Abstract
Two clusters of rat Nkrp1 genes can be distinguished based on phylogenetic relationships and functional characteristics. The proximal (centromeric) cluster encodes the well-studied NKR-P1A and NKR-P1B receptors and the distal cluster, the largely uncharacterized, NKR-P1F and NKR-P1G receptors. The inhibitory NKR-P1G receptor is expressed only by the Ly49s3+ NK cell subset as detected by RT-PCR, while the activating NKR-P1F receptor is detected in both Ly49s3+ and NKR-P1B+ NK cells. The mouse NKR-P1G ortholog is expressed by both NKR-P1D− and NKR-P1D+ NK cells in C57BL/6 mice. The rat and mouse NKR-P1F and NKR-P1G receptors demonstrate a striking, cross-species conservation of specificity for Clr ligands. NKR-P1F and NKR-P1G reporter cells reacted with overlapping panels of tumour cell lines and with cells transiently transfected with rat Clr2, Clr3, Clr4, Clr6 and Clr7 and mouse Clrc, Clrf, Clrg and Clrd/x, but not with Clr11 or Clrb, which serve as ligands for NKR-P1 from the proximal cluster. These data suggest that the conserved NKR-P1F and NKR-P1G receptors function as promiscuous receptors for a rapidly evolving family of Clr ligands in rodent NK cells.
Keywords: Comparative Immunology, Killer cell lectin-like receptor, NK cells, Rodent
Introduction
NK cells express several multi-gene families of killer cell lectin-like receptors (KLR) encoded by the NK gene complex (NKC), which is located on rat chromosome 4 and mouse chromosome 6 (Flornes et al. 2010; Yokoyama and Plougastel 2003). These include the NKR-P1, NKG2 and Ly49 families of receptors, encoded from the centromeric, central and telomeric parts of the NKC, respectively. Rat Ly49 molecules are reactive with MHC molecules from the classical class Ia (RT1-A) and non-classical class Ib (RT1-CE) regions (Naper et al. 1999, 2005). In the mouse, NKG2 together with the partner CD94 bind the class Ib molecule Qa1b (Vance et al. 1998), and in the rat tetramers of RT.BM1, the rat ortholog of mouse Qa1b stain a subset of rat NK cells low in Ly49 receptors but enriched for Nkg2a/c expression (Kveberg et al. 2010).
The NKR-P1 receptors were among the first KLRs to be discovered. Only in recent years were they shown to bind members of another NKC family, the C-type lectin-related (Clr) molecules (Carlyle et al. 2004; Iizuka et al. 2003). Four functional NKR-P1 molecules exist in Brown Norway (BN) strain rats, which are encoded by two pairs of genes located in the proximal (centromeric) part of the NKC and separated by a stretch of approximately 500 kb harbouring seven full-length Clr genes (Kveberg et al. 2009). The most centromeric cluster encodes the well-characterized NKR-P1A and NKR-P1B receptors. NKR-P1A is an activation receptor that has been used as the prototypic marker for rat NK cells, and it is also expressed by some T cells (Appasamy et al. 1996; Giorda et al. 1990). NKR-P1B is an inhibitory receptor that exists in two divergent variants (Kveberg et al. 2009; Voigt et al. 2007). The PVG strain allele is expressed by a major subset of NK cells expressing few Ly49 genes but enriched for Nkg2a/c transcripts. NKR-P1B+ NK cells are fully functional but are less responsive to IL-2 stimulation than the complementary population of Ly49s3+ (Ly49 stimulatory receptor 3) NK cells and are unable to mediate alloreactivity against normal lymphoblast target cells (Kveberg et al. 2006a, 2010). One of the NKR-P1B variants has been shown to mediate protection against cytomegalovirus (CMV) infection in vivo (Voigt et al. 2007). Both NKR-P1A and NKR-P1B bind the most distal Clr member, Clr11, and ligation leads to either activation or inhibition of NK cell cytotoxicity (Kveberg et al. 2006a, 2009; Voigt et al. 2007).
Much less is known about the telomeric Nkrp1 cluster encoding the NKR-P1F and NKR-P1G receptors and their orthologs in mice. NKR-P1F and NKR-P1G react with an overlapping panel of Clr molecules, but not with Clr11 (Kveberg et al. 2009). NKR-P1F mRNA has been detected in NK cells and some NKR-P1A+ T cells (Appasamy et al. 1996). Its mouse equivalent (mNKR-P1F) has been reported to react with Clrg (Clec2i) (Iizuka et al. 2003) and Clrx (Aust et al. 2009), the latter being encoded by a gene that maps in the same position as a fragment previously named Clrd (Plougastel et al. 2001), also known as Clec2g and Ocilrp1. mNKR-P1F is expressed by all NK cells, but not by T cells, in C57BL/6 mice as judged by staining with a novel mAb (Aust et al. 2009). Its activation potential is questionable, however, since it does not enhance cytotoxicity or IFN-γ production upon cross-linking. mNKR-P1G remains uncharacterized. It was considered a pseudogene (Carlyle et al. 2006), but we isolated a full-length cDNA clone based on its homology with the rat gene (accession number DQ113419). In the present study, we show that the NKR-P1F and NKR-P1G receptors are xenoreactive, with closely related expression profiles and Clr ligand-binding characteristics in mice and rats.
