Abstract
We have developed a computer program that aligns spliced sequences to genomic sequences, using local alignment algorithms and heuristics to put together a global spliced alignment. Spidey can produce reliable alignments quickly, even when confronted with noise from alternative splicing, polymorphisms, sequencing errors, or evolutionary divergence. We show how Spidey was used to align reference sequences to known genomic sequences and then to the draft human genome, to align mRNAs to gene clusters, and to align mouse mRNAs to human genomic sequence. We compared Spidey to two other spliced alignment programs; Spidey generally performed quite well in a very reasonable amount of time.
Expressed sequences are the key to the inner workings of an organism. To understand fully the function of an expressed sequence, however, it needs to be put in its genomic context. Alignment of expressed sequences to their parent genomic sequences can be used to find or confirm a gene's position, to locate potential regulatory elements, and to study paralogs, pseudogenes, and alternative splicing. With estimates of the human gene number ranging from only 30,000 to 35,000 (The Genome Sequencing Consortium 2001), alternative splicing may be an important factor in generating transcriptional diversity, so mRNA-to-genomic alignments will be crucial to our understanding of the genome.
Even though an expressed sequence should be identical to the genomic sequence from which it is derived, aligning an expressed sequence to its genomic sequence is nontrivial. A 1340-base signal (average size of a coding region according to The Genome Sequencing Consortium 2001) is minute compared to the noise of the full 3200 Mb (The Genome Sequencing Consortium 2001) of the human genome, or even compared to megabase-sized contigs. Furthermore, there are often small repeated sequences near exon boundaries, so that the exact exon boundaries cannot be determined unambiguously from alignment information alone. To complicate matters even more, nearby pseudogenes and paralogs or duplicated exons may match the mRNA sequence strongly enough to obscure the location of the true gene. Also, introns can be quite large, separating exons by tens of kilobases.
We have written a spliced alignment program that addresses these issues. Spidey is capable of generating mRNA-to-genomic alignments quickly despite very large genomic sequences and is not affected by wide variations in intron size. Spidey can also perform cross-species mRNA-to-genomic alignments. Spidey's power comes from its design; first it delineates genomic regions likely to contain gene models and then performs a three-pass search for the details of the alignments to those regions. By excluding probably irrelevant or paralogous sequence from the analysis, it can create useful alignments in regions with paralogous sequences or poor sequence conservation without joining exons from neighboring genes mistakenly.
Spidey has many useful features; it is designed to be flexible and relevant to real world biological research. Spidey can take several different kinds of input: the name of a file containing a set of FASTA sequences, a file with a set of GI or accession numbers, or a single GI or “accession.version.” The input mRNAs can be masked with lowercase letters and Spidey will retain that masking if desired. Spidey can also accept a feature table delineating repetitive or low-complexity regions and can mask the mRNA sequences in that way. Spidey can return a single gene model or as many gene models as requested; this feature is especially useful when examining a region with tandem paralogs or with alternative splicing to very similar exons. The user can restrict the search to a region of the genomic sequence and can adjust Spidey's stringency at different stages of its procedure. Also, Spidey can align an mRNA to a genomic sequence and then extract the CDS alignment from the mRNA alignment, using CDS information from a feature table or from ASN.1 records. Finally, Spidey's output (Box 1) , designed in conjunction with researchers, gives as much information as possible about different features of the alignment.
We have tested Spidey's accuracy by creating mRNAs from the annotations on real sequences and then aligning those mRNAs back to the genomic sequence. Then, we used Spidey to align 11,640 RefSeqs (Pruitt et al. 2000) to the human genome assembly. We tested Spidey's ability to discriminate between related sequences by aligning mRNAs from gene clusters to the corresponding genomic sequences. Finally, we aligned a set of mouse mRNAs to their orthologous human genomic sequences. We used two other spliced alignment programs, sim4 and est2genome, in two of these tests to compare Spidey to published alignment tools. While sim4 was marginally faster than Spidey, Spidey consistently gave more reliable results. est2genome did perform better than Spidey in one of the tests, but its long running time makes its use impractical .
Spidey is available as a standalone program and as a Web service, both of which are accessible at http://www.ncbi.nlm.nih.gov/spidey.
