biol Sci. 1990;326:207–215. doi: 10.1098/rstb.1990.0005. [DOI] [PubMed] [Google Scholar]
- Ewing B, Green P. Analysis of expressed sequence tags indicates 35,000 human genes. Nat Genet. 2000;25:232–234. doi: 10.1038/76115. [DOI] [PubMed] [Google Scholar]
- Frank D, Keshet I, Shani M, Levine A, Razin A, Cedar H. Demethylation of CpG islands in embryonic cells. Nature. 1991;351:239–241. doi: 10.1038/351239a0. [DOI] [PubMed] [Google Scholar]
- Gardiner-Garden M, Frommer M. CpG islands in vertebrate genomes. J Mol Biol. 1987;196:261–282. doi: 10.1016/0022-2836(87)90689-9. [DOI] [PubMed] [Google Scholar]
- Giannelli F, Anagnostopoulos T, Green PM. Mutation rates in humans. II. Sporadic mutation-specific rates and rate of detrimental human mutations inferred from hemophilia B. Am J Hum Genet. 1999;65:1580–1587. doi: 10.1086/302652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonçalves I, Robinson M, Perriere G, Mouchiroud D. JaDis: Computing distances between nucleic acid sequences. Bioinformatics. 1999;15:424–425. doi: 10.1093/bioinformatics/15.5.424. [DOI] [PubMed] [Google Scholar]
- Gonçalves I, Duret L, Mouchiroud D. Nature and structure of human genes that generate retropseudogenes. Genome Res. 2000;10:672–678. doi: 10.1101/gr.10.5.672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gouy M, Gautier C, Attimonelli M, Lanave C, Di Paola G. ACNUC—a portable retrieval system for nucleic acid sequence databases: Logical and physical designs and usage. Comput Appl Biosci. 1985;1:167–172. doi: 10.1093/bioinformatics/1.3.167. [DOI] [PubMed] [Google Scholar]
- Halushka MK, Fan JB, Bentley K, Hsie L, Shen N, Weder A, Cooper R, Lipshutz R, Chakravarti A. Patterns of single-nucleotide polymorphisms in candidate genes for blood-pressure homeostasis. Nat Genet. 1999;22:239–247. doi: 10.1038/10297. [DOI] [PubMed] [Google Scholar]
- Hellmann-Blumberg U, Hintz MF, Gatewood JM, Schmid CW. Developmental differences in methylation of human Alu repeats. Mol Cell Biol. 1993;13:4523–4530. doi: 10.1128/mcb.13.8.4523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hillier LD, Lennon G, Becker M, Bonaldo MF, Chiapelli B, Chissoe S, Dietrich N, Dubuque T, Favello A, Gish W, et al. Generation and analysis of 280,000 human expressed sequence tags. Genome Res. 1996;6:807–828. doi: 10.1101/gr.6.9.807. [DOI] [PubMed] [Google Scholar]
- Ioshikhes IP, Zhang MQ. Large-scale human promoter mapping using CpG islands. Nat Genet. 2000;26:61–63. doi: 10.1038/79189. [DOI] [PubMed] [Google Scholar]
- Jabbari K, Bernardi G. CpG doublets, CpG islands and Alu repeats in long human DNA sequences from different isochore families. Gene. 1998;224:123–127. doi: 10.1016/s0378-1119(98)00474-0. [DOI] [PubMed] [Google Scholar]
- Lander ES, Linton LM, Birren B, Nusbaum C, Zody MC, Baldwin J, Devon K, Dewar K, Doyle M, Fitzhugh W, et al. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- Larsen F, Gundersen G, Lopez R, Prydz H. CpG islands as gene markers in the human genome. Genomics. 1992;13:1095–1107. doi: 10.1016/0888-7543(92)90024-m. [DOI] [PubMed] [Google Scholar]
- Lin IG, Tomzynski TJ, Ou Q, Hsieh CL. Modulation of DNA binding protein affinity directly affects target site demethylation. Mol Cell Biol. 2000;20:2343–2349. doi: 10.1128/mcb.20.7.2343-2349.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacLeod D, Charlton J, Mullins J, Bird AP. Sp1 sites in the mouse aprt gene promoter are required to prevent methylation of the CpG island. Genes & Dev. 1994;8:2282–2292. doi: 10.1101/gad.8.19.2282. [DOI] [PubMed] [Google Scholar]
- MacLeod D, Ali RR, Bird A. An alternative promoter in the mouse major histocompatibility complex class II I-Abeta gene: Implications for the origin of CpG islands. Mol Cell Biol. 1998;18:4433–4443. doi: 10.1128/mcb.18.8.4433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahley RW. Apolipoprotein E: Cholesterol transport protein with expanding role in cell biology. Science. 1988;240:622–630. doi: 10.1126/science.3283935. [DOI] [PubMed] [Google Scholar]
- Matsuo K, Clay O, Takahashi T, Silke J, Schaffner W. Evidence for erosion of mouse CpG islands during mammalian evolution. Somat Cell Mol Genet. 1993;19:543–555. doi: 10.1007/BF01233381. [DOI] [PubMed] [Google Scholar]
- Mouchiroud D, D'Onofrio G, Aissani B, MacAya G, Gautier C, Bernardi G. The distribution of genes in the human genome. Gene. 1991;100:181–187. doi: 10.1016/0378-1119(91)90364-h. [DOI] [PubMed] [Google Scholar]
- Perillo NL, Marcus ME, Baum LG. Galectins: Versatile modulators of cell adhesion, cell proliferation, and cell death. J Mol Med. 1998;76:402–412. doi: 10.1007/s001090050232. [DOI] [PubMed] [Google Scholar]
- Pieper RO, Lester KA, Fanton CP. Confluence-induced alterations in CpG island methylation in cultured normal human fibroblasts. Nucleic Acids Res. 1999;27:3229–3235. doi: 10.1093/nar/27.15.3229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quentin Y. The Alu family developed through successive waves of fixation closely connected with primate lineage history. J Mol Evol. 1988;27:194–202. doi: 10.1007/BF02100074. [DOI] [PubMed] [Google Scholar]
- Razin A, Cedar H. DNA methylation and gene expression. Microbiol Rev. 1991;55:451–458. doi: 10.1128/mr.55.3.451-458.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Razin A, Shemer R. DNA methylation in early development. Hum Mol Genet. 1995;4:1751–1755. doi: 10.1093/hmg/4.suppl_1.1751. [DOI] [PubMed] [Google Scholar]
- Rubin CM, Vandevoort CA, Teplitz RL, Schmid CW. Alu repeated DNAs are differentially methylated in primate germ cells. Nucleic Acids Res. 1994;22:5121–5127. doi: 10.1093/nar/22.23.5121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmutte C, Jones PA. Involvement of DNA methylation in human carcinogenesis. Biol Chem. 1998;379:377–388. doi: 10.1515/bchm.1998.379.4-5.377. [DOI] [PubMed] [Google Scholar]
- Tazi J, Bird A. Alternative chromatin structure at CpG islands. Cell. 1990;60:909–920. doi: 10.1016/0092-8674(90)90339-g. [DOI] [PubMed] [Google Scholar]