Materials and methods
Cell lines and animals
Animals were housed in compliance with institutional guidelines and sacrificed at 8–12 weeks of age. PVG-RT7 b rats possess a CD45 allotype (RT7.2) but are otherwise used interchangeably with the standard PVG strain (RT7.1) and were maintained at the Institute of Basic Medical Sciences, University of Oslo. Albino Oxford (AO) rats were from Harlan U.K., C57Bl/6 mice from Taconic (Denmark) and Harlan Olac (U.K.), and BALB/c/Sca mice were from NOVA SCB. The following cell lines were used: cc531s (rat colon carcinoma), RNK-16 (rat NK cell leukaemia), R2 (rat macrophage), YB2/0 (rat B cell myeloma), NS0 (mouse myeloma), EL-4 and YAC-1 (mouse T cell lymphomas), P388.D1 (mouse macrophage-like lymphoma), P815 (mouse mastocytoma), Ltk (mouse fibroblasts), RMA-S (mouse lymphoma with antigen-processing defect), RAW 264.7 (mouse leukemic monocyte macrophage cell line) and 293T (human kidney fibroblast). Cell lines were maintained in complete RPMI (cRPMI; RPMI 1640 supplemented with 25 mM HEPES, 10% FBS, 1 mM MEM sodium pyruvate, 5 × 10−5 M 2-ME and antibiotics). The BWN3G reporter cell line (Kveberg et al. 2009) was maintained in cRPMI supplemented with 0.5 mg/mL of hygromycin B (Invitrogen, CA).
Semi-quantitative PCR
cDNA was generated from NKR-P1B+Ly49s3− and NKR-P1B−Ly49s3+ single-positive CD3−NKR-P1A+ NK cells from spleens of PVG-RT7 b rats and from NKR-P1D+ and NKR-P1D− splenic NK cells (CD3−NKR-P1C+) from C57BL/6 mice, obtained by sorting on a FACSAria by Trizol extraction, MMLV reverse transcriptase (Promega) and random hexamer primers (Fermentas). Semi-quantitative polymerase chain reaction (PCR) on three tenfold dilutions of cDNA was performed by GoTaq polymerase (Promega), using the following forward (F) and reverse (R) primers: rat Nkrp1a, 5′-CCTCAGTTTAAAGCCATCCAAGAC-3′ (F) and 5′-CACTCGGCATGGCTCCACTGAT-3′ (R); rat Nkrp1b, 5′-AGCACAAGTCACCTCCATCTCT-3′ (F) and 5′-TCCAAAATGATCTGTCTTTCCCCT-3′ (R); rat Nkrp1f, 5′-AAGAGTCTATGGTAATGTAAAGACA-3′ (F) and 5′-CATTCTAGTATGGCTGATCTCC-3′ (R); rat Nkrp1g, 5′-TGCACCAGTGCTCTATGCTGAATT-3′ (F) and 5′-ATTCCAGTGCAAATGCCAATCTCT-3′ (R); rat Cd45, 5′-CGGGGTTGTTCTGTGCTCTGTTC-3′ (F) and 5′-CTTTGCTGTCTTCCTGGGCTTTGT-3′ (R); mNkrp1a, 5′-GCCTGTCGGTGCCCACGTTC-3′ (F) and 5′-ACCCACATAGTTGCTCAGGGTTTCA-3′ (R); mNkrp1c, 5′-GCCTGTCGGTGCCCACGTTC-3′ (F) and 5′-AGATGGAGGCACAGCTGCCA-3′ (R); mNkrp1d, 5′-CCCTCGCTGGCATCGGTTGG-3′ (F) and 5′-TGGCGGCACAGCTGCCATTT-3′ (R); mNkrp1f, 5′-TCAGCCATACTAGAGTGCCCGAG-3′ (F) and 5′-TGCCTCTTGCTGGTCTGGGCT-3′ (R); mNkrp1g, 5′-GCTGGCATCGGGTGGCTCTG-3′ (F) and 5′-CAGACGTGTTTCAGTGTCTTTTGGCA-3′ (R); mCd45, 5′-AAGAGGAAGGAGCCCAGAACTGT-3′ (F) and 5′-GCCGTGCTTTGCGTAGAGAC-3′ (R). For real-time PCR, the following primers and probes were used: mNkrp1g 5′-GACTGTAATTTGAAAGGAGCCAC-3′ (F), 5′-CCATTCATCCACTTCCAAACC-3′ (R) and 5′FAM-TGCAGAACTTCTCAAAGGGAAAAGGACA-3′TAM (probe); mPmca4 5′-AGATGTCGGGTTTGCTATGG-3′ (F), 5′-GAACTGTAGGAACTTGGAAATGC-3′ (R) and 5′FAM-TGACCGATGACAACTTCACCAGCA-3′TAM (probe); mCd45 5′-GACAGAGTTAGTGAATGGAGACC-3′ (F), 5′-AAAAGTTCGGAGAGTGTAGGC-3′ (R) and 5′FAM-TCTGCTTTCCTTCGCCCCAGT-3′TAM (probe).