RESULTS
Aligning to RefSeqs
We compared Spidey to two other popular spliced alignment tools, sim4 (Florea et al. 1998) and est2genome (Mott 1997).
We used genomic records corresponding to RefSeqs; each genomic record had one or more annotated mRNAs and CDSs. mRNAs were extracted from the annotations on the genomic sequence and then aligned back to the sequences from which they were derived. The mRNAs are therefore 100% identical to their parent genomic sequences, but are still real examples of spliced sequences. Starting from 941 genomic records, we found 368 records that had annotated mRNAs, from which we extracted 646 mRNA sequences. These mRNAs had 3915 exons. Of these exons, 4.4% were <51 bases long, 24.5% had 51–100 bases, 27.9% had 101–150 bases, 17.7% had 151–200 bases, 7% had 201–250 bases, and 18.1% had >250 bases.
All three programs were run with default parameters; Table 1 shows the results.
Table 1.
Results of Aligning Annotated mRNAs to RefSeqs
| No. of exons predicted | No. of exons correct | False negatives | True positives | |
|---|---|---|---|---|
| Spidey | 3926 | 3873 | 42/3915 (1.1%) | 3873/3926 (98.7%) |
| sim4 | 3909 | 3827 | 88/3915 (2.3%) | 3827/3909 (97.9%) |
| est2genome | 3716 | 3621 | 294/3915 (7.5%) | 3621/3716 (97.4%) |
For the 3915 annotated exons, Spidey's alignments produced 3926 exons, of which 3873 were correct, giving a true positive rate (the percentage of exons predicted that were correct) of 98.7% and a false negative rate (the percentage of annotated exons that were missed) of 1.1%. sim4 got 3827 of its 3909 predicted exons correct, for a 97.9% true positive rate and a 2.3% false negative rate. est2genome did slightly worse, as 3621 of its 3716 predicted exons were correct, a true positive rate of 97.4% and a false negative rate of 7.5%.
Spidey split several of the annotated exons into two or more exons separated by short introns, to produce more than the actual number of exons. Of the 42 annotated exons that Spidey did not find, 39 had no good splice sites at either the donor or acceptor splice junctions, so Spidey could not place those junctions unambiguously. The RefSeq splice junctions are determined by a variety of methods, including experimental work and sequence alignment, so it is not clear whether the junctions are noncanonical splice sites or just annotated incorrectly. Two exons were missed because there were sequences only four bases from the true splice sites that were scored higher than the annotated splice sites by the probability matrix in Spidey. The final exon was lost due to a probable misannotation; the exon is only five bases long and is purportedly 70 kb from the rest of the exons of the gene.
Alignments to the Human Genome Assembly
We decided to test Spidey on real-world data by aligning >11,000 reference sequences to the NCBI human genome assembly based on the April 1, 2001, data freeze.
Starting with 11,640 RefSeqs, we generated alignments between the RefSeqs and the genomic contigs as described in Methods. We obtained 7846 models that fit our criteria; namely, at least 90% of the mRNA must be aligned, with the worst exon having 95% identity or greater and with at least one exon at 99% identity or greater. Of those models, 7664 represented unique mRNAs (in the original set, a single mRNA could have more than one alignment). In this experiment, we found alignments for only 68% of the RefSeqs. Using less stringent criteria, we get only slightly more models; at lower cutoffs, the models start to accumulate internal gaps (and are excluded).
We looked at the alignments in the 7846 models to determine how often Spidey had found canonical splice sites. Only 2% of the models (184 alignments) were missing one or more splice sites.
We also looked at the RefSeqs that aligned to multiple contigs. Most RefSeqs (7506) aligned to only one contig, but 138 aligned to two contigs, 18 aligned to three contigs, none aligned to four contigs, and two aligned to five contigs. Of the 158 RefSeqs that align to multiple contigs, 46 have all their alignments on one chromosome and 112 align to two or more chromosomes. In 20 cases one of the alignments has a multi-exon gene structure; the rest have only one exon, possibly indicating the presence of pseudogenes. Four RefSeqs each align to two contigs such that both alignments have exactly the same structure and exactly the same percent identity, even in the untranslated regions. These may be cases of artificial duplication within the genome assembly.