cDNA cloning, chimeric receptor constructs and transfections
The extracellular and transmembrane domains of mNKR-P1G (DQ113419) and parts of the intracellular (24 aa), the transmembrane and extracellular domains of mNKR-P1F (BALB/c, identical to DQ237931) were amplified and inserted into the EMCV.SRα and pMX vectors, respectively, both containing an inverted intracellular part of mouse CD3ζ and a 3′-terminal FLAG-tag. The following primers were used: mNkrp1f, 5′-GACTCGAGGCATCTTCTCCATCTTTCTCC-3′ (F) and 5′-GATCTAGAGACATGTATCAGGGTCTTTTG (R); mNkrp1g, 5′-GCATTCTAGACTCGGCTGTGCT-3′ (F) and 5′-GCATTCTAGAGACGTGTTTCAGTGTCT-3′ (R). Transfection with EMCV.SRα was performed as previously detailed (Ryan et al. 2000), using 20 μg of ScaI-linearized construct and a Bio-Rad GenePulser Xcell (120 V, 950 μF, Ω = ∞) followed by selection in 1 mg/ml of active G418 sulphate. A virus transduction protocol was used for pMX to improve transfection efficiency. Plat-E cells were transfected with 2 μg mNKR-P1F cDNA in a pMX vector and 1 μg pCL-Eco plasmid with FuGENE 6 (Roche, manufacturer’s protocol). Virus-containing supernatants were harvested and filtrated (0.2-μm filter) on day 4, and 1.5 ml was transferred to 105 BWN3G cells in 0.5 ml RPMI 1640 supplemented with polybrene (Sigma; to a final concentration of 8 μg/ml). Cells were spin-infected at 2,000 rpm at 32°C for 2 h, incubated for 2 h at 37°C and 5% CO2, and then the transduction solution was replaced by 2 ml cRPMI followed by selection in puromycine.
The 293T cells were transiently transfected with cloned mouse Clr constructs inserted into the EMCV.SRα vector with an external FLAG tag (except Clrd/x which was used without a tag in pMX) and with the previously published rat Clr constructs, including Clr10 inserted into a pBK-CMV vector containing an internal EYFP-encoding sequence (Kveberg et al. 2009). Clr10 contains three alternative start codons, and the shortest version was used in these experiments. Transfection was performed by FuGENE 6 24 h prior to co-incubation with reporter cells. Surface expression of transfected molecules was tested by flow cytometry for EYFP or FLAG expression using mAb M2 (Sigma). The presence of an extracellular FLAG epitope on rat Clr molecules (Clr2, Clr4, Clr7 and Clr11) did not affect ligand recognition by rat NKR-P1 receptors, as judged by a comparison with Clr molecules containing an intracellular EYFP tag instead (Kveberg et al. 2009).
Full-length mouse Clr and Nkrp1g clones were amplified from cDNA obtained from mononuclear BALB/c splenocytes by Trizol extraction, MMLV reverse transcriptase (Promega), and random hexamer primers (Fermentas), using Pwo SuperYield DNA polymerase (Roche) and the following forward (F) and reverse (R) primers, restriction sites underlined: mClrb (HQ875062), 5′-GCATCTCGAGTCTGAGATGTGTGTCAC-3′ (F) and 5′-GGCCTCTAGAGGAAGGAAAAAAAGGAG-3′ (R); mClrc (HQ875060), 5′-CCCTCGAGATGACAGCCTCACAG-3′ (F) and 5′-CCTCTAGAGCACTGTAACATATAGTTGC-3′ (R); mClrf (HQ875061), 5′-GCATCTCGAGTCACAGATGAATGCTGCAA-3′ (F) and 5′-GGCCTCTAGACACATGCCTTGGTAC-3′ (R); mClrg (identical to NM_020257), 5′-GCATCTCGAGCCTCCCATGCCAGATTG-3′ (F) and 5′-GCATTCTAGAGACAGGAGGAGTTTGGCA (R); mNkrp1g (JF429832), 5′-TCTCCCATCTCTAATGGATGC-3′ (F) and 5′-CAGATCTGGCGTGAATAGTGA-3′ (R). Clrd/x was previously cloned from mouse C57BL/6 spleen cells (Aust et al. 2009). Clra (identical to AF320600S1) was a generous gift from NOVO Nordic and was amplified from vector with the following primers: 5′-GACTCGAGACCATGCTGAAAACCGACCTGA-3′ (F) and 5′-AGTCTAGAGCCAGGGTTCACGATCAGAG-3′ (R).