We ran these alignments using Spidey's automatic CDS extraction function, which computes the CDS alignment from GenBank annotation and the mRNA alignment (see Methods). This feature is not found in sim4 or est2genome. Using these programs, one would have to generate a separate sequence for the CDS and run that separately. Although our analysis only considers the mRNA alignments, we also generated a CDS alignment for each model. We found, to our interest, that the first exon of the CDS alignment was often quite small; 161 of the 7846 models have an initial exon with length less than seven nucleotides. These small exons would not have been aligned correctly had we just run the CDS like an mRNA sequence (the exons are shorter than the minimum BLAST wordsize); only with the extraction function can we get them correct. This has implications for those who wish to align only the CDS, as the alignment may be incorrect if the CDS is aligned as a separate sequence. This also has interesting biological implications, as the first two codons are often separated from the rest of the coding sequence, meaning that splicing has to occur correctly for the translation start signal to be joined with the rest of the protein.
Avoiding Entanglement in Gene Clusters
Integral to Spidey's design is the idea that the program must be able to align an mRNA to its correct genomic position in spite of nearby paralogs. We tested whether we had succeeded in this goal by aligning mRNAs from gene clusters back to their parent genomic sequences.
We looked at four separate gene clusters: intracellular adhesion molecules ICAM1, ICAM3, ICAM4, and ICAM5 on chromosome 19; defensins DEFA3, DEFA4, DEFA5, DEFA6, and DEFB1 on chromosome 11; the hemoglobins HBB, HBD, HBG2, and HBE1 on chromosome 11; and DAZ, DAZ2, and DAZ3 on chromosome Y. We also looked at a region on chromosome 5 that has duplications and attempted to align the mRNA for BIRC1 (which should be in this region) to its genomic locus. Table 2 gives the accession numbers for the sequences used.
Table 2.
Accession Numbers for Sequences in Gene Clusters
| Genomic accession | mRNAs | No. of exons |
|---|---|---|
| NT_011130 | ICAM1: XM_008897 | 7 |
| ICAM3: XM_008894 | 7 | |
| ICAM4: XM_008896 | 3 | |
| ICAM5: XM_008895 | 11 | |
| NT_008268 | DEFA3: XM_005294 | 3 |
| DEFA4: XM_005295 | 3 | |
| DEFA5: XM_005293 | 2 | |
| DEFA6: XM_005296 | 2 | |
| DEFB1: XM_005297 | 2 | |
| NT_009409 | HBB: XM_006558 | 3 |
| HBD: XM_006557 | 3 | |
| HBG2: XM_006556 | 3 | |
| HBE1: XM_006555 | 3 | |
| AC006990 | DAZ:AF271087 | 17 |
| DAZ2:AF248480 | 11 | |
| DAZ3:AF248481 | 19 | |
| NT_006497 | BIRC1: XM_003847 | 17 |
These gene clusters consist of related genes in tandem across the chromosome. The related genes were on average 65–98% identical over 25–100% of their length.
Spidey was able to place correctly all 16 mRNAs from the gene clusters and all splice junctions were exactly correct when compared with the splice junctions annotated on the RefSeq records.
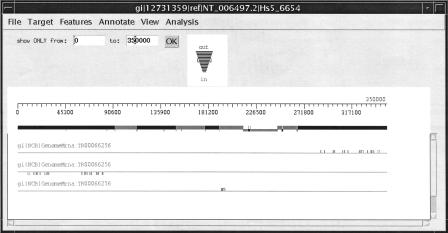
We ran Spidey in its multiple gene model mode when we ran BIRC1. This mode allows Spidey to return as many alignments as requested and the secondary models often elucidate the structure of the region being studied. For BIRC1 versus NT_006497, Spidey's top three models were all reasonable (Fig. 1). The first model had 17 exons, all 100% identical to the genomic sequence, covering 100% of the mRNA length. This is the actual BIRC1. The second model had 20 exons, 17 of which were 99.8% or more identical to the genomic sequence, and 3 exons with varying conservation, for an overall 99.7% identity and 89% coverage. This model is approximately 20 kb from the first. The third model sits between the first two and consists of 6 exons, four with 100% or higher identity and two >99.6% identity, for an overall 99.7% identity (but only 30% coverage). These two regions may be related to BIRC1 by duplication or other evolutionary events. The BIRC1 example emphasizes one of Spidey's strengths; namely, given no other information about a region, Spidey can generate useful sets of alignments that reflect the complex evolutionary history of the sequence. When using other alignment programs, one must first use local alignment tools to narrow down candidate regions and then do alignments in those regions.