Reporter cell stimulation
Reporter cells, 5 × 104, and 5 × 104 stimulator cells were resuspended in RPMI 1640 supplemented with HEPES, 5% FBS, antibiotics and 2 ng PMA/mL, spun down and incubated overnight at 37°C in round-bottomed, 96-well microtiter plates (100 μL/well). PMA (2 ng/ml) was added to the medium to enhance the specific reporter cell signal without increasing the non-specific background signal. Duplicate samples were mixed into one tube and analysed for induction of enhanced green fluorescent protein (EGFP) expression by flow cytometry (FACSCalibur, Beckton Dickinson). All target cells were excluded from the analysis by pre-labelling with SNARF-1 (Molecular Probes, Invitrogen) according to the manufacturer’s protocol.
Results and discussion
Nkrp1g is mainly expressed by the Ly49s3+NKR-P1B−NK cell subset in rats
Previous studies in PVG strain rats have demonstrated that the majority of splenic NK cells are comprised of two complementary subsets of NK cells, one expressing the NKR-P1B receptor and the other the Ly49s3 receptor (Kveberg et al. 2006a). cDNA was retrieved from sorted, single-positive NKR-P1B+Ly49s3− and Ly49s3+NKR-P1B− NK cells (NKR-P1A+CD3−), and the distribution of various Nkrp1 transcripts was evaluated by RT-PCR using locus-specific primers. Nkrp1b was mainly detected in the NKR-P1B+ cells, confirming the purity of the sorted cells. A striking finding was the almost exclusive expression of Nkrp1g in the Ly49s3+ NK subset. This non-overlapping expression pattern of Nkrp1b and Nkrp1g was confirmed by staining with a novel NKR-P1G-specific mAb (unpublished results) and is not specific for the PVG strain as it was also observed in AO strain rats. AO is closely related to PVG in the NKC and expresses the same allelic variant of Nkrp1b, which is stained by mAb STOK27 (Kveberg et al. 2006a, b). Nkrp1f, on the other hand, was expressed in both the NKR-P1B+ and Ly49s3+ subsets, but with a slight skewing towards the latter (Fig. 1a). The data indicate that the two inhibitory NKR-P1 receptors, i.e. NKR-P1B and NKR-P1G are expressed by two complementary subsets of rat NK cells, with NKR-P1G expression being concentrated in the Ly49s3+ NK subset. In contrast, the activating NKR-P1A and NKR-P1F receptors are expressed by most rat NK cells across the two subsets.
Fig. 1.
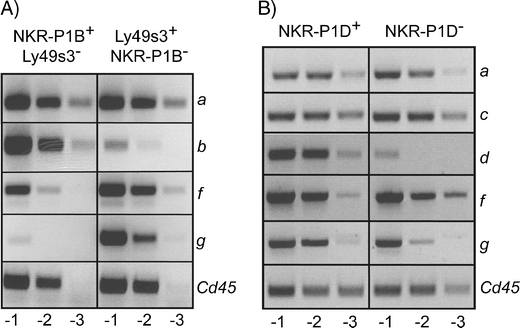
Expression of Nkrp1f and Nkrp1g in NK cell subsets in PVG-RT7 b rats and C57BL/6 mice. a Sorted single-positive NKR-P1B+Ly49s3− and Ly49s3+NKR-P1B− NK cells (CD3−NKR-P1A+) from spleens of PVG-RT7 b rats were analysed for expression of the indicated Nkrp1 transcripts by semi-quantitative RT-PCR, performed at tenfold dilutions. Cd45 was used to normalize cDNA concentration from the two subsets. Note that expression of Nkrp1g is confined to the Ly49s3+ NK subset, while Nkrp1f is present in both subsets. b Sorted NKR-P1D+ and NKR-P1D− NK cells (CD3−NKR-P1C+) from spleens of C57BL/6 mice were analysed for expression of the indicated Nkrp1 transcripts. Cd45 was used to normalize cDNA concentration from the two subsets
Nkrp1g (mNkrp1g) is expressed by NKR-P1D− and NKR-P1D+ NK cells in C57BL/6 mice
The rat and mouse Nkrp1b genes appear to be functional homologs, and mNkrp1d is an allele of mNkrp1b (Carlyle et al. 2006). In both species, these receptors recognize the most telomeric Clr molecule, named Clrb in the mouse and Clr11 in the rat. NKR-P1D positive and negative NK cells (NKR-P1C+CD3−) were sorted from C57BL/6 spleen cells stained with the 2D9 anti-NKRP1D mAb (Aust et al. 2009). The purity of the sorted cells was >85% (data not shown). Nkrp1d was detected in the NKR-P1D+ fraction confirming the purity of the sorted cells, while Nkrp1a, Nkrp1c and Nkrp1f were equally expressed by both subsets. mNkrp1g was expressed by both NKR-P1D− and NKR-P1D+ cells, and the difference between the subsets appeared to be small, but with a slight skewing towards the NKR-P1D+ subset. By contrast, a real-time PCR analysis suggested that expression of mNkrp1g, when compared with Cd45 or the house keeping gene Pmca4, was slightly enhanced in the NKR-P1D− subset (data not shown), which would be concordant with the situation in the rat. Determination of its precise distribution on mouse NK cells will have to await the generation of a specific staining reagent.
Rat and mouse NKR-P1F and NKR-P1G reporter cells react with an overlapping panel of tumour target cells
We used the BWN3G reporter cell system which allows visualization of ligand binding by transcription of EGFP upon incubation with stimulator cells. During the course of these studies, we noted that the rat NKR-P1G reporter (Kveberg et al. 2009) reacted strongly with mouse stimulator cells. To systematically evaluate xenoreactivity by NKR-P1F and NKR-P1G, we generated FLAG-tagged reporters also for the mouse equivalents. Surface expression of NKR-P1 molecules on reporter cells is shown in Fig. 2a. A collection of rat and mouse tumour cell lines was tested for their stimulatory properties. Both rat and mouse NKR-P1F and G reporters were promiscuous and reacted with most rat target cells (Fig. 2b), with the exception of the NK cell line RNK16 which was recognized mainly by the NKR-P1F reporters. This might be related to the fact that RNK-16 cells express the strong NKR-P1F ligand Clr4 (Flornes et al. 2010).
Fig. 2.
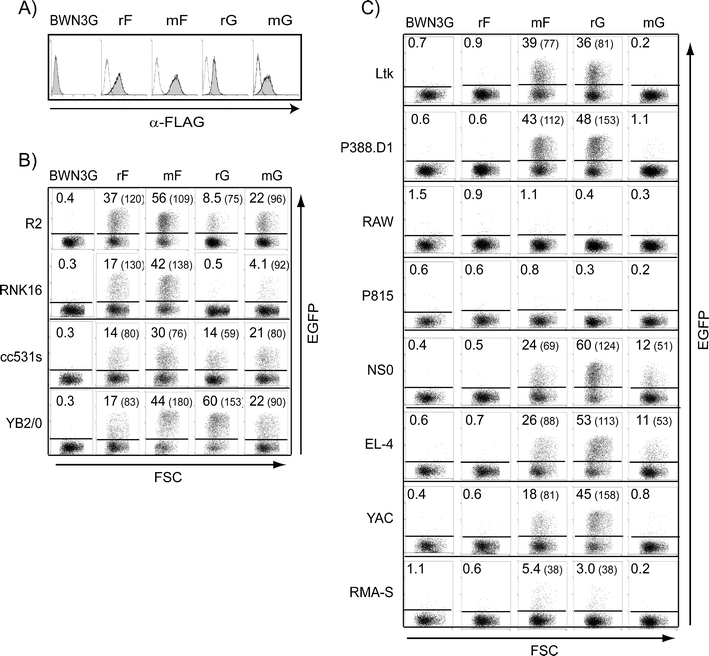
Rat and mouse NKR-P1F and NKR-P1G reporters show overlapping specificities against a panel of tumour target cells. a Expression of FLAG-tagged chimeric molecules was detected by staining with the anti-FLAG mAb, M2 (shaded histograms). Open histograms show control staining with only secondary antibody. Reporter cells (r rat and m mouse) were co-incubated overnight with rat b or mouse c tumour cell lines. Ligand-induced stimulation of chimeric receptors was measured as an EGFP signal by flow cytometry. Numbers represent percentage and MFI values (in parenthesis) of positive cells. One out of three representative experiments is shown
In general, the mouse reporters responded more firmly to rat stimulator cells than their rat counterparts. The opposite was the case when using NKR-P1G reporters and mouse target cells (Fig. 2c). While mNKR-P1G responded only to NS0 and EL-4, rat NKR-P1G showed robust responses to Ltk, P388.D1, NS0, EL-4 and YAC and a weak response to RMA-S. mNKR-P1F reacted with the same target cells as rat NKR-P1G. While it is difficult to correlate reporter stimulation with expression of individual Clrs, the redundancy in target cell recognition corresponds well with the promiscuity in Clr ligand binding observed below. Rat NKR-P1F did not react with any of the mouse stimulators but responded well with mouse Clrc and Clrg when expressed in 293T cells (see below). This suggests that endogenous Clr ligands must be expressed at a sufficiently high level for efficient reporter stimulation, and that this threshold was not reached by the mouse tumour cells in combination with the rat NKR-P1F reporter.