Figure 1.
The top three Spidey models for the alignment of BIRC1 to its genomic sequence, as seen in Ingenue (a sequence and alignment workbench by F. Aklilu, unpubl.)
Alignments to Mouse
We chose to use mouse as the organism for our interspecies test. Mouse orthologs were found, as described in the Methods section, for 88 of the RefSeqs. We assumed that the mouse gene structure would be the same as the human gene structure in most cases. Therefore, we used the human annotation as our guide for scoring the mouse–human alignments.
Spidey, sim4, and est2genome were used to align the mouse mRNAs to the human genomic sequence. Spidey was run in its interspecies mode, whereas the other two programs were run with default parameters.
Table 3 shows the outcome on the mouse sequence. est2genome had the highest true positive and lowest false negative rates. Spidey's true positive rate is quite similar to est2genome's, but Spidey does miss more exons.
Table 3.
Results of Aligning Mouse mRNA Sequences to Human RefSeqs
| No. of exons predicted | No. of exons correct | False negatives | True positives | |
|---|---|---|---|---|
| Spidey | 361 | 294 | 57/351 (15.8%) | 294/361 (81.4%) |
| sim4 | 458 | 247 | 104/351 (29.6%) | 247/458 (53.9%) |
| est2genome | 383 | 334 | 17/351 (4.8%) | 334/383 (37.2%) |
Spidey missed 67 exons in the mouse–human comparison. Of these, 32 were missed because a higher-scoring splice site lay within the overlap of the adjacent exons, 15 were missed because the mouse sequence had insertions near the splice junction, 13 were missed because the mouse sequence was much longer or shorter than the human ortholog and appeared to have a slightly different intron/exon structure, all three exons of one mouse mRNA were incorrectly aligned to the wrong gene in a human gene cluster, and 4 exons were missed because they were each split into two exons in the alignment. Without further study, it is not clear how many of these errors are truly problems with the program and how many of them are simply real results of evolutionary divergence.
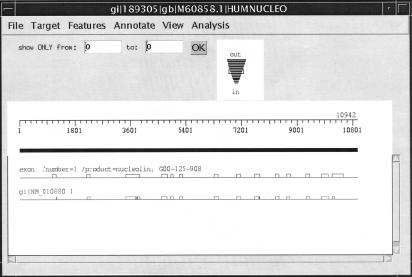
Figure 2 shows a typical orthologous mouse–human alignment, as well as the annotated gene on the human sequence. The human and mouse intron/exon structures are quite similar. A few exons are shifted slightly relative to the genomic sequence and a few others are split, but for the most part the two models are identical.
Figure 2.
Mouse NM_010880 (nucleolin) aligned to human M60858, shown in Ingenue with the annotated mRNA from the human sequence. The top line shows the human sequence, the center line represent the annotated human mRNA, and the bottom line shows the Spidey model for the mouse mRNA.
Time Considerations
Spidey runs quickly enough for practical use on a modern personal computer or workstation, even using contig-sized pieces of genomic DNA. To align a 5164 bp mRNA to a 1.03 Mb contig, Spidey took 14.43 sec on a workstation (a Sun Ultra-10 with a 300 MHz cpu and 192 megabytes of memory). To do the same task, sim4 took 2.01 sec and est2genome took 1 h 21 min.
To process 35 mRNAs against their parent RefSeqs on the same workstation, Spidey took 1 min 11 sec, sim4 took 25.3 sec, and est2genome took 2 h 56 min.
Running Spidey in multiple gene model mode does not increase the running time appreciably, as most of that time is spent in the initial BLAST run, which is done only once.
METHODS
Spidey — Design and Overview
Spidey is written in C and is incorporated in the NCBI Toolkit (Ostell 1996). It relies heavily on the alignment manager (Wheelan and Ostell, unpubl.), which is an indexing system used for easy management of and quick access to alignments and sets of alignments. Spidey was written with two main goals in mind, finding good alignments regardless of intron size and avoiding getting confused by nearby pseudogenes and paralogs. Toward the first goal, Spidey uses BLAST and DotView (another local alignment tool; F. Aklilu, unpubl.) to find its alignments. Because these are both local alignment tools, Spidey does not intrinsically favor shorter or longer introns and has no maximum intron size. To avoid including exons from paralogs and pseudogenes mistakenly, Spidey first defines windows on the genomic sequence and then performs the mRNA-to-genomic alignment separately within each window. Because of the way the windows are constructed, neighboring paralogs or pseudogenes should be in separate windows and should not be included in the final spliced alignment.
Initial Alignments and Construction of Genomic Windows
All parameters are default unless otherwise specified.
Spidey takes as input a single genomic sequence and a set of mRNA accession numbers or FASTA sequences. All processing is done one mRNA sequence at a time. The first step for each mRNA sequence is a high-stringency BLAST (e = 10−6, masked at hash for low-complexity sequence and repeats) against the genomic sequence. The resulting hits are analyzed to find the genomic windows.
The BLAST alignments are sorted by score and then assigned into windows by a recursive function which takes the first alignment and then goes down the alignment list to find all alignments that are consistent with the first (same strand of mRNA, both the mRNA and genomic coordinates are nonoverlapping and linearly consistent). On subsequent passes, the remaining alignments are examined and put into their own nonoverlapping, consistent windows, until no alignments are left. All windows are retained at this point and they go on to the next step one by one until the requested number of models has been generated. Because the windows are nonoverlapping, and because each window should contain most of a gene model, Spidey is able to generate accurate models without mixing up exons from adjacent genes.
Aligning in Each Window
Once the genomic windows are constructed, the initial BLAST alignments are freed and another BLAST search is performed, this time with the entire mRNA against the genomic region defined by the window and at a lower stringency (e = 10−3, masked at hash for low-complexity sequence and repeats) than the initial search. Spidey then uses a greedy algorithm to generate a high-scoring, nonoverlapping subset of the alignments from the second BLAST search. This consistent set is analyzed carefully to make sure that the entire mRNA sequence is covered by the alignments. When gaps are found between the alignments, the appropriate region of genomic sequence is searched against the missing mRNA, first using a very low-stringency BLAST (e = 1) and, if the BLAST fails to find a hit, using DotView functions to locate the alignment. When gaps are found at the ends of the alignments, the BLAST and DotView searches are allowed to extend past the boundaries of the window. If the 3′ end of the mRNA does not align completely, it is examined first for the presence of a poly(A) tail. No attempt is made to align the portion of the mRNA that seems to be a poly(A) tail. Sometimes there is a poly(A) tail that does align to the genomic sequence and these are noted because they indicate the possibility of a pseudogene.
Spidey looks through the alignments now present to determine whether two alignments are close enough together on the mRNA and on the genomic sequence that they should be merged. After this check, each alignment should correspond to one exon. Now, the boundaries of the alignments are adjusted so that the alignments abut each other precisely and so that they are adjacent to good splice donor and acceptor sites. Commonly, two adjacent exons' alignments overlap by as much as 10 or 15 bp on the mRNA sequence, meaning that a part of the mRNA sequence is duplicated identically in the genomic sequence surrounding the intron-exon boundaries (Box 2) . If two adjacent exons do not overlap, but in fact “underlap” (a few bases are missing between them), a simple alignment algorithm is used to extend each alignment so that the two alignments will overlap. The true exon boundary may lie anywhere within the overlap. To position the exon boundaries, the overlap is examined for splice donor sites, using functions that have different splice matrices depending on the organism chosen. The top few splice donor sites (by score) are then evaluated as to how much they affect the original alignment boundaries. The site that affects the boundaries the least is chosen (so as to retain as much as possible of the original alignments) and is evaluated as to the presence of an acceptor site. The alignments are truncated or extended as necessary so that they terminate at the splice donor site and so that they do not overlap.