The orthologous NKR-P1F and NKR-P1G receptors in rats and mice show a striking conservation of specificity for Clr ligands
Mouse Clr molecules were transiently expressed in 293T cells and tested for their stimulatory properties, together with the rat Clr transfectants previously described, including Clr10 (Kveberg et al. 2009). Our analysis included six of the seven functional Clr genes existing in the C57BL/6 genome, including Clrc which has not been previously isolated at the cDNA level, corresponding with the predicted mRNA of the Clec2f gene. A schematic representation of this part of the mouse NKC and a conversion table of the Clr and Clec2 nomenclature are given in Fig. 3a, b. Surface expression of FLAG-tagged mouse Clr molecules was verified by staining with an anti-FLAG (Fig. 3c). While the Clrg and Clrd/x transfectants induced brisk EGFP responses in both the rat and mouse NKR-P1F and NKR-P1G reporters, Clrc and Clrf specifically stimulated the NKR-P1F and NKR-P1G reporters, respectively (Fig. 3d). The results confirmed the previously reported interactions of mNKR-P1F with Clrg (Iizuka et al. 2003) and Clrd/x (Aust et al. 2009), and in addition, identified Clrc as a novel ligand for mNKR-P1F and Clrf, Clrg, and Clrd/x for NKR-P1G. It should be noted that Clrc, which is located next to NKR-P1F in the genome, is preferentially recognized by this receptor, and the same is true for Clrf and NKR-P1G. The lack of response towards Clrb was as expected since this molecule is the ligand of mNKR-P1B/D (Carlyle et al. 2004; Iizuka et al. 2003). Clra was not recognized by any of our reporter cell lines.
Fig. 3.
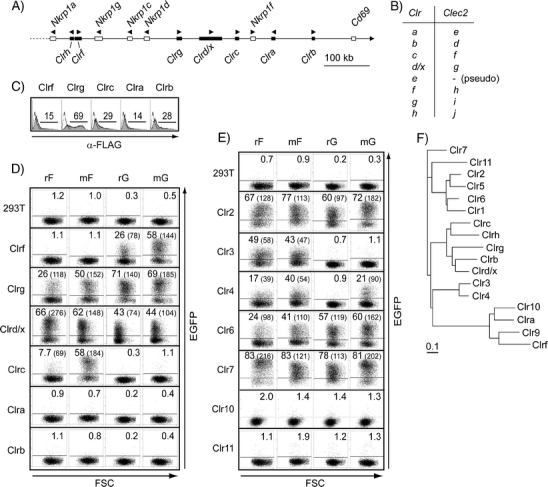
Rat and mouse NKR-P1F and NKR-P1G reporters show overlapping specificities against a panel of Clr transfectants. a Schematic map of the centromeric part of the mouse NKC based on a C57BL/6J chromosome 6 genomic contig (NT_039353.7). The distance from Nkrp1a to Clrb is approximately 577 k. Only functional genes are shown; the predicted mRNA sequence of Clrh has not been confirmed. b The Clr nomenclature has previously been confused by erroneously assembled genomic clones. A conversion table of the suggested Clr and Clec2 nomenclature is shown. c Surface expression of the FLAG-tagged mouse Clr molecules (filled histograms) was verified by staining with mAb M2 and flow cytometry, and staining of untransfected 293T control cells is shown as unfilled histograms. An untagged version of Clrd/x was tested in a separate set of experiments. Reporter cells (r rat and m mouse) were co-incubated overnight with 293T stimulator cells transiently transfected with the indicated d mouse and e rat Clr molecules and stimulation measured as an EGFP signal by flow cytometry. Numbers represent percentage and MFI values (in parenthesis) of positive cells. Transfection efficiency of the rat Clr constructs was verified by EYFP staining (data not shown). Clr10 was tested in a separate set of experiments. One representative out of three to five experiments is shown. f Phylogenetic tree based on the extracellular aa sequence of various mouse and rat Clr molecules, accession numbers given in parenthesis: Clr1 (NP_001019508), Clr2 (ABX54835), Clr3 (ACJ23591), Clr4 (ACJ23593), Clr5 (ABO15828), Clr6 (ACJ23590), Clr7 (ABX54838), Clr9 (XP_232399), Clr10 (ACJ23592), Clr11 (ABX54837), Clra (AAL37200), Clrb (AAK70357), Clrc (ADX42723), Clrd/x (also called ocilrp1, BAE23491), Clrf (AAK70358), Clrg (AAK70359), Clrh (XP_001480526)
Most rat Clr transfectants, with the exception of Clr10 and Clr11, induced a brisk EGFP signal in the rat and mouse NKR-P1F reporters (Fig. 3e). Stimulation of the NKR-P1G reporters was more restricted and limited to Clr2, Clr6 and Clr7 while Clr4 stimulated the mNKR-P1G reporter only. This difference is probably quantitative rather than qualitative, as Clr4 induced a weak positive response by rat NKR-P1G when using a more sensitive LacZ-based reporter cell line (data not shown).