Final Result
The windows are examined carefully to get the percent identity per exon, the number of gaps per exon, the overall percent identity, the percent coverage of the mRNA, presence of an aligning or non-aligning poly(A) tail, number of splice donor sites, the presence or absence of splice donor and acceptor sites for each exon, and the occurrence of an mRNA that has a 5′ or 3′ end (or both) that does not align to the genomic sequence. If the overall percent identity and percent length coverage are above the user-defined cutoffs, a summary report is printed and, if requested, a text alignment showing identities and mismatches is also printed.
Interspecies Alignments
Spidey is capable of performing interspecies alignments. The major differences in interspecies alignments are that the mRNA–genomic identity will not be close to 100% as it is in intraspecies alignments and that the alignments have numerous and lengthy gaps. If Spidey is used in its normal mode to do interspecies alignments, it produces gene models with many, many short exons. When the interspecies flag is set, Spidey uses different BLAST parameters (gap_open 5, gap_extend 1, mismatch penalty −1, and gap_x_dropoff_final 100) to encourage longer and more gaps and to not penalize as heavily for mismatches. This way, the alignments for the exons are much longer and more closely approximate the actual gene structure.
Extracting CDS Alignments
When Spidey is run in network-aware mode or when ASN.1 files are used for the mRNA records, it is capable of extracting a CDS alignment from an mRNA alignment and printing the CDS information also. Since the CDS alignment is just a subset of the mRNA alignment, it is relatively straightforward to truncate the exon alignments as necessary and to generate a CDS alignment. Furthermore, the untranslated regions are now defined, so Spidey calculates the percent identity for the 5′ and 3′ untranslated regions.
Obtaining Mouse Orthologs For Human RefSeqs
Mouse orthologs were obtained for the human RefSeqs in two ways. First, orthologous pairs determined at the Jackson Laboratory were obtained by FTP. Next, orthologous pairs were computed at NCBI using a reciprocal best hit scheme; mouse and human non-EST mRNA sequences found in LocusLink were searched against each other, mouse sequences against all human sequences as a database, and then human sequence queries against all mouse sequences. Pairs in which the mouse sequence's top human hit had the mouse sequence as its top hit were retained as orthologs.
Aligning RefSeqs to Genomic Contigs
Starting with 11,640 RefSeqs, we performed a MEGABLAST search with each RefSeq against all the contigs. A hit was accepted if it covered a minimum of 75 bases at 97% identity. This generated a list of all potential mRNAs for each contig. Spidey was run on each contig, given these lists as input. Spidey alignments were accepted when they covered at least 90% of the mRNA, had at least one exon with 99% or higher identity, and had no exons below 95% identity.
User Options
When run as an executable, Spidey has many user-controlled parameters that can tune its performance as needed. The E values for the first (stringent), second (less stringent), and third (very relaxed) BLAST searches are changeable. Also, the user can specify that the program return multiple gene models, can set a minimum percent identity or percent length coverage cutoff, can submit mRNAs with lowercase masking, and can request that the program fetch the coding sequence for the mRNA given and compute the alignments for that sequence also.
Options available from both the Web page and the executable include interspecies comparisons as well as various output options, notably a multiple alignment option (if more than one mRNA is submitted).
Table.
Box 1.
| —SPIDEY version 0.81— |
| Genomic: IcI|Hs19_25288_22:NT_025132/chr=19/len=283034, 283034 bp |
| CDS: gi|8923093|ref|NM_017660.1|Homo sapiens hypothetical protein FLJ20085 (FLJ20085), mRNA, 62–655 |
| Strand: minus |
| Number of exons: 4 |
| Exon 1: 219519-219710 (gen) 0–191 (CDS) id 100.0% gaps 0 splice site (d a): 1 0 |
| Exon 2: 218892-218966 (gen) 192–266 (CDS) id 100.0% gaps 0 splice site (d a): 1 1 |
| Exon 3: 218409-218604 (gen) 267–462 (CDS) id 100.0% gaps 0 splice site (d a): 1 1 |
| Exon 4: 215461-215591 (gen) 463–593 (CDS) id 100.0% gaps 0 splice site (d a): 0 1 |
| Number of splice sites: 3 |
| CDS coverage: 100% |
| overall percent identity: 100.0% |
| Missing DCS ends: neither |
| 5′ UTR id 98.4% |
| 3′ UTR id 99.6% |
| Genomic : IcI|Hs19_25288_22:NT_025132/chr=19/len=283034, 283034 bp |
| mRNA: gi|8923093|ref|NM_017660.1|Homo sapiens hypothetical protein FLJ20085 (FLJ20085), mRNA, 2450 bp |
| Strand: minus |
| Number of exons: 4 |
| Exon 1: 219519-219772 (gen) 0–253 (mRNA) id 99.6% gaps 0 splice site (d a): 1 0 |
| Exon 2: 218892-218966 (gen) 254–328 (mRNA) id 100.0% gaps 0 splice site (d a):11 |
| Exon 3: 218409-218604 (gen) 329–524 (mRNA) id 100.0% gaps 0 splice site (d a):11 |
| Exon 4: 215461-215591 (gen) 463–593 (mRNA) id 99.6% gaps 3 splice site (d a): 0 1 |
| Number of splice sites: 3 |
| mRNA coverage: 100% |
| overall percent identity: 99.6% |
| Missing mRNA ends: neither |
| Aligning poly(A)+ tail length: 18 |
Sample Spidey Output (Summary Only) Showing Results for CDS and mRNA Alignment
Table.
Box 2.
| Genomic: gi\8134255|ref|NT_001128.7|Hs22_541 Homo sapiens 22q13.33 sequence |
| mRNA: gi|7662251|ref|NM_014678.1|Homo sapiens KIAA0685 gene product (KIAA0685), mRNA |
| Exon 7: (showing only the 3′ boundary) |
| // AGACCCAGGTGCGGGGCC |
| ∥∥∥∥ |
| // AGACCCAGGT |
| exon →|← intron |
| Exon 8: (showing only the 5′ boundary) |
| TCTCCCCCAGGTTTGGA// |
| ∥∥∥| |
| CCCAGGTTTGGA // |
| intron →|← exon |
Typical Overlap between Adjacent Exons Before Processing
The sequence in italics is part of the mRNA, and is shown twice to emphasize the ambiguity fo the location of the intron–exon boundaries from alignments alone.
Acknowledgments
We thank David Lipman for many helpful discussions, Jonathan Kans for programming advice, Greg Schuler for the idea of aligning in windows, Donna Maglott for helpful advice on choosing gene clusters, and Kim Pruitt and Bhanu Rajput for their wonderful comments and suggestions for the Web version of Spidey. We would also like to thank Chris Burge for sharing his consensus splice site data and Fasika Aklilu for her help with figures.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL wheelan@ncbi.nlm.nih.gov; FAX (301) 480-2288.
Article published on-line before print: Genome Res., 10.1101/gr.195301.
Article and publication are at http://www.genome.org/cgi/doi/10.1101/gr.195301.
REFERENCES
- Florea L, Hartzell G, Zhang Z, Rubin G, Miller W. A computer program for aligning a cDNA sequence with a senomic DNA sequence. Genome Res. 1998;8:967–974. doi: 10.1101/gr.8.9.967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The Genome Sequencing Consortium. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- Mott R. EST_GENOME: A program to align spliced DNA sequences to unspliced genomic DNA. CABIOS. 1997;13:477–478. doi: 10.1093/bioinformatics/13.4.477. [DOI] [PubMed] [Google Scholar]
- Ostell JM. The NCBI software tools. In: Bishop M, Rawlings C, editors. Nucleic Acid and Protein Analysis: A Practical Approach. Oxford: IRL Press; 1996. pp. 31–43. [Google Scholar]
- Pruitt KD, Katz KS, Sicotte H, Maglott D. Introducing RefSeq and LocusLink: Curated human genome resources at the NCBI. Trends Genet. 2000;16:44–47. doi: 10.1016/s0168-9525(99)01882-x. [DOI] [PubMed] [Google Scholar]