Relatively low levels of expression were observed for Clrf, Clra and Clr10 in 293T cells (Fig. 3 and Kveberg et al. 2009), which constitute a separate phylogenetic clade with Clrf and Clra being orthologs of Clr9 and Clr10, respectively. Their low expression levels might be related to some structural properties or to expression programs common to this group. The fact that Clrf was effective in stimulating the NKR-P1G reporters suggests that the negative results obtained for the two orthologs Clra and Clr10 are related to conserved biological properties and not to limitations of the reporter assay (Fig. 3b). The result also predicts that Clr9 is recognized by NKR-P1G, but this assumption could not be tested as attempts to clone Clr9 have been unsuccessful.
The striking promiscuity in ligand recognition displayed by the xenoreactive NKR-P1F and NKR-P1G receptors suggests that the influence of allelic variation is of less importance. Variation appears to be very limited at the receptor level, with NKR-P1F being identical at the protein level between the C57BL/6 and BALB/c strains, while NKR-P1G shows a conservative substitution of its terminal amino acid only (Nkrp1g from BALB/c amplified as specified in “Materials and methods”). No allelic differences have, to date, been observed for the rat NKR-P1F and NKR-P1G receptors, with identical sequences being derived from NKC-deviant PVG and DA or BN strains (Kveberg et al. 2006b, 2009). Allelic differences also appear to be limited at the level of the ligand, both in the rat (Kveberg et al. 2009) and the mouse. Clrb, Clrc and Clrg are identical in C57BL/6 and BALB/c, while Clrf differs between C57BL/6 and BALB/c by only three amino acids in the extracellular region.
The rat NKR-P1A or NKR-P1B reporter cells did not respond to any of the mouse tumour targets or mouse Clr transfectants, only to rat Clr11 (data not shown). This suggests that xenoreactivity is a unique property of the distal cluster of NKR-P1 receptors in the rat and of their mouse orthologs, i.e. NKR-P1F and NKR-P1G. Preservation of ligand-binding specificities supports the notion that the two receptors were present before speciation of rats and mice in a common rodent ancestor (Kveberg et al. 2009).
Concluding remarks
These studies have shown that the inhibitory NKR-P1G receptor is expressed in an NK subset-specific fashion, which may be a general principle for inhibitory NKR-P1 receptors. The present data have extended our previous observation, that the NKR-P1F and NKR-P1G receptors are functionally related, sharing overlapping specificities for Clr ligands. The most striking finding was the conservation of ligand specificity between the orthologous NKR-P1F and NKR-P1G receptors in rats and mice, which explains the potent xenoreactivity observed in both ways. The interactions between the inhibitory NKR-P1 receptors and their Clr ligands may allow for “missing self” surveillance of neoplastic and otherwise stressed cells (Fine et al. 2010; Voigt et al. 2007). The activating receptors should also be taken into consideration since they may react with the same ligand(s) (Kveberg et al. 2009). The broad reactivity patterns observed for NKR-P1F and NKR-P1G receptors suggest that they can be seen as sensors of the global status for Clr expression in cells.
Acknowledgements
This work was supported by grants from the Norwegian Cancer Society, Solveig og Ove Lunds legat and the South-Eastern Norway Regional Health Authority. We thank Christian Naper for the discussions and Stine Martinsen for the excellent technical assistance and NOVO Nordic, Copenhagen for providing Clra cDNA.
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
References
- Appasamy PM, Kenniston TW, Brissette-Storkus CS, Chambers WH. NKR-P1dim/TCRαβ+ T cells and natural killer cells share expression of NKR-P1A and NKR-P1D. Nat Immun. 1996;15:259–268. [PubMed] [Google Scholar]
- Aust JG, Gays F, Mickiewicz KM, Buchanan E, Brooks CG. The expression and function of the NKRP1 receptor family in C57BL/6 mice. J Immunol. 2009;183:106–116. doi: 10.4049/jimmunol.0804281. [DOI] [PubMed] [Google Scholar]
- Carlyle JR, Jamieson AM, Gasser S, Clingan CS, Arase H, Raulet DH. Missing self-recognition of Ocil/Clr-b by inhibitory NKR-P1 natural killer cell receptors. Proc Natl Acad Sci USA. 2004;101:3527–3532. doi: 10.1073/pnas.0308304101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlyle JR, Mesci A, Ljutic B, Belanger S, Tai L-H, Rousselle E, Troke AD, Proteau M-F, Makrigiannis AP. Molecular and genetic basis for strain-dependent NK1.1 alloreactivity of mouse NK cells. J Immunol. 2006;176:7511–7524. doi: 10.4049/jimmunol.176.12.7511. [DOI] [PubMed] [Google Scholar]
- Fine JH, Chen P, Mesci A, Allan DSJ, Gasser S, Raulet DH, Carlyle JR. Chemotherapy-induced genotoxic stress promotes sensitivity to natural killer cell cytotoxicity by enabling missing-self recognition. Cancer Res. 2010;70:7102–7113. doi: 10.1158/0008-5472.CAN-10-1316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flornes LM, Nylenna Ø, Saether PC, Daws MR, Dissen E, Fossum S. The complete inventory of receptors encoded by the rat natural killer cell gene complex. Immunogenetics. 2010;62:521–530. doi: 10.1007/s00251-010-0455-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giorda R, Rudert WA, Vavassori C, Chambers WH, Hiserodt JC, Trucco M. NKR-P1, a signal transduction molecule on natural killer cells. Science. 1990;249:1298–1300. doi: 10.1126/science.2399464. [DOI] [PubMed] [Google Scholar]
- Iizuka K, Naidenko OV, Plougastel BFM, Fremont DH, Yokoyama WM. Genetically linked C-type lectin-related ligands for the NKRP1 family of natural killer cell receptors. Nat Immunol. 2003;4:801–807. doi: 10.1038/ni954. [DOI] [PubMed] [Google Scholar]
- Kveberg L, Bäck CJ, Dai K-Z, Inngjerdingen M, Rolstad B, Ryan JC, Vaage JT, Naper C. The novel inhibitory NKR-P1C receptor and Ly49s3 identify two complementary, functionally distinct NK cell subsets in rats. J Immunol. 2006;176:4133–4140. doi: 10.4049/jimmunol.176.7.4133. [DOI] [PubMed] [Google Scholar]
- Kveberg L, Dai K-Z, Dissen E, Ryan JC, Rolstad B, Vaage JT, Naper C. Strain-dependent expression of four structurally related rat Ly49 receptors; correlation with NK gene complex haplotype and NK alloreactivity. Immunogenetics. 2006;58:905–916. doi: 10.1007/s00251-006-0154-x. [DOI] [PubMed] [Google Scholar]
- Kveberg L, Dai K-Z, Westgaard IH, Daws MR, Fossum S, Naper C, Vaage JT. Two major groups of rat NKR-P1 receptors can be distinguished based on chromosomal localization, phylogenetic analysis and Clr ligand binding. Eur J Immunol. 2009;39:541–551. doi: 10.1002/eji.200838891. [DOI] [PubMed] [Google Scholar]
- Kveberg L, Jiménez-Royo P, Naper C, Rolstad B, Butcher GW, Vaage JT, Inngjerdingen M. Two complementary rat NK cell subsets, Ly49s3+ and NKR-P1B+, differ in phenotypic characteristics and responsiveness to cytokines. J Leukoc Biol. 2010;88:87–93. doi: 10.1189/jlb.0110039. [DOI] [PubMed] [Google Scholar]
- Naper C, Dai K-Z, Kveberg L, Rolstad B, Niemi EC, Vaage JT, Ryan JC. Two structurally related rat Ly49 receptors with opposing functions (Ly49 stimulatory receptor 5 and Ly49 inhibitory receptor 5) recognize nonclassical MHC class Ib-encoded target ligands. J Immunol. 2005;174:2702–2711. doi: 10.4049/jimmunol.174.5.2702. [DOI] [PubMed] [Google Scholar]
- Naper C, Ryan JC, Kirsch R, Butcher GW, Rolstad B, Vaage JT. Genes in two major histocompatibility complex class I regions control selection, phenotype, and function of a rat Ly-49 natural killer cell subset. Eur J Immunol. 1999;29:2046–2053. doi: 10.1002/(SICI)1521-4141(199906)29:06<2046::AID-IMMU2046>3.0.CO;2-Z. [DOI] [PubMed] [Google Scholar]
- Plougastel B, Dubbelde C, Yokoyama WM (2001) Cloning of Clr, a new family of lectin-like genes localized between mouse Nkrp1a and Cd69. Immunogenetics 53:209–214 [DOI] [PubMed]
- Ryan JC, Niemi EC, Nakamura MC. Functional analysis of natural killer cell receptors in the RNK-16 rat leukemic cell line. Methods Mol Biol. 2000;121:283–295. doi: 10.1385/1-59259-044-6:283. [DOI] [PubMed] [Google Scholar]
- Vance RE, Kraft JR, Altman JD, Jensen PE, Raulet DH. Mouse CD94/NKG2A is a natural killer cell receptor for the nonclassical major histocompatibility complex (MHC) class I molecule Qa-1b. J Exp Med. 1998;188:1841–1848. doi: 10.1084/jem.188.10.1841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voigt S, Mesci A, Ettinger J, Fine JH, Chen P, Chou W, Carlyle JR. Cytomegalovirus evasion of innate immunity by subversion of the NKR-P1B:Clr-b missing-self axis. Immunity. 2007;26:617–627. doi: 10.1016/j.immuni.2007.03.013. [DOI] [PubMed] [Google Scholar]
- Yokoyama WM, Plougastel BFM. Immune functions encoded by the natural killer gene complex. Nat Rev Immunol. 2003;3:304–316. doi: 10.1038/nri1055. [DOI] [PubMed] [Google Scholar]


